Abstract
Accurate temporal identification and normalization is imperative for many biomedical and clinical tasks such as generating timelines and identifying phenotypes. A major natural language processing challenge is developing and evaluating a generalizable temporal modeling approach that performs well across corpora and institutions. Our long-term goal is to create such a model. We initiate our work on reaching this goal by focusing on temporal expression (TIMEX3) identification. We present a systematic approach to 1) generalize existing solutions for automated TIMEX3 span detection, and 2) assess similarities and differences by various instantiations of TIMEX3 models applied on separate clinical corpora. When evaluated on the 2012 i2b2 and the 2015 Clinical TempEval challenge corpora, our conclusion is that our approach is successful – we achieve competitive results for automated classification, and we identify similarities and differences in TIMEX3 modeling that will be informative in the development of a simplified, general temporal model.
Introduction
Accurate temporal identification and normalization is imperative for many biomedical and clinical tasks including generating timelines1, identifying phenotypes2, and creating problem lists3. Natural language processing (NLP) can help enrich event, phenotype, and problem detection by providing temporal context to event descriptions extracted from clinical texts. A major NLP challenge is developing and evaluating a generalizable temporal modeling approach that performs well across corpora and institutions. Few NLP system studies evaluate the generalizability of clinical temporal models and information extraction approaches that were developed for a particular use case or institution. To date, several NLP community challenges – 2015 Clinical TempEval4, based on the THYME corpus5, 2012 i2b26, and the 2014 ShARe/CLEF eHealth challenges7 – have attempted to encourage benchmarking and system evaluation for temporal extraction by applying adapted versions of the TimeML model8 to clinical texts of various report types from different institutions (Beth Israel Deaconess Medical Center, Partner’s Healthcare, and Mayo Clinic). The TimeML model enables automated temporal reasoning from text by defining a number of components, of which three components are central: events (e.g., diseases), time expressions (TIMEX3s, e.g., dates), and time relations (e.g. disease after date). Through this, it is possible to order events in time with respect to each other, and with respect to time expressions in the texts. Correctly identifying time expressions within a temporal reasoning system is necessary to capture specific mentions of time, and thus enable time anchoring and positioning of relevant events from narratives. These community challenges have established the state-of-the-art performance for NLP identification of these three core components in the adapted TimeML models.
For temporal modeling, overall performance has been published for each community challenge; however, there are few studies reporting system performance across different clinical corpora, and there has not been any in-depth analysis of performance on different temporal model subtypes. Our long-term goal is to create a generalizable temporal reasoning model that leverages and simplifies existing models. We initiate our work on reaching this goal by focusing only on TIMEX3 identification. In particular, we present a systematic approach to 1) generalize existing solutions for automated TIMEX3 span detection, and 2) assess similarities and differences in different instantiations of TIMEX3 models applied on different clinical corpora.
In the aforementioned challenges, successful approaches for TIMEX3 span detection, that is, correctly identifying and extracting specific mentions of time expressions in text such as “tomorrow”, “14 April 1998”, “for two weeks”, have either employed rule-based systems or machine learning approaches using lexical and syntactic features in combination with rule-based information. In the 2012 i2b2 challenge, the top performing approaches resulted in F1 scores between 0.8–0.91 for overlapping TIMEX3 spans6. The results in the 2014 ShARe/CLEF eHealth challenge were lower: 0.37 overlap F17. Our research lab, the Biomedical Language Understanding Laboratory (BluLab), recently participated in the 2015 Clinical TempEval challenge9. Similar to other successful, high-performing NLP approaches, we developed a machine learning approach using local contextual features and information from a rule-based system to identify TIMEX3 spans. Our team ranked first on the TIMEX3 span detection task in this challenge, resulting in an overall exact F1 of 0.72. However, official scores in these challenges only report overall results for TIMEX3 detection. Moreover, to our knowledge, there are few studies reporting system performance across different clinical corpora, along with an in-depth analysis of similarities and differences in TIMEX3 annotations.
Methods
We performed our study on two corpora1: the 2015 Clinical TempEval corpus, and the 2012 i2b2 challenge corpus. The Clinical TempEval corpus was created within the THYME project5 consisting of 440 clinical notes and pathology reports for colon cancer patients from the Mayo clinic (in total 293 documents for training, 147 for test). The i2b2 corpus consists of 310 discharge summaries (190 training, 120 test) from Partners Healthcare and the Beth Israel Deaconess Medical Center. We chose these two corpora for this study because they employ similar, but not identical, temporal models, and were specifically developed for temporal reasoning evaluation. Three main steps were employed: 1) TIMEX3 model mapping, with the aim of generating a common, comparable model, 2) state-of-the-art TIMEX3 classification assessment, with the aim of evaluating top performing TIMEX3 system approaches across corpora, and 3) qualitative error analysis, feature ablation evaluation and TIMEX3 type characterization, with the aim of completing an in-depth analysis.
Common TIMEX3 type model
In our attempt to characterize and analyze similarities and differences in separate instantiations of TIMEX3 models applied on different corpora, the first step was to create a common TIMEX3 type model. The two corpora employ slightly different TIMEX3 definitions and types. The Clinical TempEval corpus contains six TIMEX3 types: DATE, TIME, DURATION, PREPOSTEXP, QUANTIFIER and SET, in contrast to the i2b2 corpus which defines four types: DATE, TIME, DURATION and FREQUENCY. To enable comparison and analysis, we map the more fine-grained Clinical TempEval TIMEX3 types to the i2b2 types in the following way: PREPOSTEXP annotations are merged to DATE, and QUANTIFIER and SET annotations are merged to FREQUENCY. Examples are provided in Table 1. For our experiments, we maintained the training and test splits from each respective challenge corpus.
Table 1.
Mapped TIMEX3 types and examples from the 2012 i2b2 and THYME annotation guidelines.
| TIMEX3 type | Example |
|---|---|
| DATE | 05-04-1998, 29, February 2005, next week |
| PREPOSTEXP | Postoperative |
| DURATION | the first few days, the next 12 hours |
| FREQUENCY | several times a day, daily |
| QUANTIFIER | twice |
| SET | three times weekly |
| TIME | 5.30 PM |
Time expression extraction using ClearTK
To generalize existing solutions for automated TIMEX3 span detection, we applied the top-performing TIMEX3 span detection approach from the 2015 Clinical TempEval challenge9. This is a UIMA-based machine learning solution using the ClearTK framework10. A separate support vector machine (SVM: Liblinear) sequence label classifier (CleartkSequenceAnnotator) was created for each TIMEX3 type, where parameter setting (C-value) was determined by a grid search on a subset of the training data and set manually on the final model. The data was preprocessed with cTAKES v. 3.2 to extract lexical, morphological, and syntactic features11. For the TIMEX3 types, gazetteer information specific to each type was also added as features, based on an adapted version of HeidelTime12. A Begin-Inside-Outside chunking representation was used, with the following features: the current token, the current token with the two last characters stripped (decades ➔ decad), part-of-speech tag, numeric type (e.g. digit, alphanumeric), capital type (e.g. all in upper case, mixed case), lower case, context tokens2, and whether or not the token is found in the TIMEX3 type-specific gazetteer. Compared to the successful approaches in the 2012 i2b2 challenge that used machine learning, this approach is similar, but not identical – for instance, Xu et al.13 employed the conditional random fields (CRF) algorithm and slightly different features. Roberts et al.14 used CRF for TIMEX3 boundary detection and SVM for type classification. Most other solutions were entirely rule-based6.
Three SVM models were trained using the datasets: 1) i2b2 training data, 2) Clinical TempEval training data, and 3), merged model with the two training data sets combined. System performance was evaluated on the i2b2 test data and the Clinical TempEval test data releases, using exact and overlapping precision, recall, and F1 as main outcome measures. We used the official evaluation script provided in the 2015 Clinical TempEval challenge3.
Qualitative error analysis, feature ablation evaluation and TIMEX3 type characterization
A qualitative error analysis to identify commonalities and differences was performed. We analyzed true positives on the results obtained by training a model on one corpus and evaluating on the other, to identify commonalities between the corpora and to characterize “core” TIMEX3 types. We also analyzed false positives and negatives, to identify and characterize differences between the annotations. Error analysis was performed on exact match only. Additionally, we performed a feature ablation study on the results obtained by this training/testing setup as well as on the results obtained when training a merged model with both corpora, evaluated on each test set. We built separate models with one feature removed each time, to identify which features seem to contribute the most for this task (i.e., if results are worse when removing one feature compared to using all features, this is an indication of an important feature).
Results
We present classification results on the i2b2 and Clinical TempEval test sets and results from the qualitative error analysis. First, we evaluate performance on within-corpus models. Second, we analyze performance on the two test sets on applied on the model trained on 1) i2b2 training data, 2) Clinical TempEval training data, and 3) merged training data. Finally, we present results from the qualitative error analysis, feature ablation evaluation and TIMEX3 type characterization, using the outputs from applying each test set on the model created from the other corpus’ training data. Overall corpus statistics are presented in Table 2.
Table 2.
TIMEX3 count (prevalence) statistics per type and corpus – Clinical TempEval and 2012 i2b2 datasets.
| TIMEX3 Type | Clinical TempEval | i2b2 | ||
|---|---|---|---|---|
| Training data | Test data | Training data | Test data | |
| DATE | 2892 (75.7%) | 1594 (76.7%) | 1640 (69.4%) | 1222 (67.1%) |
| DURATION | 434 (11.4%) | 200 (9.6%) | 406 (17.2%) | 341 (18.7%) |
| FREQUENCY | 377 (9.8%) | 225 (10.9%) | 249 (10.5%) | 197 (10.9%) |
| TIME | 118 (3.1%) | 59 (2.8%) | 69 (2.9%) | 60 (3.3%) |
| Total | 3821 (100%) | 2078 (100%) | 2364 (100%) | 1820 (100%) |
Temporal Information Extraction using ClearTK
In Table 3, classification results on the two test datasets when applied on the model built from their corresponding training data are shown, strict and overlap. Results for the majority TIMEX3 type DATE are best overall. Results for DURATION and FREQUENCY are considerably higher when using overlapping evaluation criteria, in particular for DURATION, from 0.60 to 0.76 F1 for i2b2, and from 0.47 to 0.70 for Clinical TempEval. Precision is considerably higher for FREQUENCY when using overlapping evaluation, from 0.71 to 0.84 for i2b2 and 0.56 to 0.78 for Clinical TempEval. Results for the least frequent TIMEX3 type TIME differ between the corpora: for the i2b2 corpus, precision is high when using overlap evaluation, but results are low overall for the Clinical TempEval corpus.
Table 3.
Classification results for the mapped TIMEX3 types on the Clinical TempEval (CTE) and the i2b2 test datasets using the top-performing 2015 Clinical TempEval approach on the corresponding corpus training data. P=precision, R=recall.
| TIMEX3 type | strict – CTE/i2b2 | overlap – CTE/i2b2 | ||||
|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | |
| span | 0.80/0.79 | 0.66/0.70 | 0.72/0.74 | 0.92/0.93 | 0.76/0.81 | 0.83/0.87 |
| class | 0.78/0.77 | 0.65/0.69 | 0.71/0.72 | 0.88/0.87 | 0.74/0.78 | 0.80/0.82 |
| DATE | 0.82/0.80 | 0.76/0.78 | 0.79/0.79 | 0.90/0.89 | 0.83/0.86 | 0.86/0.88 |
| DURATION | 0.55/0.65 | 0.41/0.55 | 0.47/0.60 | 0.81/0.83 | 0.61/0.70 | 0.70/0.76 |
| FREQUENCY | 0.56/0.71 | 0.25/0.46 | 0.35/0.56 | 0.78/0.84 | 0.36/0.54 | 0.49/0.66 |
| TIME | 0.42/0.6 | 0.08/0.25 | 0.14/0.35 | 0.42/0.92 | 0.08/0.38 | 0.14/0.54 |
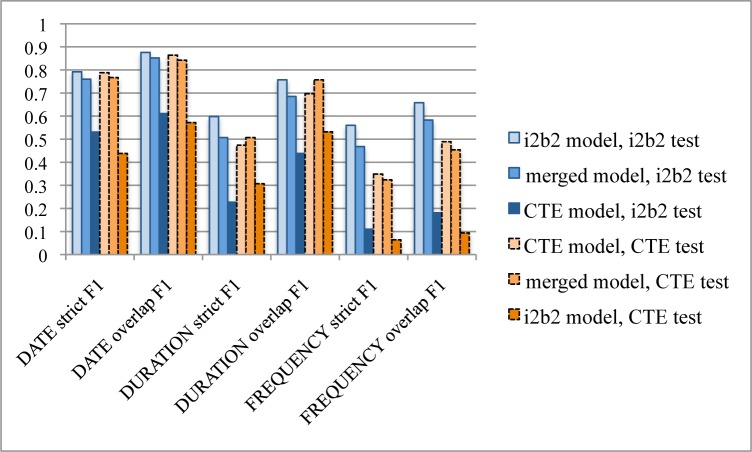
To further analyze differences and commonalities, the two test sets were also evaluated on all three created models. The least frequent TIMEX3 type (TIME) is not included since the number of training instances is insufficient. Not surprisingly, results are worst when applying each test set on the model created from the other corpus (e.g. training on Clinical TempEval and testing on i2b2), Figure 1. Specifically, results for DATE drop from 0.79 strict F1 (0.88 overlap F1) to 0.53 (0.61) on the i2b2 corpus, and from 0.79 (0.86) to 0.44 (0.57) on the Clinical TempEval corpus. For DURATION and FREQUENCY, the drop is from 0.60 (0.76) to 0.23 (0.44) and 0.56 (0.66) to 0.11 (0.18) on the i2b2 corpus. On the Clinical TempEval corpus, the drop is from 0.47 (0.70) to 0.31 (0.53) for DURATION and from 0.35 (0.49) to 0.06 (0.10) for FREQUENCY. Moreover, creating a merged model from the two training sets did not improve overall results for either test set (0.69 overall span strict F1 when evaluated on the i2b2 test set, and 0.69 when evaluated on the Clinical TempEval test set). However, results for DURATION on the Clinical TempEval test set are slightly improved when applied on the merged model (0.51 strict F1, 0.76 overlap).
Figure 1.
Classification results per TIMEX3 type DATE, DURATION and FREQUENCY, strict and overlap. Three models: i2b2 model, Clinical TempEval (CTE) model, and merged (the two training sets combined) model, each evaluated on the i2b2 and CTE test sets.
Qualitative error analysis, feature ablation evaluation and TIMEX3 type characterization
We perform a qualitative error analysis on true positives, false negatives, and false positives (exact match) from the system output of each test set applied on the model created from the other corpus’ training data. This way, we are able to characterize corpus-specific annotations, and also commonalities between the two corpora.
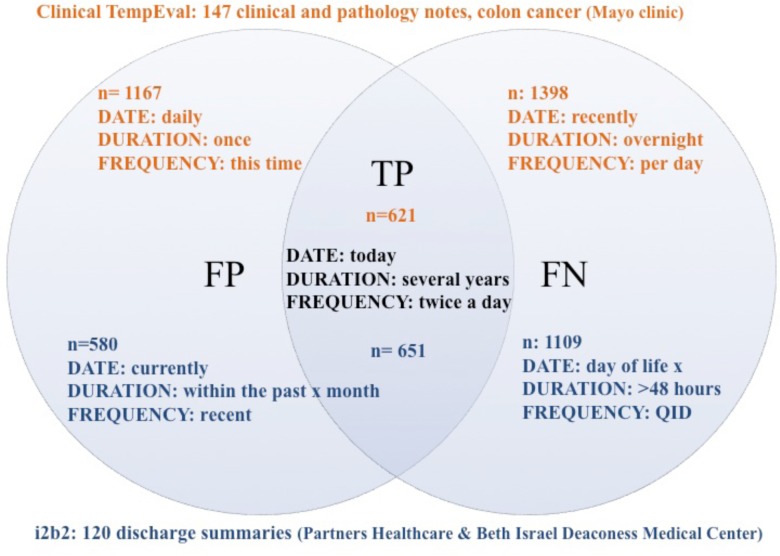
Most true positives on both test sets for DATE cover explicit dates such as “9 November, 2010”. These are also the most common DATE annotations in both corpora. However, DATE annotations such as “today”, “this time”, “many years ago”, “the same time” are also correctly classified in both test sets. For DURATION and FREQUENCY, common annotations include expressions like “several years” and “every x hours”. Examples are given in Figure 2.
Figure 2.
Venn diagram depicting error types and examples for each test set, based on system predictions for the i2b2 test set applied on the model built on the Clinical TempEval training data, and vice versa.
However, the differences outweigh the commonalities between the corpora, as evidenced by the results. The Clinical TempEval DATE annotations contain a larger number of pre-/post-operative expressions that are not covered in the i2b2 corpus, as well as relative DATE instances such as “recent” and “now”. The i2b2 corpus, on the other hand, contains more DATE expressions typical for the document type (discharge summaries), such as “day of life x”, “hospital day x”, “day of delivery”, “day of transfer”, “the time of discharge”, that are not covered in the TempEval model. This is reflected also in the false positive results: when evaluating the i2b2 test data annotations on the Clincal TempEval model, expressions such as “currently”, “recent”, “intraoperatively”, “the future”, “the time”, “that time” are erroneously predicted as DATEs.
For DURATION, the i2b2 corpus contains expressions such as “>48 hours”, “the entire night”, “one months time”, “overnight”, “the next several hospital days” that are missed by the Clinical TempEval model, while the i2b2 model fails to represent Clinical TempEval DURATION annotations such as “quite a few years”, “a while”, “lifelong”, “overnight”.
The i2b2 FREQUENCY annotations contain expressions not covered in the Clinical TempEval model, such as “q24”, “QID”, “qhs”, “bid”, “x 1”, while expressions such as “per day” and “xx/min” are common in the Clinical TempEval FREQUENCY annotations.
In addition to the lexical variations in the two corpora, the feature ablation analysis reveals that surrounding words are the most informative feature for all TIMEX3 types both when evaluating a trained model built on one corpus using the other test set, and when evaluating the merged model on each test set, Table 4. All other features have limited individual impact overall.
Table 4.
Feature ablation study for error analysis. Each feature was removed when building the TIMEX3 model, keeping all remaining features. Results are presented for the TIMEX3 types DATE, DURATION and FREQUENCY, once when building a model on one corpus (Clinical Tempeval (CTE) or i2b2) and evaluating on the other (cross-corpus), and once when building a model with both training sets (merged).
| DATE F1 | DURATION F1 | FREQUENCY F1 | ||||
|---|---|---|---|---|---|---|
| cross-corpus CTE/i2b2 | merged CTE/i2b2 | cross-corpus CTE/i2b2 | merged CTE/i2b2 | cross-corpus CTE/i2b2 | merged CTE/i2b2 | |
| all features | 0.53/0.44 | 0.77/0.76 | 0.23/0.31 | 0.51/0.51 | 0.11/0.06 | 0.32/0.47 |
| -current token | 0.48/0.48 | 0.73/0.69 | 0.18/0.23 | 0.28/0.39 | 0.07/0.08 | 0.22/0.46 |
| -gazeteer | 0.54/0.33 | 0.76/0.76 | 0.18/0.26 | 0.51/0.50 | 0.11/0.05 | 0.33/0.47 |
| -pos | 0.50/0.41 | 0.77/0.76 | 0.29/0.32 | 0.51/0.50 | 0.1/0.05 | 0.35/0.45 |
| -context | 0.38/0.16 | 0.58/0.46 | 0.00/0.04 | 0.04/0.03 | 0.00/0.07 | 0.11/0.33 |
| -capitaltype | 0.53/0.44 | 0.77/0.76 | 0.23/0.31 | 0.49/0.50 | 0.10/0.06 | 0.33/0.46 |
| -numerictype | 0.51/0.42 | 0.77/0.76 | 0.23/0.30 | 0.51/0.51 | 0.11/0.06 | 0.33/0.49 |
| -lowercase | 0.54/0.44 | 0.77/0.76 | 0.24/0.29 | 0.50/0.50 | 0.11/0.05 | 0.32/0.45 |
| -ngram2 | 0.53/0.43 | 0.77/0.76 | 0.23/0.32 | 0.52/0.50 | 0.11/0.06 | 0.33/0.47 |
Discussion and Conclusion
In this study, we present a systematic, simplistic, and scalable approach to 1) generalize existing solutions for automated TIMEX3 span detection, and 2) assess similarities and differences in different instantiations of TIMEX3 models applied on different clinical corpora.
Retraining and applying the top-performing TIMEX3 span detection solution from the 2015 Clinical TempEval challenge on the i2b2 corpus produces competitive results: 0.74 strict F1 and 0.87 overlapping F1. The top ten performing systems in the 2012 i2b2 challenge resulted in strict F14 between 0.67–0.80 and overlapping F1 between 0.8–0.916. Combining the two training data sets to create a merged model slightly decreases overall results when evaluating on each test set, but results for DURATION on the Clinical TempEval test set are improved, indicating that the addition of new training data from a different corpus was informative.
Enabling characterization and analysis of commonalities and differences between different time expression instantiations by evaluating system output errors from a model trained on one corpus and evaluating on another is informative. In particular, this method reveals that some TIMEX3 expressions can be accurately covered across the two corpora, in particular for the majority type DATE. The main differences between the studied corpora can be categorized into two types: lexical (and document-type specific) variants (e.g. “qid”) and structural (document type). Further, the feature ablation study sheds light on the informativeness of individual features, where surrounding tokens provide context that is clearly crucial for improved performance.
Rule-based systems were successful in the TIMEX3 span extraction subtask of the 2012 i2b2 challenge. Such systems generally produce high recall, while machine learning-based systems generally produce high precision results. Given the new knowledge about lexical variants for different TIMEX3 types in different corpora, we also want to study how to incorporate this information and how to best make use of a combination approach for all elements needed in a temporal reasoning solution.
Contributions and limitations
The main contribution of our approach is that it is systematic and simple in acquiring and building a high-performing task model for TIMEX3 classification. Moreover, our results do not necessarily depend on the system approach for TIMEX3 detection – one could choose to apply any of the previous successful machine learning-based approaches for comparison or further development. Second, we achieve competitive results for automated classification on a new corpus. Third, we identify similarities and differences in TIMEX3 modeling which could be informative in the development of a simplified, general temporal model. Finally, our in-depth analysis identifies lexical variants of time expressions that could be useful for several clinical information extraction use-cases and feature engineering. However, we are planning to tackle the following potential future points:
Our proposed approach is evaluated only on two corpora annotated with similar temporal models. Other models have been proposed for the clinical narrative domain, such as the Temporal Constraint Structure15. Applying our proposed approach on this model would probably require more mapping steps, as well as access to annotated documents for system development and assessment. An alternative approach to our mapping strategy could be to keep more granular types separate and build hierarchical structures (e.g. PREPOSTEXP is-a DATE) instead of mapping to broad TIMEX3 types – this way, one would get a richer representation while still keeping a generalizable overall model.
Document structure information, such as sections, is not taken into consideration when mapping annotations or for the feature engineering of the machine learning models, which is a central part of clinical narratives. We plan to incorporate this information in future extensions.
Our analysis of commonalities and differences is performed only on exact matched expressions: a richer overall picture would be obtained if also studying overlapping annotations. For successful unification of annotations, more training data, hierarchical ontologies, and annotation layers are needed, in particular for a more accurate and detailed analysis and model of less frequent types, such as FREQUENCY and TIME. The feature ablation study reveals that surrounding words are the most informative features. We intend to further study the impact of different context window sizes, and also whether the contextual information could be de-lexicalized in some way, to address generalizability across document types.
We will continue our work on developing a simplified, common temporal reasoning model for core elements needed in a full-scale temporal reasoning system. As a first step, we will employ our suggested approach on other available annotated corpora for further analysis and development. In particular, we plan to use the 2014 ShARe/CLEF eHealth corpus that contains disorder mention annotations along with semantic attributes, including time expression annotations. Since this corpus was created with a focus on disorder annotations, not overall temporal information, we expect that our proposed approach will have high coverage on extracting relevant time expressions, but that tailoring will be necessary for disorder context. We will also extend our work to include temporal expression normalization, as well as event span and attribute classification, and temporal relations.
Acknowledgments
We thank the Mayo clinic and the challenge organizers for providing access to the 2015 Clinical TempEval corpus, and i2b2 and the 2012 challenge organizers for providing access to their corpus. This work was partially funded by Swedish Research Council (350-2012-6658), NLM R01 LM010964, and United States Department of Veteran Affairs CREATE Information Extraction and Visualization Toolkit (IE-Viz) CRE 12-312.
Footnotes
University of Utah Institutional Review Board approval was obtained for using these corpora for research
different context windows of surrounding tokens were employed for each TIMEX3 type as a result of experiments on the Clinical TempEval training data: 5 preceding, 3 following for DATE, 3/3 for TIME, 4/4 for DURATION and 5/3 for FREQUENCY
results for strict matching were recalculated using the official 2012 i2b2 evaluation script on system submission outputs provided by the challenge organizers
References
- 1.Irvine AK, Haas SW, Sullivan T. TN-TIES: A System for Extracting Temporal Information from Emergency Department Triage Notes. AMIA Annual Symposium Proceedings. 2008;2008:328–332. [PMC free article] [PubMed] [Google Scholar]
- 2.Raghavan P, Lai AM. Leveraging natural language processing of clinical narratives for phenotype modeling; Proceedings of the 3rd workshop on Ph.D. students in information and knowledge management (PIKM ′10); New York, NY, USA: ACM; 2010. pp. 57–66. [Google Scholar]
- 3.Mowery DL, Jordan P, Wiebe J, Harkema H, Dowling J, Chapman WW. Semantic Annotation of Clinical Events for Generating a Problem List. AMIA Annual Symposium Proceedings. 2013;2013:1032–1041. [PMC free article] [PubMed] [Google Scholar]
- 4.Bethard S, Derczynski L, Pustejovsky J, Verhagen M. SemEval-2015 Task 6: Clinical TempEval; Proc 9th International Workshop on Semantic Evaluation (SemEval 2015); ACL; 2015. [Google Scholar]
- 5.Styler W, Bethard S, Finan S, Palmer M, Pradhan S, de Groen P, Erickson B, Miller T, Chen L, Savova GK, Pustejovsky J. Temporal annotations in the clinical domain. Transactions of the Association for Computational Linguistics. 2014;2:143–154. [PMC free article] [PubMed] [Google Scholar]
- 6.Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. JAMIA. 2013;20:806–813. doi: 10.1136/amiajnl-2013-001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mowery DL, Velupillai S, South BR, Christensen L, Martinez D, Kelly L, Goeuriot L, Elhadad N, Pradhad S, Savova GK, Chapman WW. Task 2: ShARe/CLEF ehealth evaluation lab 2014; CEUR Workshop Proceedings on CLEF; 2014. pp. 31–42. [Google Scholar]
- 8.Pustejovsky J, Lee K, Bunt H, Romary L. ISO-TimeML: An International Standard for Semantic Annotation; Proc 7th Intl Conference on Language Resources and Evaluation (LREC’10); ELRA; 2010. [Google Scholar]
- 9.Velupillai S, Mowery DL, Abdelrahman S, Christensen L, Chapman WW. BluLab: Temporal Information Extraction for the 2015 Clinical TempEval Challenge; Proc 9th International Workshop on Semantic Evaluation (SemEval 2015); ACL; 2015. [Google Scholar]
- 10.Bethard S, Ogren P, Becker L. ClearTK 2.0: Design Patterns for Machine Learning in UIMA; Proc 9th Intl Conference on Language Resources and Evaluation (LREC’ 14); ELRA; 2014. [PMC free article] [PubMed] [Google Scholar]
- 11.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Ass. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strötgen J, Gertz M. Multilingual and Cross-domain Temporal Tagging. Language Resources and Evaluation. 2013;47(2):269–298. [Google Scholar]
- 13.Xu Y, Wang Y, Liu T, Tsujii J, Chang EI. An end-to-end system to identify temporal relation in discharge summaries: 2012 i2b2 challenge. J Am Med Inform Assoc. 2013;20:849–58. doi: 10.1136/amiajnl-2012-001607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts K, Rink B, Harabagiu S. A flexible framework for recognizing events, temporal expressions, and temporal relations in clinical text. J Am Med Inform Assoc. 2013;20:867–75. doi: 10.1136/amiajnl-2013-001619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou L, Melton GB, Parsons S, Hripcsak G. A temporal constraint structure for extracting temporal information from clinical narrative. Journal of Biomedical Informatics. 2006;39:424–439. doi: 10.1016/j.jbi.2005.07.002. [DOI] [PubMed] [Google Scholar]