Abstract
Using longitudinal data in electronic health records (EHRs) for post-marketing adverse drug event (ADE) detection allows for monitoring patients throughout their medical history. Machine learning methods have been shown to be efficient and effective in screening health records and detecting ADEs. How best to exploit historical data, as encoded by clinical events in EHRs is, however, not very well understood. In this study, three strategies for handling temporality of clinical events are proposed and evaluated using an EHR database from Stockholm, Sweden. The random forest learning algorithm is applied to predict fourteen ADEs using clinical events collected from different lengths of patient history. The results show that, in general, including longer patient history leads to improved predictive performance, and that assigning weights to events according to time distance from the ADE yields the biggest improvement.
Introduction and Motivation
Drug safety is a critical public health issue, with the prevalence of hospital admissions caused by adverse drug events (ADEs) ranging from 2.4% to 12.0%1,2,3. Although the safety of a drug must be evaluated in clinical trials prior to its launch, there are potentially many undiscovered side effects due to limitations in terms of both the number of participants and follow-up time. In fact, many drugs have been withdrawn from the market for causing fatal ADEs, such as Vioxx for its doubled risk of causing myocardial infarction4 and Cerivastatin for causing fatal rhabdomyolysis5. The continued surveillance of drug safety, post marketing, is therefore of utmost importance. Currently, this activity is almost exclusively dependent on spontaneous reports6, i.e., voluntary reporting of ADEs by patients or clinicians. Research on using alternative data sources, such as electronic health records (EHRs), are, however, emerging7,8,9. The use of EHRs for pharmacovigilance has several advantages that can complement and compensate for some of the weaknesses of spontaneous reports, such as under-reporting of ADEs and limited access to patient history10. An important property, and advantage, of EHRs is that they record longitudinal observations of patients and their treatment, including drug use. This opens up great opportunities for analyzing the temporal relationships between ADEs and other clinical events, which, in turn, can aid in the detection of ADEs.
In comparison to spontaneous reports, the reporting of ADEs in EHRs is better regulated, yet the problem of under-reporting remains11. To improve the reporting of ADEs, given the large amounts of data archived in EHR systems, manually screening all records to identify ADEs that were not reported is obviously very costly and even unrealistic. Instead, systems that employ machine learning methods are more efficient – and perhaps not less effective – by learning patterns from patients who have ADEs encoded in their medical history and detecting patients that have suffered an ADE, yet in whose records it has not, but should have, been encoded. Employing such systems could result in improved reporting of ADEs and, in the long run, potentially reduce the unsafe use of drugs.
There are two parallel approaches for such systems to facilitate the reporting of ADEs: (1) to prospectively predict whether there are ADEs to report in new health records; and (2) to retrospectively detect ADEs that are missing in earlier health records. Both approaches require the underlying models to include information from patients’ medical history, given that the first approach needs to use patient history to train the predictive models. The problem thus boils down to making meaningful use of patient history information in EHRs in order to detect ADEs. This problem has previously been studied from different perspectives, such as how best to represent clinical events12,13. In this study, we focus on the temporal aspect, i.e., which time periods should be considered when collecting clinical events for predicting ADEs. This aspect is expected to have a high impact on the resulting predictive performance, as the probability of an adverse event of a drug occurring at a certain point in time is highly dependent on when the drug was taken, e.g., an ADE will typically not occur for the first time years after a drug has been taken. On the one hand, considering longer time periods may lead to the ADE signal drowning in the volumes of collected information, while, on the other hand, shorter time periods may exclude information that is crucial for making accurate predictions.
In previous research, a few studies have touched upon or analyzed temporal relationships in EHRs for detecting ADEs. Norén et al. monitor clinical events that have occurred before and after a drug is prescribed to look for abnormalities as signals indicating ADEs14. Their study is, however, in a different genre as it does not involve a learning procedure. At the same time, Jin et al. transform the temporal problem onto a cross-section problem by defining hazard period, effect period and reference period after a drug is prescribed15. In a recent study, Eriksson et al. extract clinical notes and structured prescription data between drug introduction and discontinuation and then filter ADEs based on time stamp inconsistency between structured data and notes16. However, all of these studies mainly look at the temporality on a rather detailed level, such as between specific events or predefined periods; how to handle the temporality in EHRs in general still remains unanswered. In this study, the scope is set to detecting ADEs that should be reported in EHRs by employing machine learning methods to learn patterns using clinical events, and hence the essence of the problem is how to model the events that occurred at different time points in the patient history. To the best of our knowledge, this is the first study to explore ways of handling the temporality of patient history prior to an ADE. The aim of this study is therefore two-fold: (1) to investigate various ways of handling the temporality of clinical events; (2) to explore the importance of clinical events at different time points for detecting ADEs.
Materials and Methods
In this section, we first introduce the strategies of handling temporality that are proposed in this study. Then, we describe a series of empirical experiments that are conducted to evaluate the proposed strategies, starting by describing the data source and subsequently the details of each experiment.
- Strategies to handle temporality
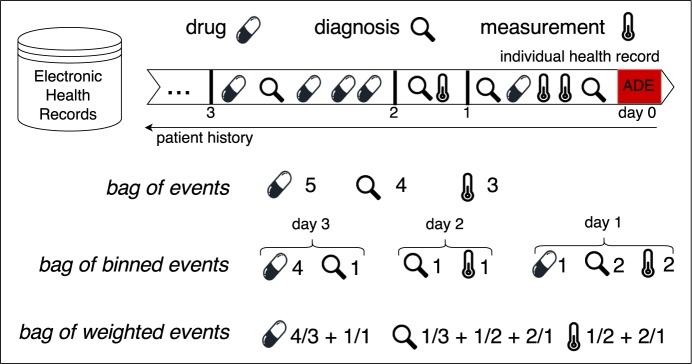
Three strategies – bag of events, bag of binned events, and bag of weighted events – to handle the temporality of clinical events in patients’ medical history for the detection of ADEs were proposed and evaluated in this study. In each of the strategies, only clinical events that occurred prior to the target ADE were included. A toy example of handling the temporality of one drug, one diagnosis and one clinical measurement in a time period of three days is demonstrated in Figure 1.
Figure 1:
Strategies to handle temporality of three clinical events during three days
Here, let x denote each unique event in the whole EHR database, d denote the number of days prior to the occurrence of the target ADE, d ≤ D, and nd denote the number of times x occurred in day d.
Bag of Events (BE) This strategy counts the number of occurrences of event x within D days. In this case, the value of feature x is .
Bag of Binned Events (BBE) This strategy counts the number of occurrences of event x in each day within D and represents x as x1, x2, …, xD. The value of the corresponding feature here is n1, n2, …, nD.
Bag of Weighted Events (BWE) This strategy assigns different weights to event x that occurred at different days d and takes into account the weights when counting the number of occurrences of x. The proportional weights are assigned according to the time distance between the event and the target ADE: events that occurred further in time from the target ADE receive proportionally less weight. In this case, the time distance between x at day d and the target ADE is d, and hence the value of feature x is .
- Data source
In this study, data was extracted from the Stockholm EPR Corpus17, which contains health records of around 700,000 (anonymized) patients over a two-year period (2009–2010), obtained from Karolinska University Hospital in Stockholm, Sweden. This database contains large amounts of diagnosis information, drug administrations, clinical measurements and clinical notes in free-text. In this study, we used only the structured data, i.e., diagnoses, drugs and clinical measurements, as features. In the Stockholm EPR Corpus, diagnoses are encoded by the International Statistical Classification of Diseases and Related Health Problems, 10th Edition (ICD-10) and drugs are encoded by the Anatomical Therapeutic Chemical Classification System (ATC). This research has been approved by the Regional Ethical Review Board in Stockholm (Etikprövningsnämnden i Stockholm) with permission number 2012/834-31/5.
The targeted use case in this study is to detect patients who should, but do not, have a specific ADE reported in their health records by retrospectively analyzing clinical events in their medical history. Among the diagnosis codes that indicate ADEs18, category A.1 (a drug-related causation was noted in the diagnosis code) and category A.2 (a drug-or other substance-related causation was noted in the diagnosis code) were considered in this study. We selected the 14 most frequent A.1 and A.2 codes in the Stockholm EPR Corpus, and thus created 14 datasets, where the existence of the diagnosis code indicating a particular ADE was used as the class label in each dataset. The classification task is hence binary: positive or negative towards a specific ADE. Both inpatients and outpatients are included in this study, where the positive examples are patients whom have been assigned an ADE-specific diagnosis code and the negative examples are patients whom have been assigned a similar diagnosis code to the diagnosis code indicating ADE. Here, similarity is defined as two codes sharing the same first three levels of the ICD-10 concept hierarchy. For instance, if the positive examples are patients with diagnosis code G44.4 (drug-induced headache), the negative examples are patients with any diagnosis code starting with G44 (other headache syndromes), but not G44.4. Table 1 lists the basic descriptions of each dataset, including the diagnosis code that indicates the corresponding ADE (dataset name), the description of this code and the number of positive and negative examples.
Table 1:
Dataset description
| Dataset | Corresponding diagnosis code description | Positive | Negative |
|---|---|---|---|
| D642 | Secondary sideroblastic anemia due to drugs and toxins | 113 | 4234 |
| G240 | Drug-induced dystonia | 16 | 44 |
| G444 | Drug-induced headache, not elsewhere classified | 31 | 1102 |
| G620 | Drug-induced polyneuropathy | 19 | 367 |
| I952 | Hypotension due to drugs | 38 | 480 |
| L270 | Generalized skin eruption due to drugs and medicaments | 174 | 291 |
| L271 | Localized skin eruption due to drugs and medicaments | 58 | 407 |
| O355 | Maternal care for (suspected) damage to fetus by drugs | 334 | 373 |
| T782 | Adverse effects: anaphylactic shock, unspecified | 136 | 1467 |
| T783 | Adverse effects: angioneurotic oedema | 147 | 1448 |
| T784 | Adverse effects: allergy, unspecified | 984 | 612 |
| T808 | Other complications following infusion, transfusion and therapeutic injection | 353 | 59 |
| T886 | Anaphylactic shock due to correct drug or medicament properly administered | 53 | 607 |
| T887 | Unspecified adverse effect of drug or medicament | 472 | 277 |
Around 10,000 unique ICD-10 diagnosis codes, 1,500 unique ATC codes and 730 unique clinical measurements exist in the Stockholm EPR Corpus. Since most clinical events only occurred to a small group of patients, the datasets where clinical events are used to describe each patient are consequently both high-dimensional and very sparse, i.e., for a single observation (or example) the vast majority of the features have a value of 0. The employed algorithm for generating predictive models, i.e., the random forest algorithm19, is rather efficient in handling high-dimensional data, as only a small random sample of the available features is considered when determining the best way of partitioning the training examples during tree growth. However, for highly sparse data, it is not unlikely that all sampled features are uninformative, i.e., lead to no separation. Unless specifically handled, the tree growth will terminate prematurely and lead to an overall low predictive performance. Rather than employing any of the more sophisticated approaches to handle this20, we here employed the quite straightforward approach of removing features that are more sparse than 99%, i.e., the ones for which non-zero values were observed in fewer than 1% of the patients; for the datasets with fewer than one hundred observations, features with only one non-zero value were also removed. The motivation behind this is simply that for features with very few non-zero values, the impact of applying different strategies to handle temporality will be negligible, even though some of these features might be valuable indicators.
In this study, we defined the following 12 thresholds from patient history: 1, 2, 3, 4, 5, 6, 7, 14, 21, 30, 60, 90 days before the target ADE, where, for each threshold n, clinical events that occurred n days before the target ADE are studied. The choice was made with the assumption that more clinical events would have occurred in the days closer to the ADE and the difference between days closer to the ADE has a higher impact on detecting ADEs, especially on the ones that are dose-independent. We therefore studied clinical events that occurred in each day of the first week, followed by every week in the first month, and finally every month up to 3 months (90 days) before the occurrence of the target ADE. In the bag of binned events strategy, clinical events are binned within a time threshold. For example, all events that occurred one day before ADE are binned in day 1, and all events that occurred from 60 days to 90 days before the ADE are binned in day 90. Here, the same event that occurred at different times is treated as different events, the number of features in bag of binned events is hence much higher than the other two strategies. To reduce the number of potential models, we do not distinguish between the events that occurred in different days within, e.g., the third month before the ADE. The average number of features and sparsity over the 14 datasets, before and after removing the extremely sparse features at each time threshold, are illustrated in Figure 2.
Figure 2:
Average number of features and sparsity before and after removing extremely sparse features
- Experiments
In this study, a series of experiments was conducted to evaluate the strategies to handle temporality of clinical events in patient history and explore the impact of using different lengths of patient history on detecting ADEs. In the first experiment, the three strategies – bag of events, bag of binned events and bag of weighted events – were compared by employing the random forest19 learning algorithm to features (clinical events) generated by each of them respectively. When more than two competing models are compared, the Friedman test was employed for statistical testing of the null hypothesis that all models perform equally, followed by a post-hoc test using the Bergman-Hommel procedure21.
The choice of random forest was made due to its reputation of achieving high accuracy, its ability to handle high-dimensional data, as well as the possibility of obtaining estimates of variable importance. The algorithm constructs an ensemble of decision trees, which together vote for what class label to assign to an example that is to be classified. Each tree in the forest is built from a bootstrap replicate of the original instances, and a subset of all features is sampled at each node when building the tree, in both cases to increase diversity among the trees. When the number of trees in the forest increases, the probability that a majority of trees makes an error decreases, given that the trees perform better than random and that the errors are made independently. Although this condition can only be guaranteed in theory, the algorithm has often been shown in practice to result in state-of-the-art predictive performance. In this study, we used random forest with 500 trees. In all experiments, models were built and evaluated using stratified 5-fold cross validation with two iterations, where the original class distribution was retained in each fold.
The considered performance metrics are accuracy, area under the ROC curve (AUC), precision, recall, F-score and area under the precision-recall curve (AUPRC). Accuracy corresponds to the percentage of correctly classified instances. AUC depicts the performance of a model without regard to class distribution or error costs by estimating the probability that a model ranks a randomly chosen positive instance ahead of a negative one. Precision measures the fraction of true positives among all the predicted positives, while recall, also known as sensitivity, measures the fraction of true positives among all the positives in the gold standard. In the case of detecting ADEs, high precision means that the algorithm is able to detect more true ADEs than false ones, while high recall means that the algorithm is able to detect most true ADEs. F-score describes the harmony between precision and recall by calculating 2 × (precision × recall)/(precision + recall). Only both high precision and high recall can yield a high F-score. AUPRC depicts the probability that precision is higher than recall for each recall threshold. It is considered to be a more careful measurement compared to AUC, since high AUC often leads to high AUPRC, but not necessarily the other way around22.
In a subsequent set of experiments, variable importance generated with random forest using bag of binned events was analyzed in order to obtain a deeper understanding of in which time period clinical events are more informative. Variable importance can be estimated in different ways19. In this study, Gini importance was chosen as the variable importance metric, where high Gini importance means that a variable plays a greater role in splitting the data into the defined classes. A Gini importance of zero indicates that a variable is considered useless or is never selected to build any tree. Here, we ranked clinical events from different days according to their Gini importance, and then aggregated all events from the same time threshold by averaging their ranks. Given that each time threshold now has an average rank, we calculated the relative importance among the 12 time thresholds: D = {1, 2, 3, 4, 5, 6, 7, 14, 21, 30, 60, 90}; for each dataest, the average rank for each time threshold d is rd, rd ∈ R, and then the relative importance ri for time threshold d is rid = (max(R) − rd)/(max(R) − min(R)), rid ∈ (0, 1). For example, if the relative importance of day 5 is 0 and of day 30 is 0.12, it means that day 30 is 12% more important than day 5.
Results
Here, we first report the results on comparing the proposed strategies for handling temporality: bag of events, bag of binned events and bag of weighted events, and then on the impact of clinical events from different days of patient history for each specific ADE.
- Comparing strategies to handle temporality
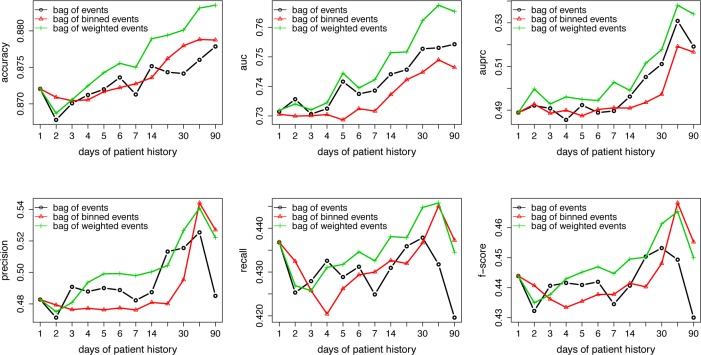
The predictive performance of random forest with each strategy, in terms of accuracy, AUC, AUPRC, precision, recall and F-score, is presented in Figure 3. We can see that in general, bag of weighted events (in green) outperforms the other two and the predictive performance improves overall with increasing number of days of patient history.
Figure 3:
Classification results by models using the proposed strategies for each time threshold
Friedman tests were conducted to compare the three strategies at each time threshold over the 14 datasets (p-values are shown in Table 2). P-values are not available for time threshold 1 day since the three strategies are identical for clinical events that occur one day before the ADE. From this table we can see that the p-values generally decrease with more days of patient history, and the null hypothesis that the three strategies perform equally well is rejected with most evaluation metrics towards the end, i.e., using 90 days of patient history. This indicates that when we include more days of patient history in our predictive models, the impact of how temporality is handled increases. Note that although there seems to be a performance drop from 60 days to 90 days in Figure 3, if we take a closer look at the performance on each dataset (e.g., Figure 4 for bag of weighted events), we can see that for most datasets, the predictive performance improves from 60 days to 90 days, except for a big drop on two (G620 and L271), which explains most of the overall performance drop observed in Figure 3.
Table 2:
P-values of statistical significance of differences among the three strategies with each time threshold
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 14 | 21 | 30 | 60 | 90 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy | – | 0.54 | 0.84 | 0.66 | 0.37 | 0.56 | 0.3 | 0.048 | 0.74 | 0.12 | 0.07 | 0.14 |
| AUC | – | 0.93 | 0.81 | 0.22 | 0.22 | 0.61 | 0.17 | 0.26 | 0.11 | 0.07 | 0.002 | 0.001 |
| AUPRC | – | 0.75 | 0.75 | 0.07 | 0.9 | 0.61 | 0.08 | 0.61 | 0.08 | 0.07 | 0.003 | 0.002 |
| Precision | – | 0.93 | 0.42 | 0.61 | 0.02 | 0.1 | 0.1 | 0.04 | 0.59 | 0.04 | 0.029 | 0.01 |
| Recall | – | 0.83 | 0.84 | 0.72 | 0.84 | 0.27 | 0.61 | 0.9 | 0.53 | 0.54 | 0.57 | 0.32 |
| F-score | – | 0.64 | 0.49 | 0.16 | 0.12 | 0.18 | 0.7 | 0.64 | 0.13 | 0.04 | 0.27 | 0.025 |
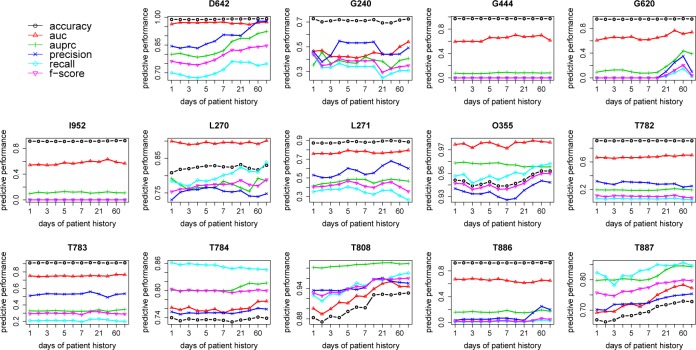
Figure 4:
Predictive performance of random forest with bag of weighted events for each ADE
Given these results, a post-hoc analysis comparing the strategies pairwise was carried out with 90 days as threshold; the results are shown in Table 3. The left half of the table shows the average rank – the lower the rank, the better the performance – among the three strategies, and we can see that bag of weighted events is consistently better than the other two. The pairwise p-values in the right half of the table indicate that bag of weighted events is significantly better than bag of events for AUPRC, precision and F-score; bag of weighted events is significantly better than bag of binned events for AUC and AUPRC; and bag of binned events is significantly better than bag of events for AUC.
Table 3:
Average ranks and p-values from the post-hoc analysis of Friedman tests with 90 days as threshold
| Average rank
|
Pairwise p-value
|
|||||
|---|---|---|---|---|---|---|
| Metric | BE | BBE | BWE | BBE vs. BWE | BE vs. BWE | BBE vs. BE |
| Accuracy | 2.14 | 2.29 | 1.57 | 0.176 | 0.176 | 0.705 |
| AUC | 1.96 | 2.71 | 1.32 | 0.0007 | 0.089 | 0.047 |
| AUPRC | 2.36 | 2.43 | 1.21 | 0.004 | 0.004 | 0.85 |
| Precision | 2.57 | 1.89 | 1.54 | 0.34 | 0.018 | 0.07 |
| Recall | 2.28 | 1.96 | 1.75 | 0.57 | 0.47 | 0.47 |
| F-score | 2.5 | 1.96 | 1.54 | 0.26 | 0.032 | 0.156 |
- Impact of temporality on specific adverse drug events
The predictive performance of the best observed strategy – bag of weighted events – for each specific ADE is presented in Figure 4. As described in the previous section, the predictive performance overall increases on a small scale towards using more days of patient history, which is especially clear for D642, T808 and T887. Even with an obvious drop at the end on e.g. G620, the predictive performance with 90 days is still higher than it is with 1 day. Apparently, the choice of threshold has different degrees of impact on different ADEs.
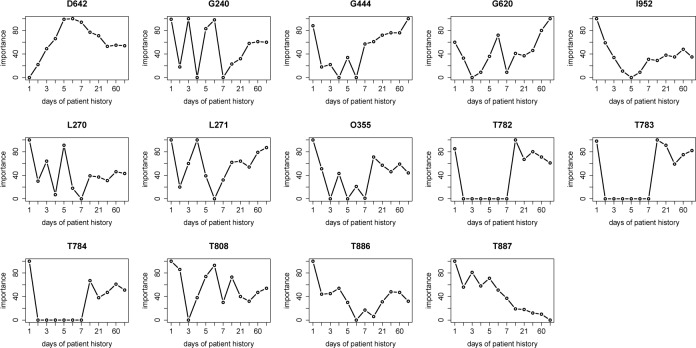
To obtain a deeper understanding of the contribution of clinical events from different days of patient history to the predictive performance, variable importance analysis was conducted using bag of binned events, where they are treated as different variables. Figure 5 shows the relative importance of clinical events occurring in different days of patient history for detecting each specific ADE. For most ADEs, it is clear that clinical events occurring in the late stage of patient history, e.g., just one day before the ADE, is of highest importance; for some ADEs, such as D642 and G620, clinical events occurring in a much earlier stage of patient history are relatively more important.
Figure 5:
Relative importance of clinical events in different days of patient history
Discussion
For the task of detecting ADEs using clinical events in EHRs, the predictive performance with the three strategies differs from each other within a fairly small range, even though there are significant differences observed. This can be explained by the difficulty of the task itself. Here, in each dataset, the positive examples are patients who have a specific ADE, while the negative examples are patients who have a disease that is in the same family as the ADE (they share the first three levels of ICD-10). In many cases, these patients are very similar to each other in terms of having similar drugs and clinical measurements; therefore it is difficult for the learning algorithm to distinguish between them. Hence it is difficult to observe big changes in predictive performance with different strategies since they are all bounded with the similar clinical events these patients share.
Based on our results, the random forest built with bag of events has the worst predictive performance, followed by bag of binned events, and both of them are, in most cases, outperformed by the random forest built with bag of weighted events. This finding is consistent with the granularity of each of the strategies: the model with bag of events completely ignores temporality, and hence is the crudest model; although the model with bag of binned events treats events from different time periods as different features, such a representation also results in increased dimensionality and sparsity that have a negative impact on the predictive performance; and, finally, the model with bag of weighted events tackled the problems with the other two strategies – temporality, dimensionality and sparsity – by aggregating the events from different time periods according to their time distance from the target ADE.
The predictive performance and variable importance for each specific ADE to some extent reflect the different nature of each ADE. Adverse drug events are typically divided into two types: dose-dependent or not. The former is related to the accumulation of toxics from drugs or medications; therefore, when predicting ADEs of this type, we would intuitively expect that using events from longer patient history contributes to improved predictive performance compared to using only events from the most recent days. This assumption is supported by the results on predicting D642 – drug induced anemia, where the predictive performance improves monotonically with the use of more days of patient history (see Figure 4 – D642) and the clinical events from seven days prior to the ADE are much more important compared to one day (see Figure 5 – D642). Another example is G620 – drug induced polyneuropathy, which is an ADE known to be caused by drugs used in the treatment of very severe or chronic diseases like cancer or tuberculosis. Such patients are normally exposed for a long period until the side effect is observed. Here, again, a similar performance trend is observed to support our hypothesis (see Figure 4 – G620 and Figure 5 - G620). On the other hand, the second type of ADE is related mainly to idiosyncratic or immunological nature, such as an allergic reaction, which indicates that most likely, with a very marginal assumption, they are more or less instant side effects from taking drugs or medications. The results on predicting T886 – anaphylactic shock due to correct drug or medicament properly administered is a good example that agrees with this assumption. The predictive performance almost remains the same as using events from only one day before the ADE (see Figure 4 – T886) and the events from one day before are absolutely the most important ones (see Figure 5 – T886).
However, the assumptions that are made above are not always reflected by our results in this study, which can be explained by several reasons. First, only clinical events from the structured EHRs are used as features here, while ADEs are often described in the notes rather than reported as diagnoses (given that ADEs are heavily under-reported as we described in the introduction). Especially for patients who suffer from severe diseases, ADEs become less important to report. Therefore, using only the structured clinical events can hardly capture a holistic and precise picture for detecting ADEs. Second, in the real clinical setting, where the EHR data is collected, there is a lack of controls on when clinical events, especially diagnosis codes, are reported in the database. Sometimes events that occurred weeks ago are reported together with the ones that occurred one day ago when the patients are discharged. This, unfortunately, results in noise in the training data and also inaccurate weights based on the time distance from the target ADE. Third, diagnosis codes, i.e., the ICD-10 codes, are themselves heterogeneous. For example, T782–T784 cover not only ADEs but also adverse reactions to substances that are not drugs.
One limitation of this study concerns the fact that the events are represented as a bag in all of the strategies, which neglects the temporality between the events in the same day or period. This has a potential impact on the predictive performance, especially for predicting ADEs that are not dose-dependent, since the temporal relationship between events within a short period is important to capture when the ADEs are mostly instant effects. To take this into account, techniques that enable mining event sequences would be useful. In addition, the use of ICD-10 codes to select patients with and without ADEs should be proceeded with caution due to the fact that they are coded also for billing purposes and other reasons. For future work, it would also be interesting to see the impact of the proposed strategies on the task of distinguishing patients who have ADEs from patients who do not, i.e., the negative examples would be randomly selected from the population. The way of assigning weights in bag of weighted events is fairly crude and harsh; a follow-up study could focus on exploring alternative ways of assigning weights to clinical events that occur at different time points. To improve the predictive performance, including free-text clinical notes in the models and handling the temporality of information in notes are indeed worth investigating.
Conclusions
It is advantageous to use longitudinal data from electronic health records for the detection of adverse drug events. This study proposed and evaluated three strategies to handle temporality of clinical events: drugs, diagnosis and clinical measurements. These strategies differ from each other in how they take into account the clinical events that occur in different days of patient history: bag of events counts the number of times that an event occurred without regard to when it occurred; bag of binned events separates the patient history into predefined bins and then counts the number of times that an event occurred in each bin; and bag of weighted events counts the weighted number of times that an event occurred, where the weights are assigned proportionally according to time distance between the event and the target adverse drug event. Based on our empirical investigation, the bag of weighted events strategy yields the best predictive performance with the considered metrics. Here, we conclude that, in general, using longer patient history leads to improved predictive performance, and the temporality of clinical events matters more when using more days of patient history.
Acknowledgments
This work was supported by the Swedish Foundation for Strategic Research through the project High-Performance Data Mining for Drug Effect Detection, ref. no. IIS11-0053.
References
- 1.Schneeweiss S, Hasford J, Göttler M, Hoffmann A, Riethling AK, Avorn J. Admissions caused by adverse drug events to internal medicine and emergency departments in hospitals: a longitudinal population-based study. European Journal of Clinical Pharmacology. 2002;58(4):285–291. doi: 10.1007/s00228-002-0467-0. [DOI] [PubMed] [Google Scholar]
- 2.Pirmohamed M, James S, Meakin S, Green C, Scott AK, Walley TJ, et al. Adverse drug reactions as cause of admission to hospital: prospective analysis of 18 820 patients. BMJ. 2004;329(7456):15–19. doi: 10.1136/bmj.329.7456.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mjörndal T, Boman MD, Hägg S, Bäckström M, Wiholm BE, Wahlin A, et al. Adverse drug reactions as a cause for admissions to a department of internal medicine. Pharmacoepidemiology and Drug Safety. 2002;11(1):65–72. doi: 10.1002/pds.667. [DOI] [PubMed] [Google Scholar]
- 4.Sibbald B. Rofecoxib (Vioxx) voluntarily withdrawn from market. Canadian Medical Association Journal. 2004;171(9):1027–1028. doi: 10.1503/cmaj.1041606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furberg CD, Pitt B. Withdrawal of cerivastatin from the world market. Curr Control Trials Cardiovasc Med. 2001;2(5):205–207. doi: 10.1186/cvm-2-5-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Puijenbroek EP, Bate A, Leufkens HG, Lindquist M, Orre R, Egberts AC. A comparison of measures of disproportionality for signal detection in spontaneous reporting systems for adverse drug reactions. Pharmacoepidemiology and Drug Safety. 2002;11(1):3–10. doi: 10.1002/pds.668. [DOI] [PubMed] [Google Scholar]
- 7.Schuemie MJ, Coloma PM, Straatman H, Herings RM, Trifirò G, Matthews JN, et al. Using electronic health care records for drug safety signal detection: a comparative evaluation of statistical methods. Medical Care. 2012;50(10):890–897. doi: 10.1097/MLR.0b013e31825f63bf. [DOI] [PubMed] [Google Scholar]
- 8.Karlsson I, Zhao J, Asker L, Boström H. Artificial Intelligence in Medicine Lecture Notes in Computer Science. Springer; 2013. Predicting adverse drug events by analyzing electronic patient records; pp. 125–129. [Google Scholar]
- 9.Harpaz R, Haerian K, Chase HS, Friedman C. Mining electronic health records for adverse drug effects using regression based methods; 1st ACM International Health Informatics Symposium; ACM; 2010. pp. 100–107. [Google Scholar]
- 10.Goldman SA. Limitations and strengths of spontaneous reports data. Clinical Therapeutics. 1998;20:C40–C44. doi: 10.1016/s0149-2918(98)80007-6. [DOI] [PubMed] [Google Scholar]
- 11.Classen DC, Resar R, Griffin F, Federico F, Frankel T, Kimmel N, et al. Global trigger tool shows that adverse events in hospitals may be ten times greater than previously measured. Health Affairs. 2011;30(4):581–589. doi: 10.1377/hlthaff.2011.0190. [DOI] [PubMed] [Google Scholar]
- 12.Zhao J, Henriksson A, Boström H. Detecting adverse drug events using concept hierarchies of clinical codes. IEEE International Conference on Healthcare Informatics. 2014:285–293. [Google Scholar]
- 13.Zhao J, Henriksson A, Asker L, Boström H. Detecting adverse drug events with multiple representations of clinical measurements; IEEE International Conference on Bioinformatics and Biomedicine; 2014. pp. 536–543. [Google Scholar]
- 14.Norén GN, Hopstadius J, Bate A, Star K, Edwards IR. Temporal pattern discovery in longitudinal electronic patient records. Data Mining and Knowledge Discovery. 2010;20(3):361–387. [Google Scholar]
- 15.Jin H, Chen J, He H, Kelman C, McAullay D, O’Keefe CM. Signaling potential adverse drug reactions from administrative health databases. IEEE Transactions on Knowledge and Data Engineering. 2010;22(6):839–853. [Google Scholar]
- 16.Eriksson R, Werge T, Jensen LJ, Brunak S. Dose-specific adverse drug reaction identification in electronic patient records: temporal data mining in an inpatient psychiatric population. Drug Safety. 2014;37(4):237–247. doi: 10.1007/s40264-014-0145-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalianis H, Hassel M, Henriksson A, Skeppstedt M. Stockholm EPR Corpus: a clinical database used to improve health care; Swedish Language Technology Conference; 2012. pp. 17–18. [Google Scholar]
- 18.Stausberg J, Hasford J. Drug-related admissions and hospital-acquired adverse drug events in Germany: a longitudinal analysis from 2003 to 2007 of ICD-10-coded routine data. BMC Health Services Research. 2011;11(1):134. doi: 10.1186/1472-6963-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breiman L. Random forests. Machine Learning. 2001;45(1):5–32. [Google Scholar]
- 20.Karlsson I, Boström H. Handling sparsity with random forests when predicting adverse drug events from electronic health records; IEEE International Conference on Healthcare Informatics; 2014. pp. 17–22. [Google Scholar]
- 21.Garcia S, Herrera F. An extension on ”statistical comparisons of classifiers over multiple data sets” for all pairwise comparisons. Journal of Machine Learning Research. 2008;9(12) [Google Scholar]
- 22.Davis J, Goadrich M. The relationship between Precision-Recall and ROC curves; 23rd International Conference on Machine Learning; ACM; 2006. pp. 233–240. [Google Scholar]