Abstract
In this paper we sought to reproduce, as a computational retrospective study in an EHR database (MIMIC-II), a recent large prospective clinical study: the 2013 publication, by the Japanese Association for Acute Medicine (JAAM), about disseminated intravascular coagulation, in the journal Critical Care (PMID: 23787004). We designed in SQL and Java a set of electronic phenotypes that reproduced the study’s data sampling, and used R to perform the same statistical inference procedures. All produced source code is available online at https://github.com/fabkury/paamia2015. Our program identified 2,257 eligible patients in MIMIC-II, and the results remarkably agreed with the prospective study. A minority of the needed data elements was not found in MIMIC-II, and statistically significant inferences were possible in the majority of the cases.
Introduction and objectives
The presently widening scale of production and sharing of electronic health records (EHRs) [2] increases the interest on possible secondary uses for them. In this paper, we approached one publicly available dataset of electronic health records – the MIMIC-II database [3], version 2.6 – and applied one possible study design, namely, a computational retrospective study [4].
The central objective of our retrospective study was to demonstrate how MIMIC-II could be used to reproduce, and thereby validate, a contemporary prospective clinical study. The chosen prospective clinical study was by Gando et al., for the Japanese Association for Acute Medicine (JAAM) Sepsis Registry Study Group, published in 2013 in the journal Critical Care Medicine [1], hereafter referred to as the reference study. The objective of the reference study was to validate the prognostic value of the JAAM scoring system for diagnosis of disseminated intravascular coagulation (DIC). It was performed in Japan, from June 1, 2010 to May 31, 2011, and largely succeeded in demonstrating, among other things, that the higher the JAAM DIC score of the patient on day 1 of diagnosis of severe sepsis, the higher the mortality and morbidity. Therefore, our central objective was to reproduce the reference study in the MIMIC-II data, as an inexpensive computational, retrospective, observational study.
Scoring systems for disseminated intravascular coagulation
Disseminated intravascular coagulation (DIC) is a health condition characterized by the intravascular activation of the coagulation cascade [5] that is secondary to a range of disparate causes such as sepsis, pancreatitis, malignancy, heat stroke, and others. Prospective clinical studies have demonstrated that the development of DIC in patients with sepsis or severe trauma roughly doubles the risk of death, and is an independent predictor or mortality [7]. While its treatment hallmark remains to treat the provoking cause, there is interest in improving the identification of poor prognosis in DIC patients so that a more intensive or specialized therapy can be started earlier [8] or later [16] in the progression of the disease. The definition of DIC itself is not free from debate, and four scoring systems for the diagnosis of this condition have been published in the clinical literature by four institutions [5]. In this paper, we, intentionally replicating the reference study, applied and compared the one created and revised by the Japanese Association for Acute Medicine, and the one by the International Society of Thrombosis and Hemostasis (ISTH) (Table 1). Of note, our source code, available online [9], presently allows the computation of these two scoring systems as well as the other two (KSTH and JMHLW), which are outside the scope of the present study.
Table 1.
DIC scoring systems by the JAAM and the ISTH
| Japanese Association for Acute Medicine (JAAM) | International Society of Thrombosis and Hemostasis (ISTH) | ||
|---|---|---|---|
| SIRS* criteria | Platelet count | ||
| ≥3 | +1 | <50 × 109/l | +2 |
| 0 to 2 | 0 | ≥50 <100 × 109/l | +1 |
| Platelet count | ≥100 × 109/l | 0 | |
| <80 × 109/l or >50% decrease within 24 hours | +3 | Elevated fibrin-related markera | |
| ≥80 <120 × 109/l or >30% decrease within 24 hours | +1 | Strong increase | +3 |
| ≥120 × 109/l | 0 | Moderate increase | +2 |
| Prothrombin time (value of patient/normal value) | No increase | 0 | |
| ≥1.2 | +1 | Prolonged prothrombin time | |
| <1.2 | 0 | ≥6 seconds | +2 |
| Fibrin/fibrinogen degradation products | ≥3 <6 seconds | +1 | |
| ≥25 mg/l | +3 | <3 seconds | 0 |
| ≥10 <25 mg/l | +1 | Fibrinogen level | |
| <10 mg/l | 0 | <100 g/ml | +1 |
| Diagnosis | ≥100 g/ml | 0 | |
| If, ≥4, there is positive diagnosis of DIC. | Diagnosis | ||
| If >5, there is positive diagnosis of overt DIC. | |||
| If <5, suggestive (not affirmative) for non-overt DIC. | |||
Systemic Inflammatory Response Syndrome
Reproducible research
To increase transparency and to allow re-execution of our analysis on other datasets, we voluntarily sought to adhere to the criteria for reproducible research as accepted by the journal Biostatistics [10]. Therefore, we offer the complete source code produced for this paper free online [9], while remarking that all data we used – MIMIC-II v. 2.6 – is publicly available for research, for free, via its own regulations. [3] We welcome critical review, correspondence and contributions to our work.
Methods
The reference study specified a list of data elements that were collected from patients in days 1 and 4 of diagnosis of severe sepsis, including the calculated JAAM and ISTH scores for DIC, and then used that data for statistical analysis. Our retrospective study had to therefore emulate these procedures in MIMIC-II as closely as possible. We used the Java and R languages because of our expertise with them and their free availability. The process is described below.
A. Data retrieval
We analyzed the reference study for its data sampling procedures and definitions, which Gando et al. dutifully report in the Data Sampling section of their paper, and manually listed all conceptual data elements it produced from each eligible patient (Table 2). Each item in that list would become the header of a column in a table called Clinical Data Table.
Table 2:
All conceptual data elements required from each patient by the reference study
| Data element | Data type |
|---|---|
| Age | Number |
| Gender | Male/Female |
| Septic shock on day 1? | Yes/No |
| Positive blood culture on day 1? | Yes/No |
| Acute Physiology and Chronic Health Evaluation II score on day 1 | Number |
| Sequential Organ Failure Assessment score on day 1 | Number |
| Multiple Organ Dysfunction Syndrome (MODS) on day 1* | Yes/No |
| Systemic Inflammatory Response Syndrome criteria on day 1 | Number |
| JAAM DIC score on day 1 | Number |
| JAAM DIC score on day 4 | Number |
| ISTH DIC score on day 1 | Number |
| Patient deceased within 28 days of diagnosis of severe sepsis? | Yes/No |
| Patient deceased within one year of diagnosis of severe sepsis? | Yes/No |
| Patient deceased during the hospital stay? | Yes/No |
| *Defined as SOFA score >= 12, as per the reference study. | |
For knowing or calculating the value of those conceptual data elements, we knew, from their definitions, that we needed a much larger list of raw data elements. For example, for calculating the JAAM DIC score (a conceptual data element) it is necessary to know raw data elements from the patient such as platelet count, fibrinogen level, prothrombin time, and others. We manually produced a list of all those raw data elements needed, and then searched for the MIMIC-II-specific code(s) (ITEMIDs) for each. We did those searches inside MIMIC-II itself using SQL queries, and on the documentation of the database [11]. Table 3 brings examples of those raw data elements.
Table 3:
Examples of raw data elements required from MIMIC-II v. 2.6
| Data element | SQL Table | ITEMID(s) |
|---|---|---|
| Age | ADMISSIONS | – |
| Date of death if it happened | D_PATIENTS | – |
| Platelet count | LABEVENTS | 50428 |
| D-dimer plasma level | LABEVENTS | 50370 |
| Fibrin/fibrinogen degradation products plasma level | LABEVENTS | 50376 |
| PT (seconds) in blood | LABEVENTS | 50439 |
| PT (INR) in blood | LABEVENTS | 50399 |
| Temperature (Celsius) of blood | CHARTEVENTS | 676, 677 |
| Temperature (Fahrenheit) of blood | CHARTEVENTS | 678, 679 |
| Breath rate | CHARTEVENTS | 3337 |
| Heart rate | CHARTEVENTS | 211 |
| Overall SOFA Score | CHARTEVENTS | 20009 |
| APACHE II Score | Not available |
B. Electronic phenotyping
We used Java to reproduce the reference study’s inclusion criteria and data sampling procedures. Our program screened all hospital admissions of non-neonates in the database, and read the results of the laboratory tests and charted data in chronological order. For each case where the patient met the eligibility criteria, the program created one row in the Clinical Data Table, as defined in section A, and populated it with the required study data elements. In cases where MIMIC-II did not contain the required input data at the required point in time, which are common in retrospective studies [4], the Java program generated “Not Available” (NA) as output. The program eventually produced a CSV file that was imported into R for subsequent statistical analysis.
C. Statistical analysis
The reference study provided clear descriptions of the statistical analyses, which were reproduced using R. Groups were compared using Welch’s t-tests for data with unequal variances and Mann–Whitney U tests for data with equal variances. Effect of independent variables predicting mortality was calculated by stepwise logistic regression. For comparing patient survival, we simply counted how many patients were alive at the end of the follow-up period.
Results
A. Data retrieval
We identified 15 data elements produced by the Data Sampling methodology of the reference study, listed in Table 2. A few of them could potentially be derived from others, but were kept separate for simplicity. In order to produce all of them for each patient, we identified 49 raw data elements from MIMIC-II. Most of the raw data elements were MIMIC-II-specific ITEMIDs of charted data such as laboratory test results, but others were demographic or related to workflow such as patient age, gender, hospital admission date and date of death if it happened. Table 3 shows examples of raw data elements; the complete list can be found online together with this paper’s accompanying source code.
B. Electronic phenotyping
The eligibility criteria of our reference study consisted of the presence of diagnosis of severe sepsis [1]. Although not specified in the paper, we assumed as an additional, implied eligibility criterion that the patient be not a neonate. We based this assumption on the expectation that neonatal patients would have been mentioned in the reference study if they were included, since the usual setting in hospitals is to have separate ICUs for neonates and for non-neonates. In MIMIC-II, a specialized variable – ICUSTAY_AGE_GROUP – informs you whether the patient is an “adult”, “neonate”, or “middle”. The eligibility criteria of our retrospective study consisted thus of the patient not being labeled “neonate”, and a diagnosis of severe sepsis be established.
Table 4 shows the definitions we used for identifying Systemic Inflammatory Response Syndrome (SIRS), sepsis, severe sepsis and organ failure from raw clinical data elements such as white blood cell count, partial pressure of carbon dioxide in arterial blood, urinary output in the last 24 hours, and others. They derive from the original definitions used in the reference study [13][14]. We translated our definitions into algorithms in Java that read each data element and compared it to its threshold(s) (e.g. Temperature < 36 C or >38.3C = one SIRS criteria).
Table 4.
SIRS, sepsis and severe sepsis diagnostic criteria
| SIRS: 2 or more of the following: |
| • Temperature < 36 C or > 38.3 C. |
| • Heart rate > 90 beats per minute |
| • Breath rate > 20 breaths per minute. |
| • Abnormal white blood cell count: |
| ○ > 12,000/μL, |
| ○ or < 4,000/μL, |
| ○ or > 10% immature neutrophil (band) forms. |
| Sepsis: SIRS + known or documented infection. |
| ➢ Hospital admission must contain ICD-9 code for sepsis or septicemia. |
| Severe sepsis: Sepsis + organ dysfunction. |
| • Overall SOFA score ≥ 3. |
Due to unavailability of ready means to know, from the data available in MIMIC-II, the moment when a patient first had a known or documented infection, which is a requirement for establishing a diagnosis of sepsis [13], in our electronic phenotype we adopted an alternate criterion for diagnosing severe sepsis. We accepted that, given the satisfaction of the remaining criteria of severe sepsis (Table 4), the requirement of known or documented presence of infection was satisfied by the presence of an ICD-9 code for either sepsis (ICD-9 = 995.91 or 995.92 [severe sepsis]) or septicemia (ICD-9 = 038.X) on the hospital admission. Our electronic phenotyping algorithm for severe sepsis thereby consisted as follows. First, SQL queries retrieved patients from MIMIC-II by their ICD-9 code for sepsis, severe sepsis or septicemia. For each of those patients individually, the Java program ingested and processed all pertinent EHR data. If the program found together two or more SIRS criteria, with at least one Overall SOFA score ≥ 3 in the same day, the patient was deemed eligible for our retrospective study, starting from the chart time of the data element with the latest chart time.
For identifying groups of data elements together (e.g. the presence of two SIRS criteria together with a SOFA score above 2), we had to implement the concept of time windows, because each data element is charted at a potentially different time. In a prospective study there is no necessity for such windows, because patient data (e.g. laboratory tests, vital sign measurements) is collected on-demand, according to the study protocol, virtually all at the same planned time. In our retrospective study, the essential question to be answered was: how far from each other, in time, can patient data elements be (e.g. a platelet count, a fibrinogen level measurement, and prothrombin time measurement) and still be considered effectively simultaneous?
The definitions of the DIC scoring systems also assume that all needed tests are performed on-demand, virtually at the same time, thus they also do not provide such specifications of time windows. Therefore, to adapt the prospective study’s design to a retrospective study design, we arbitrated two separate time windows.
Time window one: for answering whether one data element, such as one record of overall SOFA score, was present at a point in time T, the algorithm returned the record closest to that time T, in the future or in the past, within 24 hours of distance of T, that is, within the interval T ± 24 hours.
Time window two: for the calculation of scores of DIC or SIRS, which are aggregations of raw data elements, at a given point in time T, we considered only the EHR data charted until that time T and within the 48 hours immediately before T. That is, we considered only data charted within the interval from “T – 48 hours” until T.
Inside any time window, and for any data element, the record of that data element that was closest to time T was always the one used.
Table 6 exposes the characteristics of the patients after the screening for eligibility and calculation of DIC scores, reproducing the composition of Table 3 of the reference study. The difference is the column “Missing data”, which informs the percentage of patients that, on day 1, did not have that data element available.
Table 6.
Characteristics of JAAM DIC and non-DIC patients on the day of inclusion (day 1)*
| JAAM DIC (n = 395) | Non-DIC (n = 1565) | P value | Missing data (%) | |
|---|---|---|---|---|
| Age (years) | 59.84 ± 16.84 | 66.95 ± 16.39 | <0.0001 | 0 |
| Gender (male/female) | 214/181 | 925/640 | 0.0790† | 0.03 |
| JAAM DIC score | 4.74 ± 1.22 | 1.26 ± 0.52 | <0.0001 | 13.16†† |
| Platelet count (× 10^9/l) | 103.98 ± 101.56 | 224.83 ± 132.03 | <0.0001 | 0.1 |
| Prothrombin time (seconds) | 16.28 ± 4.2 | 15.67 ± 4.85 | 0.0125 | 1.48 |
| Prothrombin time ratio | 1.87 ± 1.24 | 1.7 ± 1.05 | 0.0122 | 2.45 |
| Fibrinogen (g/l) | 4.4 ± 2.44 | 4.38 ± 2.1 | 0.9139 | 61.94 |
| FDP (mg/l) | 91.81 ± 181.01 | 53.48 ± 152.85 | 0.0624 | 83.21 |
| SIRS score | 2.42 ± 0.67 | 2.37 ± 0.66 | 0.1720 | 0 |
| SAPS I score | 16.82 ± 5.5 | 16.31 ± 5.19 | 0.1093 | 9.03 |
| SOFA score | 10.72 ± 4.61 | 8.27 ± 3.89 | <0.0001 | 0 |
| MODS (%) | 41.5% | 22.4% | <0.0001 | 0 |
| 28-day mortality (%) | 44.1% | 27.5% | <0.0001 | 0 |
| 1-year mortality (%) | 59.7% | 49.1% | 0.00013 | 0 |
The count of patients add up not to the number of total eligible patients in the study, but only to the number of patients whose DIC status was known on day 1, that is, 13.16% less than all eligible patients.
From a t-test comparing the proportion of male/female between the two groups.
This cell differs from the others in the column because here the denominator is the number of eligible patients, rather than the number of eligible patients with known DIC status.
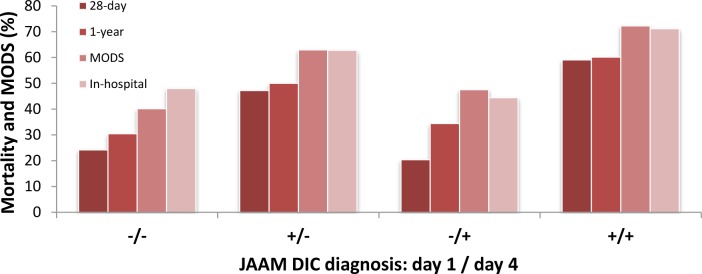
Figure 1, below, illustrates how the JAAM DIC diagnosis on days 1 and 4 correlate with mortality and Multiple Organ Dysfunction Syndrome (MODS). It is possible to identify a relative drop in the numbers in the case where the patients did not have JAAM DIC on day 1, then acquired it by day 4, which differs from this same graph in the reference study.
Figure 1.
Mortality and Multiple Organ Dysfunction Syndrome (MODS) in JAAM DIC patients
The charting time of calculated scores – SIRS and DIC
The specific points in time of any score assessment or diagnosis (and suspension of thereof) naturally coincided with the chart times of the data in MIMIC-II. We considered the chart time to be the moment when, ideally, each new piece of data about the patient data became known in the original clinical setting that originated MIMIC-II, such as a new platelet count, or a new measurement of body temperature. All patient data was read and processed in chronological order according to the chart time.
C. Statistical analysis
Our results agreed with those of the reference study. As the JAAM DIC score at day 1 increased, so did the mortality at day 1, as well as the SAPS I and organ dysfunction scores, as can be seen in Table 7. One intriguing exception was the score of 7, which did not follow the trend.
Table 7.
Disease severity, organ dysfunction and mortality for JAAM DIC score on inclusion day (day 1)*
| JAAM DIC score on day 1 | ||||||||
|---|---|---|---|---|---|---|---|---|
| 1 (n = 1224) | 2 (n = 276) | 3 (n = 65) | 4 (n = 254) | 5 (n = 71) | 6 (n = 8) | 7 (n = 42) | 8 (n = 20) | |
| SAPS I score | 16.18 ± 5.12 | 16.77 ± 5.61 | 16.74 ± 4.47 | 16.32 ± 5.21 | 17.41 ± 5.51 | 18.14 ± 5.21 | 17 ± 6.39** | 20.21 ± 6.21 |
| SIRS score | 2.31 ± 0.63 | 2.59 ± 0.71 | 2.4 ± 0.75 | 2.33 ± 0.62 | 2.68 ± 0.73 | 2.62 ± 0.74 | 2.26 ± 0.63 | 2.9 ± 0.72 |
| SOFA score | 8 ± 3.74 | 9.13 ± 4.17 | 9.72 ± 4.7 | 10.31 ± 4.28 | 10.79 ± 4.55 | 14.12 ± 5.46 | 11.12 ± 5.34 | 13.55 ± 5.7 |
| SOFA peak during hospital stay | 10.05 ± 4.38 | 11.2 ± 4.74 | 12.49 ± 5.43 | 12.82 ± 5.08 | 13.17 ± 5.48 | 14.62 ± 5.37 | 13.88 ± 5.01 | 16.95 ± 5.38 |
| MODS (%) | 19.7 | 32.6 | 29.2 | 36.2 | 46.5 | 75 | 45.2 | 70 |
| 28-day mortality (%) | 27 | 26.8 | 40 | 46.9 | 32.4 | 62.5 | 35.7 | 60 |
| 1-year mortality (%) | 48 | 50 | 66.2 | 64.2 | 52.1 | 62.5 | 42.9 | 65 |
Scores of zero were discarded as per study design – see Limitations section.
The bold and underline is meant to draw attention to the exception to the trends that is column 7.
Table 8 shows that a diagnosis of DIC by the JAAM scoring system increased the odds of mortality by 18.2% to 43.2% (95% confidence interval), and that a decrease in the JAAM score within 4 days was associated with a 13.3% reduction in the odds of mortality (Table 8).
Table 8.
Stepwise logistic regression analysis on day of inclusion (day 1) for prediction of 28-day mortality
| Odds ratio | P value | 95% confidence interval | Missing data (%) | |
|---|---|---|---|---|
|
|
|
|
|
|
| Age | 1.020 | 0.00013 | 1.010 to 1.030 | 0 |
| JAAM DIC score | 1.300 | <0.0001 | 1.182 to 1.432 | 13.16 |
|
|
|
|
|
|
| Delta JAAM DIC score (day 1 – day 4) | 0.867 | 0.00516 | 0.784 to 0.958 | 21.09 |
|
|
|
|
|
|
| Fibrinogen | 0.998 | 0.00014 | 0.998 to 0.999 | 64.95 |
Discussion
Our endeavor in this paper builds upon a central concept that is as follows. Given structured data from the electronic health record, and cautious design of the data processing pipeline, a computer algorithm is able to calculate some popular scoring systems used in clinical practice. The criteria for the Systemic Inflammatory Response Syndrome is one example of such a scoring system, as well as essentially all four criteria for diagnosis of disseminated intravascular coagulation published in the literature [5]. Supported by this concept, we have accepted our electronic phenotype for severe sepsis without formal validation against a gold standard. Otherwise, one could interpret the high agreement of our results with those of the reference prospective study as evidence that the electronic phenotype, and the calculations of the used disease scores, had altogether acceptable performance.
Foremost, this paper shows the value of retrospective studies for clinical research, and that MIMIC-II can be used for them. Despite not a new resource [3], and the many studies with MIMIC-II [3], to our knowledge this is the first validation of a clinical study done in this valuable database. Our results (especially Tables 8 and 9) demonstrated resilience towards the limitations of a retrospective study, and remarkably followed the direction of the results of the reference study, which was done at a different country, and across 15 hospitals instead of one (which is the case of MIMIC-II).
Table 9.
Comparison of final conclusions between the reference study and this study
| Sentence from the Results part of the Abstract of the reference study | Findings in our retrospective study | Agreement between studies? |
|---|---|---|
| The prevalence of JAAM DIC was 46.8% (292/624) | Incidence of JAAM DIC on day 1 was 17.5% (395/2,257). | No. |
| and 21% of the DIC patients were scored according to the reduction rate of platelets. | Not assessed. | |
| The JAAM DIC patients were more seriously ill | SAPS I score demonstrated a small difference (16.8 vs 16.3) and P = 0.10. | No. SAPS score was used instead of APACHE, and results were not statistically significant. |
| and exhibited more severe systemic inflammation, | Average SIRS criteria had a difference in the direction of agreement, but it was too small (2.42 vs 2.37) and not statistically significant (P = 0.17). | No. |
| a higher prevalence of multiple organ dysfunction syndrome (MODS) | Yes, 41.5% vs 22.4%, P < 0.0001. | Yes. |
| and worse outcomes than the non-DIC patients. | Yes, both 28-day and 1-year mortalities were higher and with P < 0.001: 44.1% vs 27.5%, and 59.7% vs 49.1%, respectively. | Yes. |
| Disease severity, | Average SAPS I score | Yes, see table 7. |
| systemic inflammation, | Average number of SIRS criteria | Yes, see table 7. |
| MODS | Overall SOFA score >= 12 | Yes, see table 7. |
| and the mortality rate | Both 28-day and 1-year | Yes, see table 7. |
| worsened in accordance with an increased JAAM DIC score on day 1. | Yes, see Table 7, but notice there were exceptions. | |
| The JAAM DIC score on day 1 (odds ratio = 1.282, P <0.001) and the Delta JAAM DIC score (odds ratio = 0.770, P <0.001) were independent predictors of 28-day death. | The JAAM DIC score on day 1 (odds ratio = 1.300, P <0.0001) and the Delta JAAM DIC score (odds ratio = 0.867, P = 0.00516) were statistically significant independent predictors of 28-day death. | Yes. |
| Dynamic changes in the JAAM DIC score from days 1 to 4 also affected prognoses. | Not assessed. | |
| The JAAM DIC scoring system included all patients who met the International Society on Thrombosis and Haemostasis (ISTH) overt DIC criteria on day 1. | From the 189 patients with ISTH overt DIC on day 1, 174 also had JAAM DIC on day 1, a 92% coverage. | Not all, but a large majority: 92%. |
| The International Society on Thrombosis and Haemostasis scoring system missed a large number of nonsurvivors recognized by the JAAM scoring system. | Mortality among Non-ISTH overt DIC patients (33.8%) was higher than among Non-JAAM DIC patients (27.4%). Moreover, from the 946 Non-ISTH overt DIC patients, 221 (23.3%) had JAAM DIC, and these had 46.1% mortality. | Yes. |
Limitations
Missing data
A problem inherent to a retrospective study, the extent of missing data in our study ranged from zero to 83.21% (Fibrin/Fibrinogen Deg. Prod. [FDP] level) or 100% (“known source of infection”). We did not perform statistical artifacts on the data to fill in missing values. As can be seen in Table 6, it was of utmost importance to this study the fact that the clinical data required by the reference study is commonly collected as part of routine care. The appraisal of data availability can easily be the foremost concern to a researcher considering a retrospective study. For the “known source of infection”, which we deemed completely unavailable and translated into the presence of an ICD-9 code for sepsis or septicemia in the patient’s hospitalization data, this workaround can be expected to be imparting selection bias according to imperfections in the ICD-9 coding.
For the FDP level, as well as other charted patient information, our program understood as Not Available (NA) any needed data element that was not available within the requested time window. This approach brought the design caveat that for aggregations of raw data elements such as SIRS and DIC scores, it was not clear how to differentiate a known zero from a NA. This is because any pertinent data element can cause each score to be nonzero. However, in a retrospective study, it can easily be too restrictive to require that all possible data elements must be available before you can accept a known zero. This is because, unlike in a prospective study, you cannot collect patient data on-demand to satisfy your data needs. Our approach to this issue was a simplifying one – we chose to ignore any score below 1, and treat them as Not Available. Our understanding was that, because the diagnostic threshold is much higher than 1 (Table 1), this would be of minimal to no impact to our study. The absence of score zero can be readily noticed in Table 7.
Diagnosis of severe sepsis/DIC near death
From manual inspection of the data, we noticed that in a considerable (approx.. 10%) of the cases a diagnosis of severe sepsis and/or DIC was only ever made in the last hours before the moment of patient death – and not rarely a bit after the time of death. We rationalized that, near the moment of passing, a clear laboratorial diagnosis of severe sepsis or disseminated intravascular coagulation is clouded by a greater disruption of body homeostasis, and the computation is clouded by possible imperfections in data collection such as tests collected or charted after the moment of death. Therefore, we ruled to completely ignore any patient data charted within 12 hours of the time of patient death. Patients could be electronically phenotyped only before that point in time.
Issues with data representation
Three data elements from MIMIC-II – namely: D-Dimer level, Fibrin/Fibrinogen Degradation Products level, and Fibrinogen level – were found to be available as notations rather than only numbers, for example: “<10”, “10–40”. “160–320”, “>10000”. We decided that our Java program would thereby perform conversion of those values.
The sign “<” was interpreted as “95%”, thus “<10” became 9.5;
ranges were averaged, thus “160–320” became 240;
The sign “>” was interpreted as “105%”, thus “>10000” became 10500.
Furthermore in this issue, some scoring systems requested data in a level of granularity that was not available in MIMIC-II. That was the case of Fibrin\Fibrinogen Degradation Products (FDP) level for the JAAM DIC scoring system. The JAAM DIC scoring system defines three ranges – 0–10, 10–25 and >25 – with different values, however, that data element is available in mimic as the following ranges: 0–10, 10–40, 40–80, and so on. Our referred averaging method turned “10–40” into 25 (3 points in the JAAM DIC) and “0–10” into 5 (0 points), effectively impeding any patient to receive just 1 point for that criteria inside the JAAM DIC.
Conclusion
We reproduced a contemporary prospective clinical study as a computational retrospective study in an EHR database – MIMIC-II – which found considerably more eligible patients than the prospective study, and was executed at a fraction of the cost and time. Despite the limitations of the retrospective study design, the results demonstrated resilience and agreed with those of the prospective study in a large majority of the findings.
Our results speak in favor of the usefulness of computational retrospective studies [4] as eventual proxies to costlier clinical studies, and suggest an altogether good performance, in MIMIC-II, of our developed electronic phenotypes for SIRS, severe sepsis and two scoring systems for disseminated intravascular coagulation available in the clinical literature.
We strived to maximize the dissemination and reuse of the work by making all source code we produced available online free [9], in addition to the fact that MIMIC-II is also available free to the research community.
From the clinical perspective, we offer our results as one contribution to the body of evidence on the validity and use of scoring systems for diagnosis of disseminated intravascular coagulation. In particular, we highlight the value of our study as a positive third-party validation of the work by Gando et al.
Table 5.
Study eligibility screening results
| Reference study [1] | MIMIC-II Study | |
|---|---|---|
| Total patients screened | 14,417 | 27,579* |
| Total eligible patients found | 624 | 2,257 |
| Gender (male/female) | 391/233 | 1281/975** |
| Eligible patients with JAAM DIC diagnosis on day 1 (% of eligible patients) | 292 (46.8) | 395 (17.5) |
| 28-day mortality in patients with/without JAAM DIC diagnosis on day 1 (%) | 31.2/16.0 | 44/27.5 |
All non-neonates in MIMIC-II v. 2.6. Please note that this number differs from earlier versions of MIMIC-II.
For one patient that information was not available (NA).
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM) and Lister Hill National Center for Biomedical Communications (LHNCBC). This research was also supported in part by an appointment to the NLM Research Participation Program, administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the US Department of Energy (DoE) and the NLM.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest in this study.
References
- 1.Gando, et al. A multicenter, prospective validation study of the Japanese Association for Acute Medicine disseminated intravascular coagulation scoring system in patients with severe sepsis. Crit Care Med. 2013;17:R111. doi: 10.1186/cc12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ross MK, Wei W, Ohno-Machado L. “Big Data” and the Electronic Health Record. IMIA Yearbook. 2014. pp. 97–104. http://dx.doi.org/10.15265/IY-2014-0003. [DOI] [PMC free article] [PubMed]
- 3.Saeed M, et al. Multiparameter Intelligent Monitoring in Intensive Care II (MIMIC-II): A public-access intensive care unit database. Crit Care Med. 2011 May;39(5):952–960. doi: 10.1097/FCCM.0b013e31820a92c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anglemyer A, Horvath HT, Bero L. Healthcare outcomes assessed with observational study designs compared with those assessed in randomized trials. Cochrane Database Syst Rev. 2014 Apr 29;4:MR000034. doi: 10.1002/14651858.MR000034.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Nisio M, et al. Diagnosis and treatment of disseminated intravascular coagulation: Guidelines of the Italian Society for Haemostasis and Thrombosis (SISET) Thrombosis Research. 2012 May;129(5):e177–e184. doi: 10.1016/j.thromres.2011.08.028. [DOI] [PubMed] [Google Scholar]
- 6.Lee JH, Song JW, Song KS. Diagnosis of Overt Disseminated Intravascular Coagulation: A Comparative Study Using Criteria from the International Society Versus the Korean Society on Thrombosis and Hemostasis. Yonsei Med J. 2007 Aug 31;48(4):595–600. doi: 10.3349/ymj.2007.48.4.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levi M, ten Cate Hugo. Disseminated Intravascular Coagulation. N Engl J Med. 1999;341:586–592. doi: 10.1056/NEJM199908193410807. [DOI] [PubMed] [Google Scholar]
- 8.Wada, et al. Diagnosis and treatment of disseminated intravascular coagulation (DIC) according to four DIC guidelines. Journal of Intensive Care. 2014;2:15. doi: 10.1186/2052-0492-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.https://github.com/fabkury/paamia2015. Made available online by this paper’s authors on March 11, 2015
- 10.Peng RD. Reproducible research and Biostatistics. Biostat. 2009;10(3):405–408. doi: 10.1093/biostatistics/kxp014. [DOI] [PubMed] [Google Scholar]
- 11. [Accessed March 12, 2015]. MIMIC-II official website: http://physionet.org/mimic2/
- 12.Scott DJ, Silva I. MIMIC II SQL Cookbook. [Accessed March 12, 2015]. http://www.physionet.org/mimic2/demo/MIMICIICookBook_v1.pdf.
- 13.Richesson R, Smerek M. Electronic Health Records-Based Phenotyping. [Acessed March 12, 2015]. http://sites.duke.edu/rethinkingclinicaltrials/ehr-phenotyping/
- 14.Members of the American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference Committee American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference: definition for sepsis and organ failure and guidelines for the use innovative therapies in sepsis. Crit Care Med. 1992;20:864–874. [PubMed] [Google Scholar]
- 15.Levy, et al. SCCM/ESICM/ACCP/ATS/SIS: 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit Care Med. 2003;31:1250–1256. doi: 10.1097/01.CCM.0000050454.01978.3B. [DOI] [PubMed] [Google Scholar]
- 16.Wada H, Matsumoto T, Yamashita Y, Hatada T. Is early treatment of disseminated intravascular coagulation beneficial in septic patients? Crit Care. 2014;18(4):447. doi: 10.1186/cc13971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asche C, Said Q, Joish V, Hall CO, Brixner D. Assessment of COPD-related outcomes via a national electronic medical record database. [DOI] [PMC free article] [PubMed] [Google Scholar]