Abstract
Detection of early symptoms in cervical cancer is crucial for early treatment and survival. To find symptoms of cervical cancer in clinical text, Named Entity Recognition is needed. In this paper the Clinical Entity Finder, a machine-learning tool trained on annotated clinical text from a Swedish internal medicine emergency unit, is evaluated on cervical cancer records. The Clinical Entity Finder identifies entities of the types body part, finding and disorder and is extended with negation detection using the rule-based tool NegEx, to distinguish between negated and non-negated entities. To measure the performance of the tools on this new domain, two physicians annotated a set of clinical notes from the health records of cervical cancer patients. The inter-annotator agreement for finding, disorder and body part obtained an average F-score of 0.677 and the Clinical Entity Finder extended with NegEx had an average F-score of 0.667.
Introduction
Cervical cancer
Cervical cancer is one of the most common cancers worldwide1, frequently affecting young women below age 40. Therefore screening and early detection is essential2. While cervical cancer incidence and mortality rates have dropped in countries where screening procedures have evolved into established prevention methods3,4, the rates in less developed countries are still high.
Cervical cancer, highly mortal in advanced stages, is usually described as having few early symptoms except vaginal discharge, bleeding and pain post coitus. Early detection of cervical cancer is however crucial for treating it successfully. Novel ways to diagnose the disorder at even earlier stages would be highly valuable, as this could prevent treatable pre-cancer from turning into invasive cancer4. Early detection is yet sometimes hindered since not all women participate in cervical screening programs.
Today, electronic health records that describe the whole healthcare period of a patient are available in many countries including Sweden. To define and detect these possible early symptoms in a patient’s health record, text-mining tools capable of identifying symptoms discriminatory for cervical cancer, are needed.
The overall aim of this study is to evaluate the performance of two previously developed text mining tools on health records of patients with cervical cancer, with a further aim to detect and discover early symptoms in a patient’s medical history.
Text mining in the cervical cancer domain
Previous research in the field of text mining and cervical cancer has mostly focused on oncologic documents and pathology reports. One study used text mining of pathology reports, with the aim to transfer unstructured pathology reports to a structured database5. A review article6 regarding different approaches for clinical text mining within the cancer domain describes only two studies focused on cervical cancer. Both studies concern methods used to retrieve scientific oncology documents relevant to clinical decisions within a particular domain of cervical cancer. None of them are in the domain of clinical text mining of symptoms relating to cervical cancer. However, there are two articles on the subject of cervical cancer staging. In the first, the authors used 250 cervical patient records for their experiments. The cervical cancer tumors were assigned 15 parameters to classify the stage of the cancer and 0.73 accuracy was obtained using a neural network architecture7. In the second article, 221 cervical patient cases were used as input to a staging system using soft computing/neural networks. The cases were classified in stages I-IV and a classification score of 0.79 was obtained on test data. The C4.5 machine-learning algorithm was also applied on the data set, obtaining a score of 0.808.
In another study on tumor staging, the authors trained Naïve Bayes, Bayesian Network, Support Vector Machines, and Random Forest algorithms on manually annotated pathology reports from one hospital and evaluated the portability on another hospital’s pathology reports. They noted a decline in performance with at least 25 percent9. However, in a later study they improved their results considerably to only a few percent decline in cross-hospital evaluation, by using feature selections to tune the classifiers and simple rules to identify numeric values10.
Named entity recognition and negation detection
The Clinical Entity Finder (CEF) is a tool for entity recognition based on the machine-learning algorithm CRF++a and trained on manually annotated clinical entities in Swedish texts from an internal medicine emergency unit11 An earlier approach to detect findings and disorders in the cervical cancer domain using Clinical Entity Finder revealed that many of the entities found by the tool were negated12. Negations are important in essentially all clinical text, as many diagnoses are made based at least in part on exclusion of certain symptoms or test results. Negation detection have for example been incorporated in adverse drug reaction detection text mining workflows13 and similarly in the search for diagnostic codes in free text in electronic medical records14. The Clinical Entity Finder has no built-in negation detection and a possible tool for negation detection in clinical text is NegEx, a rule based tool. NegEx has previously been adapted to Swedish with a set of Swedish negation triggers15.
As it has been shown that different clinical subspecialties use clinical language with different distinctive features16, 17 a decrease in the performance of a natural language processing (NLP) system developed in one clinical domain is expected when applied in another clinical domain. The specific aim of this study is to investigate the efficiency of existing tools for text mining of Swedish health records, i.e. for entity recognition and negation detection, respectively, on the subdomain of clinical notes relevant for text mining studies on cervical cancer.
The research questions are:
How well will a machine learning based tool (Clinical Entity Finder) trained on clinical notes from an internal medicine emergency unit automatically annotate cervical cancer symptoms in physicians’ notes from departments of gynecology and oncology?
How well can the Swedish NegEx differentiate between negated and non-negated symptoms of cervical cancer?
Methods and Materials
Data
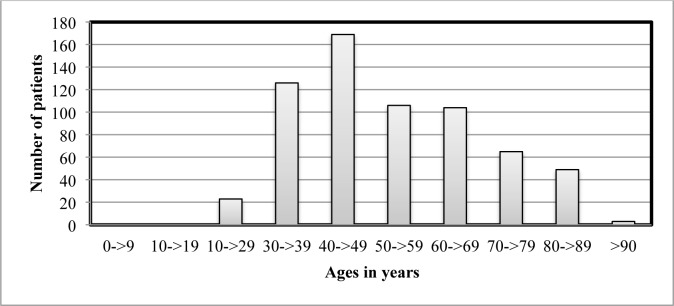
The Stockholm EPR Corpus18 is a database containing large quantities of clinical text in Swedish; over 600 000 patient records encompassing over 500 health care units from the Karolinska University Hospital. For this study, all patient recordsb from the departments of obstetrics/gynecology and oncology from the years 2009–2010 with a diagnosis code for malignant neoplasms in the cervix (ICD-10 codes C53.0, C53.1, C53.8 and C53.9), were extracted, resulting in 646 patient records. The extracted data, containing notes from nurses, physicians and other professionals, is called the Cervical Cancer Corpus. Figure 1 shows the age distribution of the patients in the Cervical Cancer Corpus. For this study the physicians’ notes, in total 17,263 notes and 776,719 tokens were used.
Figure 1.
Age distribution of patients in the Cervical Cancer Corpus
Annotation
Annotation is the process of marking relevant entities in a text. This is performed manually by domain experts, and the annotation can be used to train a machine learning tool for automatic entity recognition. Manually annotated text is also used for the evaluation of such tools.
In this study, the entity types finding, disorder and body part were annotated. Both findings and disorders can be relevant as symptoms or indications of cervical cancer. The entity type body part is also relevant since it can give the location of symptoms. A finding is an observation made at a certain point in time and it is not necessarily abnormal, it can be a symptom reported by the patient as well as a finding from a medical examination. Disorders are abnormal and can be present even when they are not observable. The annotation performed in this study follows a set of guidelines based on those used when developing the Clinical Entity Finder, with added instructions for annotating negations12.
Pre-annotation with the Clinical Entity Finder and NegEx
Pre-annotation is when a tool is used to mark relevant entities in a text before the manual annotation. This pre-annotation can then be used as a timesaving support for the annotators when doing the manual annotation. All physicians’ notes from the Cervical Cancer Corpus were pre-annotated using the Clinical Entity Finder11. The Clinical Entity Finder was used to pre-annotate instances of the named entities body part, finding and disorder. To distinguish between negated and non-negated entities, the Clinical Entity Finder was augmented with the tool NegEx, which was used to classify entities of the types finding and disorder in the corpus as negated or non-negated. NegEx uses lists of negation triggers and looks for the presence of those triggers in text surrounding the annotated instances. A trigger can be a word or a sequence of words indicating negation. An example of a sentence with a negation is Patienten har ej smärta (The patient has no pain), where the finding smärta (pain) is negated by the trigger ej. Body parts and words outside of annotations were not investigated for negation.
Manual annotation of the physicians’ notes
Two physicians annotated subsets of the physicians’ notes from the Cervical Cancer Corpus. One physician is experienced in the text mining domain and in annotation (Annotator A), and one physician is an expert in the area of cervical cancer (Annotator B). They received the pre-annotated data, agreed on the interpretation of guidelines, and revised the annotations performed by the Clinical Entity Finder, using the Brat annotation tool19. To properly evaluate the tools at least 100 manually annotated notes were considered as necessary to ensure variation and coverage. Annotator A and Annotator B annotated 180 and 100 notes respectively. The annotators removed faulty annotations, changed the span or type of annotations when needed, added missing annotations and marked annotations when negated. Figure 2 shows an example of a text pre-annotated by the Clinical Entity Finder with corrections carried out by Annotator A.
Figure 2.
Screenshot of one sentence processed by the Brat annotation tool showing the pre-annotation in green and the manual annotation in blue. (Key phrases are translated to English).
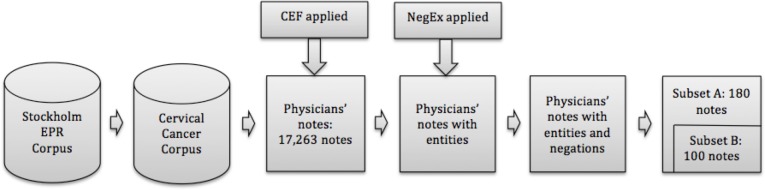
An overview of the annotation pipeline is shown in Figure 3. The 17,263 physicians notes were firstly pre-annotated using the Clinical Entity Finder. In the next step all disorders and findings found by the Clinical Entity Finder were investigated for possible negations using NegEx. Finally, 180 of these pre-annotated notes were randomly selected for manual annotation (subset A). Annotator A corrected all of them, and Annotator B corrected 100 of them (subset B). The annotator agreement was measured on subset B and the performance of the Clinical Entity Finder was measured on subset A.
Figure 3.
Annotation pipeline, note that subset B is covered by subset A, both annotators annotated subset B but Annotator A annotated another 80 notes.
Evaluation metrics
The performance of the tools was measured using precision (P), recall (R) and F1-score20. A high precision indicates that found named entities are correctly classified; a high recall shows that a large portion of named entities in the text are found, and the F1-score is the harmonic mean of the two. To calculate the scores, the number of true positives (TP), true negatives (TN) and false negatives (FN) were counted. A true positive is achieved when a tool has correctly annotated a named entity, a false positive occurs when a tool has incorrectly annotated a named entity, and a false negative when a tool has failed to detect a named entity that is present in the text.
The inter-annotator agreement between Annotator A and Annotator B was measured using the F1-score. The inter-annotator agreement can be viewed as an indicator of how well two individuals agree on this task and this can be compared to how well a tool can be expected to perform that same task.
Results
Results of the manual annotation
The annotators A and B each annotated 100 physicians’ notes from medical records of cervical cancer patients. Table 1 shows the confusion matrix for the two annotators. The rows show the annotations performed by Annotator A and the columns show the annotations performed by Annotator B. The diagonal shows the numbers of true positives – instances of named entities where the annotators agree on the type and the span of the annotation. To get a match, the two annotators must pick the same exact span of text. An example of when the annotators have marked different spans is when one annotator marked the span skivepitelcancer in situ (squamous cell carcinoma in situ) as a disorder but the other only marked the word skivepitelcancer (squamous cell carcinoma) in the same note. Such scope errors fall into the No exact match category, together with the cases of annotations carried out by only one of the annotators, either because of disagreement or by mistake. The level of inter-annotator agreement between Annotator A and Annotator B for exact and also partial match where there is at least one overlapping token (word), is shown in Table 2.
Table 1.
Confusion matrix for Annotator A and Annotator B obtained from 100 clinical notes
| Body part | Finding | Disorder | Ncg. Finding | Ncg. Disorder | No exact match | |
|---|---|---|---|---|---|---|
| Body part | 253 | 4 | 0 | 0 | 0 | 95 |
| Finding | 5 | 256 | 27 | 3 | 0 | 153 |
| Disorder | 2 | 3 | 108 | 0 | 4 | 28 |
| Ncg. Finding | 0 | 5 | 0 | 48 | 6 | 11 |
| Ncg. Disorder | 0 | 0 | 7 | 1 | 1 | 0 |
| No exact match | 36 | 141 | 62 | 16 | 5 | – |
Table 2.
F1-scores for Inter-Annotator Agreement between Annotator A and B. The first row shows the scores when only exact matches are allowed; the second row shows the scores when also including partial matches.
| Entity type | Body part | Finding | Disorder | Negated Finding | Negated Disorder |
|---|---|---|---|---|---|
| A, B exact match | 0.78 | 0.60 | 0.62 | 0.70 | 0.08 |
| A, B partial match | 0.79 | 0.76 | 0.73 | 0.81 | 0.08 |
Results of the annotation performed by the Clinical Entity Finder and NegEx
All of the physicians’ notes in the Cervical Cancer Corpus were processed by the Clinical Entity Finder and NegEx, and when evaluating the tools, the annotations carried out by Annotator A were used as a Gold Standard, assuming that Annotator A had correctly classified all named entities in the text. Annotator A and the tools were in complete agreement for 54 percent of annotations and Table 3 shows the confusion matrix for Annotator A and the tools, while Table 4 gives the precision, recall and F1-score for each of the entity types. Table 4 also demonstrates the improved scores when allowing partial matches, where there is at least one overlapping token, excluding punctuation.
Table 3.
Confusion matrix for Annotator A and the Clinical Entity Finder and NegEx combined obtained from 180 clinical notes
| Body part | Finding | Disorder | Neg. Finding | Neg. Disorder | No exact match | |
|---|---|---|---|---|---|---|
| Body part | 285 | 4 | 1 | 0 | 0 | 258 |
| Finding | 0 | 401 | 6 | 16 | 2 | 375 |
| Disorder | 1 | 3 | 181 | 0 | 2 | 62 |
| Ncg. Finding | 0 | 19 | 1 | hi) | -> | 47 |
| Ncg. Disorder | 0 | 0 | 4 | 0 | 14 | 0 |
| No exact match | 29 | 89 | 35 | 5 | 0 | – |
Table 4.
Precision, recall and the F1-score for the Clinical Entity Finder and NegEx using annotator A as Gold Standard. The evaluation was performed on 180 clinical notes and the scores for both exact and partial matches are given.
| Entity type | Body part | Finding | Disorder | Negated Finding | Negated Disorder | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
Scope Precision |
Exact | Partial | Exact | Partial | Exact | Partial | Exact | Partial | Exact | Partial |
| 0.90 | 0.96 | 0.78 | 0.89 | 0.80 | 0.85 | 0.74 | 0.77 | 0.70 | 0.70 | |
| Recall | 0.52 | 0.55 | 0.50 | 0.57 | 0.73 | 0.78 | 0.47 | 0.48 | 0.78 | 0.78 |
| F1-score | 0.66 | 0.70 | 0.61 | 0.70 | 0.76 | 0.81 | 0.57 | 0.59 | 0.74 | 0.74 |
Since NegEx only looks for negations when a finding or disorder has been annotated the performance of NegEx is directly dependent on the performance of the Clinical Entity Finder. The scores given in Table 4 for negated finding and negated disorder are therefore not perfectly suitable for evaluation of how well NegEx performs in the domain of cervical cancer. An additional evaluation has therefore been performed where all findings and disorders annotated both by Annotator A and CEF were selected. By selecting the instances already agreed on, the performance of NegEx could be isolated. There were 500 such instances and of those 57 were marked as negated by both Annotator A and NegEx. Precision, recall and F1-score for the ability of NegEx to correctly determine negations was calculated to be 0.78, 0.75 and 0.76 respectively.
Results of porting the Clinical Entity Finder
Table 5 shows the performance of the Clinical Entity Finder and NegEx in our setting compared to the performance of the Clinical Entity Finder applied to the type of data used to train on; health records from emergency units.
Table 5.
The achieved F1-scores of the Clinical Entity Finder combined with NegEx compared to the F1-scores of the Clinical Entity Finder on health records from emergency units11.
| Entity type | Body part | Finding | Disorder | Neg. Finding | Neg. Disorder | Finding + Disorder |
|---|---|---|---|---|---|---|
| CEF + NegEx, cervical cancer records | 0.66 | 0.61 | 0.76 | 0.47 | 0.78 | 0.65 |
| CEF, emergency unit records | 0.85 | 0.69 | 0.81 | – | – | 0.78 |
Error analysis
A manual error analysis has been performed on 60 of the annotated notes produced by the Clinical Entity Finder and NegEx as compared to the annotations done by Annotator A. The automatic and manual annotation most often classified the annotated entities in the same way, a recurring mistake, however, was made when a compound word for a disorder contained words describing a body part. Such compound words were sometimes wrongly classified as a body part. An example of this is the word skelettmetastaser (skeletal metastases), which contains the Swedish word for skeleton. Another source of error is the word cervixcancer (cervical cancer), a compound word in Swedish, which in many instances in the records was written as two words, cervix cancer. The CEF sometimes interpreted this as two separate entities; the body part cervix and the disorder cancer.
A second step of analysis was performed by looking at instances only annotated by either the automatic or manual annotation. The analysis found 333 false negatives, meaning annotations marked by Annotator A but missed by the tools, and 71 false positives, meaning annotations only performed by the tools. Table 6 shows the different errors sorted by type.
Table 6.
The number of false positives and false negatives found in error analysis.
| Scope Errors | Body part | Finding | Disorder | Total | |
|---|---|---|---|---|---|
| False Positives | 47 | 2 | 16 | 6 | 71 |
| False Negatives | 65 | 116 | 132 | 20 | 333 |
After excluding scope errors, about half of the cases of false negatives in all categories were caused by the Clinical Entity Finder failing to detect words and expressions associated with cervical cancer. For example in the category body part, the word cervix was missed several times. The false negatives for disorders directly related to cervical cancer included for example dysplasi (dysplasia) and cancer in situ. Several of the missed disorders were either misspellings or abbreviations, for example the abbreviation cervixca for cervixcancer (cervical cancer in Swedish) and skiveptilcancer a misspelling of skivepitelcancer (squamous cell carcinoma) were both missed by the tools.
The largest source of false positives for disorders and findings were expressions describing procedures or drugs, for example preventivmedel (contraception). For body parts, the false positives originate from expressions where a body part word was part of a non-body part expression. For example, the expression över huvud taget (at all) contains the Swedish word for head (huvud), which causes a faulty annotation.
The individual evaluation of NegEx on 500 instances of findings and disorders resulted in 16 false positives and 19 false negatives. The false positives were almost all due to instances of findings and disorders appearing close to unrelated negation triggers. The false negatives were mainly caused by negation expressions being used that were not included in the lists of negation triggers used by NegEx and there was also a few cases were the manual annotation had failed to discover a negation and borderline cases.
Named entities and negations detected in the physicians’ notes
Among the 776,719 tokens in physicians’ notes the Clinical Entity Finder identified 58,366 disorders, findings and body parts. Of these, 43,334 were either classified as findings or disorders and a total of 12 percent of findings and disorders were determined to be negated by NegEx. Table 7 shows the most common symptoms and most frequently negated symptoms in the physicians’ notes. The findings and disorders occurring more than 10 times in the text are also sorted on how large a proportion of them that is negated.
Table 7.
The most frequent findings, disorders and negations found in the physicians’ notes.
| Most frequent findings and disorders | Nbr of i ns t a nc e s | Most frequently negated findings and disorders | Nbr of negated instances | Findings and disorders with highest portion of negation | Portion negated |
|---|---|---|---|---|---|
| cervixcancer (cervical cancer) | 873 | besvär (trouble/problem) | 338 | gynekologiska besvär (gynecological problems) | 1.0 |
| besvär (trouble/problem) | 790 | feber (fever) | 243 | palpabla resistenser (palpable resistance) | 1.0 |
| illamående (nausea) | 677 | illamående (nausea) | 198 | särskilda besvär (particular problems) | 1.0 |
| mår bra (feels well) | 662 | blödningar (bleedings) | 171 | nytillkomna symtom (new symptoms) | 0.96 |
| smärta (pain) | 656 | smärta (pain) | 150 | nytillkomna besvär (new problems) | 0.92 |
| tumör (tumor) | 642 | smärtor (pains) | 126 | infektionstecken (signs of infection) | 0.89 |
| smärtor (pains) | 629 | blödning (bleeding) | 99 | biljud (murmur) | 0.86 |
| feber (fever) | 562 | infektionstecken (signs of infection) | 91 | tumörstrukturer (tumour structures) | 0.83 |
| cancer (cancer) | 508 | tumör (tumor) | 83 | tumörsuspekta förändringar (tumor suspicious changes) | 0.82 |
| blödningar (bleedings) | 491 | nytillkomna besvär (new troubles) | 79 | subjektiva besvär (subjective problems) | 0.8 |
| blödning (bleeding) | 482 | buksmärtor (pain of the abdomen) | 72 | tumörsuspekt (tumor suspicion) | 0.80 |
| skivepitelcancer (squamous cell carcinoma) | 428 | hydronefros (hydronephrosis) | 65 | spridning (spreading) | 0.78 |
Discussion
In this study, the efficiency of two existing tools for text mining of Swedish clinical text, i.e. for entity recognition and negation detection, was evaluated on a corpus of health records from patients with cervical cancer. The approach involved both manual and automated annotation of clinical entities, including negations. The investigation gave promising results and suggestions for further improvements. The issues studied relates partly to the problem of porting tools between subdomains of clinical text, and partly to the problem of inter-tool dependency when using a pipeline. As a result of this study, lists of clinical entities relevant to cervical cancer were created.
The term “sublanguage” is used to display that language in specialized domains exhibit characteristics that set them apart from general language21. However, clinical language is not homogeneous but consists of several specialized domains that exhibit the characteristics of sublanguages. The subdomain language will influence the creation of NLP tools for clinical text, as a tool relying on term statistics or semantics trained on one clinical note type have been suggested to not work as well on another. Clinical notes from different professions and specialties have been shown to cluster into readily distinguishable groups of lexical and semantic features17.
Here, a machine-learning tool called Clinical Entity Finder trained on clinical notes from a medical emergency unit was applied to notes in a Cervical Cancer Corpus. In both sub corpora, only notes written by physicians were used, as to avoid the further complexity of inter-professional sublanguages. Evaluation by comparing to manual annotation showed that precision was higher than recall for all of the annotated entity types, and manual inspection of tokens classified as findings, disorders, or body parts by the Clinical Entity Finder substantiates the precision of the tool. There were very few apparently faulty classifications; the lower recall was however often related to the tool being unable to classify expressions associated with cervical cancer.
NegEx classifies 12 percent of the findings and disorders in the data as negated; this is line with the findings of the manually annotated English clinical BioScope corpus that contained 14 percent negations22. In another annotation study on clinical text from the internal medicine emergency unit, 19 percent of diagnosis expressions were found to be negated23. The low F-scores for inter-annotator agreement on negated disorders was partly due to the disagreement between finding/disorder and not only because of disagreement on negation/non-negation. Error analysis did not indicate that the domain of cervical cancer affected the performance of NegEx. Instead the error analysis showed that NegEx gave false positives in some cases where an entity appears close to a negation trigger without actually being affected by it. False negatives where mainly the result of unusual negation triggers used in the physicians’ notes.
Findings and disorders associated with cervical cancer were often negated in the records, this is perhaps due to the fact that the patients in the corpus have known cancers and it is therefore of importance to document the presence or absence of such symptoms. Furthermore, as expected, statements regarding the general well being of the patient such as mår bra (feels fine) or orolig (worried) were frequently present in the records. These kinds of statements were actually seldom negated. Previous studies have found that affirmed and negated findings may be expressed on different levels of hierarchy24. Here, this can be exemplified by the findings gynecological problems and signs of infection that were dominantly negated. However, when the patients had such problems, they were described as more fine-grained symptoms, such as bleeding, fever or pain, as opposed to the higher hierarchy general terms.
Manual annotation of clinical text is a time consuming and costly process and there is a need for domain experts or training of non-experts to obtain good quality annotations. Crowdsourcing is not an option due to the sensitive nature of the data. A tool that makes the annotation process easier is therefore valuable. It is also inevitable for human annotators to miss some entities when annotating, and the tools described here may increase the quality of the annotation in such cases.
Future work
Future work includes to extend the training data of the Clinical Entity Finder with the manually annotated cervical cancer health records, as this could be expected to improve the results on the cervical cancer domain by reducing the number of false negatives as indicated by the error analysis. The annotations performed in this study were on a relatively small amount of text. For the retraining of the tool, more annotation is needed, and a Gold Standard needs to be constructed by reviewing the annotations. The error analysis showed that a majority of false positives for disorders and findings were incorrectly classified procedures or drugs; it could therefore perhaps be useful to include procedures and drugs as entity classes in the annotation to increase the precision of the Clinical Entity Finder. The symptoms found by the tools revealed that the same symptom was written in many different ways. This indicates the need for further usage of stemming, lemmatization and perhaps clustering of the found symptoms, to be able to draw more robust conclusions from our findings.
Conclusion
To our best knowledge, this is the first time cervical cancer narratives have been annotated both by human annotators and a machine learning based tool. By porting the Clinical Entity Finder to this new domain from the domain in which it was trained, the performance was reduced. Precision is still good, but the recall is decreased. This is expected and could be caused by the fact that the new domain contains patterns previously unseen by the tool.
The performance of negation detection was less dependent on the domain change, but since the negation detection is dependent on entities being found by the Clinical Entity Finder, its performance was more difficult to evaluate.
The Clinical Entity Finder was shown to be a useful tool for pre-annotation, since both annotators perceived it as making the annotation process more efficient as compared to annotating text without pre-annotation.
The clinical text found in health records is a promising source of information and the tools evaluated here could be a first step towards finding early symptom patterns in patients with cervical cancer.
Acknowledgments
We thank Maria Skeppstedt for letting us use the Clinical Entity Finder and we also thank Claudia Ehrentraut for her excellent initial study on cervical cancer text mining. This work was supported by the Nordic Information for Action eScience Center (NIASC); a Nordic Center of Excellence financed by NordForsk (Project number 62721). This work was also partly supported by the project High-Performance Data Mining for Drug Effect Detection at Stockholm University, funded by the Swedish Foundation for Strategic Research under grant IIS11-0053.
Footnotes
This study has been approved by the Regional Ethical Review Board in Stockholm (Etikprövningsnämnden i Stockholm), permission number 2014/1882-31/5
References
- 1.Cancer Research UK “Worldwide cancer incidence statistics,” visited: November 13th 2014. [Online]. Available: http://www.cancerresearchuk.org/cancer-info/cancerstats/world/incidence/#Common.
- 2.Sundström K. PhD dissertation. Department of Medical Epidemiology and Biostatistics, Karolinska Institutet; Stockholm, Sweden: 2013. Human papillomavirus test and vaccination – impact on cervical cancer screening and prevention. [Google Scholar]
- 3.Axelsson A, Borgfeldt C. “Cervixcancer”, internetmedicin. [Online] Jul, 2013. Available: http://www.internetmedicin.se/page.aspx?id=2735.
- 4.Society AC. “Cervical Cancer Prevention and Early Detection” [Online] 2014. Available: http://www.cancer.org/acs/groups/cid/documents/webcontent/003094-pdf.pdf.
- 5.Coden A, Savova G, Sominsky I, Tanenblatt M, Masanz J, Schuler K, Cooper J, Guan W, de Groen PC. Automatically extracting cancer disease characteristics from pathology reports into a Disease Knowledge Representation Model. Journal of biomedical informatics. 2009;42(5):937–949. doi: 10.1016/j.jbi.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Spasić I, Livsey J, Keane JA, Nenadić G. Text mining of cancer-related information: Review of current status and future directions. International journal of medical informatics. 2014;83(9):605–623. doi: 10.1016/j.ijmedinf.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Phinjaroenphan P, Bevinakoppa S. Automated prognostic tool for cervical cancer patient database; Intelligent Sensing and Information Processing, 2004. Proceedings of International Conference on; IEEE; 2004. pp. 63–66. [Google Scholar]
- 8.Mitra P, Mitra S, Pal SK. Staging of cervical cancer with soft computing. Biomedical Engineering, IEEE Transactions on. 2000;47(7):934–940. doi: 10.1109/10.846688. [DOI] [PubMed] [Google Scholar]
- 9.Martinez D, Cavedon L, Pitson G. Stability of text mining techniques for identifying cancer staging; Louhi, The 4th International Workshop on Health Document Text Mining and Information Analysis; Canberra, Australia: NICTA; 2013. [Google Scholar]
- 10.Martinez D, Pitson G, MacKinlay A, Cavedon L. Cross-hospital portability of information extraction of cancer staging information. Artificial intelligence in medicine. 2014;62(1):11–21. doi: 10.1016/j.artmed.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Skeppstedt M, Kvist M, Dalianis H, Nilsson GH. Automatic recognition of disorders, findings, pharmaceuticals and body structures from clinical text: An annotation and machine learning study. Journal of Biomedical Informatics. 2014;49:148–158. doi: 10.1016/j.jbi.2014.01.01. [DOI] [PubMed] [Google Scholar]
- 12.Ehrentraut C, Dalianis H, Sundström K. In: Krogstie J, Juel-Skielse G, Kabilan V, editors. Exploration of Known and Unknown Early Symptoms of Cervical Cancer and Development of a Symptom Spectrum – Outline of a Data and Text Mining Based Approach; Proceedings of the CAiSE-2015 Industry Track co-located with 27th Conference on Advanced Information Systems Engineering (CAiSE 2015); Stockholm, Sweden: CEUR; Jun 11, 2015. 2015. pp. 34–44. urn:nbn:de:0074-1381-0. [Google Scholar]
- 13.Eriksson R, Werge T, Jensen LJ, Brunak S. Dose-specific adverse drug reaction identification in electronic patient records: temporal data mining in an inpatient psychiatric population. Drug Saf. 2014 Apr;37(4):237–47. doi: 10.1007/s40264-014-0145-z. Erratum in: Drug Saf. 2014 May;37(5):379.PMID: 24634163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roque FS, Jensen PB, Schmock H, Dalgaard M, Andreatta M, Hansen T, Søeby K, Bredkjær S, Juul A, Werge T, Jensen LJ, Brunak S. Using electronic patient records to discover disease correlations and stratify patient cohorts. PLoS Comput Biol. 2011 Aug;7(8):e1002141. doi: 10.1371/journal.pcbi.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skeppstedt M. Negation detection in Swedish clinical text: An adaption of NegEx to Swedish. J Biomedical Semantics. 2011;2(S-3):S3. doi: 10.1186/2041-1480-2-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith K, Megyesi B, Velupillai S, Kvist M. Professional Language in Swedish Clinical Text – Linguistic Characterization and Comparative Studies. Nordic Journal of Linguistics. 2014;37(2):297–323. [Google Scholar]
- 17.Patterson O, Hurdle JF. Document Clustering of Clinical Narratives: A Systematic Study of Clinical Sublanguages; AMIA Annual Symposium Proceedings; American Medical Informatics Association; 2011. pp. 1099–1107. [PMC free article] [PubMed] [Google Scholar]
- 18.Dalianis H, Hassel M, Henriksson A, Skeppstedt M. Stockholm EPR Corpus: A Clinical Database Used to Improve Health Care; Proceedings of the Fourth Swedish Language Technology Conference, (SLTC-2012); October 25–26; Lund, Sweden: 2012. pp. 17–18. [Google Scholar]
- 19.Stenetorp P, Pyysalo S, Topić G, Ohta T, Ananiadou S, Tsujii JI. BRAT: a web-based tool for NLP-assisted text annotation; Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics; Association for Computational Linguistics; 2012. pp. 102–107. [Google Scholar]
- 20.van Rijsbergen CJ. Information Retrieval. Butterworth: 1979. [Google Scholar]
- 21.Harris ZS. Mathematical structures of language. Interscience Publishers; 1968. [Google Scholar]
- 22.Vincze V, Szarvas G, Farkas R, Móra G, Csirik J. The BioScope corpus: biomedical texts annotated for uncertainty, negation and their scopes. BMC bioinformatics. 2008;9(Suppl 11):S9. doi: 10.1186/1471-2105-9-S11-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanushi H, Dalianis H, Duneld M, Kvist M, Skeppstedt M, Velupillai S. Negation scope delimitation in clinical text using three approaches: NegEx, PyConTextNLP and SynNeg; 19th Nordic Conference of Computational Linguistics (NODALIDA 2013); May 22–24; Oslo, Norway: Linköping University Electronic Press; 2013. pp. 387–474. [Google Scholar]
- 24.Kvist M, Skeppstedt M, Velupillai S, Dalianis H. In: Fensli, Dale, editors. Modeling human comprehension of Swedish medical records for intelligent access and summarization systems – Future vision, a physician’s perspective; Proc 9th Scandinavian Conf. on Health Informatics, SHI2011; Oslo: Tapir Academic Press; 2011. pp. 31–35. [Google Scholar]