Abstract
The secondary use of electronic health records opens up new perspectives. They provide researchers with structured data and unstructured data, including free text reports. Many applications been developed to leverage knowledge from free-text reports, but manual review of documents is still a complex process.
We developed FASTVISU a web-based application to assist clinicians in reviewing documents. We used FASTVISU to review a set of 6340 documents from 741 patients suffering from the celiac disease.
A first automated selection pruned the original set to 847 documents from 276 patients’ records. The records were reviewed by two trained physicians to identify the presence of 15 auto-immune diseases. It took respectively two hours and two hours and a half to evaluate the entire corpus. Inter-annotator agreement was high (Cohen’s kappa at 0.89).
FASTVISU is a user-friendly modular solution to validate entities extracted by NLP methods from free-text documents stored in clinical data warehouses.
Introduction
The worldwide trend toward secondary use of health data for research opens up new and exciting perspectives for researchers in all the realms of medicine. The large adoption of EHRs provides a steady flow of data, structured and unstructured (and particularly free-text medical reports). Today, almost all the preeminent clinical research institutions have deployed Clinical Data Warehouses (CDW)1–4, to store and integrate all the data produced in routine care.
Our institution, the European Hospital Georges Pompidou (HEGP) is a 730 beds public research hospital in Paris. An i2b2 CDW has been installed and used since 2008. It integrates data from more than 700,000 patients5. The large majority of the data stored is structured data with more than 100 million data points of lab results, but the data warehouse also integrates more the 4 million free-text reports in French ranging from discharge summaries, letters, to imaging and pathology reports.
Structured data, by their organized nature, are a target of choice for secondary use. However, free-texts are at least equally important. In medicine, free-text has always been the support of medical records. Free-text reports collect any information that the physician considers of any importance and that took a role in a medical decision. Text reports also include data that might be difficult to find elsewhere such as family history, results of exams performed outside of the institution, rejected hypotheses. Nevertheless, the extraction and reuse of data from free-text reports requires processing and dedicated tools and resources.
Structured and unstructured data capture different views on the patient and her/his disease. Billing codes, often coded with ICD-9 CM or ICD-10 capture the medico-economical aspect of a medical encounter. The main purpose is to evaluate the cost of the treatment provided; lab results coded in LOINC provide standardized biology lab results. Issues might occur when researchers attempt to leverage structured data in a context for which they were not designed. For example billing codes are not aiming at providing a global coverage of comorbidities of a patient. In their article, Li et al.6 compared information found in structured data to information found in free medical text. They reported two types of differences: information completeness and concept granularity, and concluded that data extracted from free-text complement structured data.
Medical documents come in a large variety of type of texts. The biomedical literature reports text mining of radiology reports7,8 medical observations9,10, nurse narratives11 – documents produced for care activity – but also from documentations and guidelines12. Most of these clinical documents are stored in CDWs. However they provide information of heterogeneous quality: for example a discharge summary is validated by a senior physician while observations or notes may be written by medical students.
Standardized text annotation is required to enable automated processing. Terms in the text are annotated using standard terminologies. The Unified Medical Language System® (UMLS®) is often used as a lingua franca for biomedical domains. Tools have been developed to assist annotators, such as the BRAT Rapid Annotation Tool13 (BRAT is not dedicated to medical applications). However, manual annotation is a tedious and time consuming process that requires to follow guidelines, and therefore to be trained. The Natural Language Processing community has developed over the last decades many applications for virtually all the fields of medicine. For example, for the detection of incidental findings in chest x-ray14, pneumonia or rheumatoid arthritis identification from narrative reports15–17. There is a large variety of tools available for different purposes.
Medical concept recognizers (MER) leverage medical terminologies and linguistic resources to identify medical entities in free-text (popular MERs include MetaMap18 developed by the National Library of Medicine, the Bioportal Annotator developed by the National Center for Biomedical Ontologies – NCBO, or cTakes developed by the Apache Software Foundation). In addition to recognizing medical concepts, it is often needed to capture the context of the information. The meaning of concept can drastically change depending of the context (e.g. negation, potentiality, family history). Some of the previously cited tools include some context recognition. The most commonly used context recognizer is probably NegEx19 for negation20 and its extension CONText21. Expert validation is often required to secure further use of the results. I2b2 itself proposes NLP tools through its ‘Natural Language Processing cell’ based on a HITEx22 core. This functionality is encapsulated into i2b2 interface where the user can retrieve different types of concepts detected in text: diagnoses, discharge medications, smoking status.
Shivade & al.20 reviewed 97 articles describing approaches to identifying patient phenotype cohorts using EHRs. Forty-six articles used NLP based approach.
A common task in a clinical research is the selection of eligible patients for a clinical study. It is a tedious and costly task that requires time and highly skills personal. In the case of retrospective studies, it is often required to browse through the entire record archive of a patient to identify relevant information. Tools have been developed to assist in the selection process. They provide a connection between CDW and NLP tools, and help the user in the selection process. Many systems use selection systems inspired by the “basket” from online-shopping websites23, i.e. the clinician browse through available documents and identify those of interest.
However, it is often needed to be, not only able to identify patients that fit a set of criteria and are eligible for a study, but also to identify and annotated the presence, or the status of phenotypes in their record. We did not find a publicly available solution that would allow a flexible entity selection, and allow phenotypic annotation by one or more users.
Moreover, most of the documents available for research in our hospital are in French. Non-English languages lack tools and terminology, and despite substantial effort toward better NLP coverage simple solution are still needed. We needed a solution with good performance, reducing physician time involved reviewing and classifying features on medical records for different tasks from patient eligibility to phenotypes extraction.
We have designed FASTVISU as an application that connects NLP outputs with free-text from CDW and provides an interface to efficiently select a set of interesting features. FASTVISU allows a single or multi-user evaluation of several features at the same time at the patient, visit or document level, based on a voting mechanism. In the next sections, we will describe the architecture of FASTVISU, and present a use case on the annotation of a cohort of patients with the Celiac Disease.
Material and Methods
Architecture
The modular FASTVISU architecture is based on a Web Service delivered as a PHP REST API. The API provides abstraction layers to:
Retrieve documents from an i2b2 CDW,
Create, modify and retrieve corpora of documents (stored in an Oracle database)
Vote on features to classify patients (e.g. to select patients having a diagnosis of interest), encounters (e.g. to differentiate normal follow up encounters from relapses) or documents (e.g. classify legally conform documents from non-conform during an administrative audit). Votes are stored in an Oracle database.
The clients using this API are of two types:
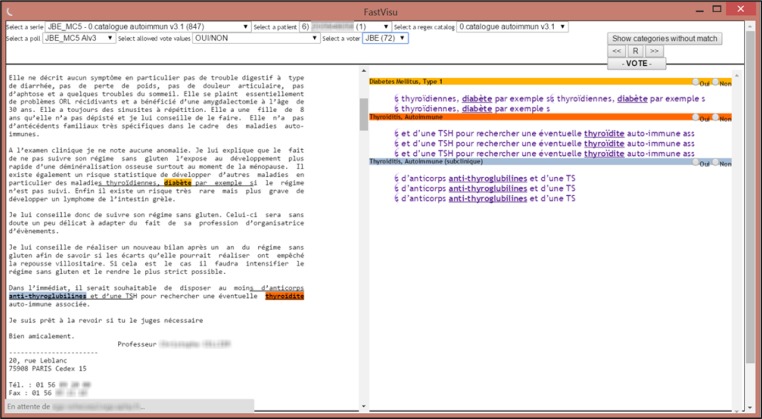
A user interface for human interaction (Figure 2)
bots who automatically go through every document and vote according the computation from text mining modules.
Figure 2.
Screenshot of the web client visualizing a whole patient record on the left panel (using regular expressions from the text processing module to highlights concepts). The panel on the right summaries instances of concepts (color coded) and provide the user with voting options for the different categories of concepts (e.g. ‘presence of Type 1 Diabetes Mellitus: Yes/No’).
Clinical Data Warehouse
I2b2 is a widely used CDW which became de facto a standard over the years. The database of the CDW at HEGP contains data for more than 700,000 unique patients. Data available include full medical and administrative records (for in- and out-patients), diagnoses and procedures codes, EHR, structured observations, free-texts (discharges, letters), laboratory tests and pathology results, as well as every drugs prescription done in the hospital.
Evaluation
We evaluated the performance of FASTVISU to help identifying the autoimmune comorbidities associated with celiac disease in the HEGP i2b2 data warehouse. Celiac disease (CD) is an autoimmune disorder induced by the intake of proteins found in gluten. Autoantibodies such as anti-transglutaminase characterize the disease as well as intestinal villous atrophy. Several autoimmune comorbidities are known to be associated with CD, such as dysthyroidism or type 1 diabetes. More precisely, the final objective was to estimate the prevalence of the 15 most frequent autoimmune diseases (AID) in a cohort of CD patients followed at HEGP, which is a French national reference center for CD.
Corpus and data processing
Selecting comorbidities from the medical literature
We used the MeSH® (Medical Subject Headings®) co-occurrence file provided by the U.S. National Library of Medicine (NLM) to establish a list of the most frequent comorbidities in CD. We retrieve co-occurrence frequencies of the MeSH main heading Celiac Disease (D002446) with all main headings children of ‘Autoimmune Disease’ (D001327), corresponding to all terms with MeSH Tree Number starting with C20.111.*. We restrained our search to the fifteen most frequent comorbidities.
Selecting the CD corpus
We extracted a CD dataset from our data warehouse. CD cases were identified via an i2b2 query based on the combination of the three following criteria: having had an hospitalization stay with ICD-10 code for CD (K90) in billing claims; one or more stay, or consultation in the gastroenterology department; and at least one text document (discharge or letter) containing the term ‘celiac disease’ or its synonyms. The final corpus was made of all available medical documents for the patient identified by the previous query
Identifying AID comorbidities in structured data
We leveraged two sources to identify comorbidities in patient: ICD codes from billing system, drugs prescribed and dispensed at the hospital (limited to insulin and levothyroxin). We used the UMLS to map the MeSH terms to the controlled vocabularies used in the CDW, namely ICD10 for diagnoses and ATC for drugs. We used an R script interacting with the voting module from FASTVISU to automatically record identified comorbidities with the corresponding patient.
Preprocessing free text for AID comorbidities identification with FASTVISU
We used a regular expression module for entity recognition. We elaborated a list of regular expressions (regex) for each auto-immune disease. The voting bot using the regex text processing module filtered out documents without any regex match. Expressions and keywords associated with the diseases targeted by the regex were broad to avoid any decrease in sensitivity. We expect the specificity to be near 100% because the set of documents is manually reviewed by experts (establishing a gold-standard).
Review using FASTVISU web interface
Two trained physicians, with a background in epidemiology and medical informatics, independently reviewed the corpus of documents using the FASTVISU web client. FASTVISU presents all documents from a patient in a chronological order in the left-hand side panel of the screen (see Figure 2). Keywords and expression detected by the regex modules are highlighted. These highlighted concepts are also summarized on the right-hand side panel of the application (and provided with a clickable link to the corresponding occurrence in the text). Each occurrence (e.g. insulin) is associated with a broader category (in this study an auto-immune disease, e.g. diabetes).
After a careful review of set of documents, the reviewer can vote for the presence or the absence of phenotypes and diagnosis. At the end of this step, each patient is characterized by a vote for the presence (or absence) of each of the fifteen diseases, from each of the reviewer (and from the automatic bot voting process).
The next step is the obtention of a consensus between reviewers. This is done in FASTVISU by visualizing only the differences between the two reviewers’ votes. Reviewers will analyze and discuss their choice and agree upon a final decision.
For analysis, the API can export votes. The R statistical software can retrieve poll results directly from the API and can compute prevalence, compare sources of vote (types of codes, human text review).
Results
Our cohort contained 741 patients, constituting a corpus of 6340 text documents. The collection is mainly made of consultation summaries and letters (44.9%), and discharge summaries (15.3%). Median number of documents per patient was 5, IQR [3; 10] with a maximum of 146.
The bot filtered the initial corpus, using the regex module, into a corpus containing 847 documents from 276 patients’ records.
Manual review using the web client interface took two hours and two and half hours respectively for each physician. The mutual agreement on the identification of the presence of the 15 auto-immune diseases for the 276 patients was excellent (with a Cohen’s kappa of 0.89).
Discussion
Elements contributing to decrease the review duration
The human expert time required to manually review was very low thanks to two mechanisms: the automatic selection and pre-identification of relevant entities through regular expressions. First the voting bot filtered records without mention of any of the diseases or concepts related to the disease. Manual review on the remaining documents is fast because the interface presents whole the records on one single page, with links to highlighted occurrences of medical terms of interest. The UI is also conceived to integrate all functionality on a single page to allow fast transition from a patient to the next.
The time spent is neither comparable to retrospective review of paper records, nor to review from the interface of the EHR software, for which the physician would have to find patients one by one, opening every document and voting on a separate application. The best gold standard method, blinded double reviewing24, is accessible with very reasonable experts effort involved.
Compared to fully automated NLP solutions, human time spent is higher, but human interpretation remains the gold standard, thus confidence in the results is higher with FASTVISU.
Inter-annotator agreement between the two physicians was high. Differences were mostly pointed associated with ambiguous text.
Technical significance
The architecture of FASTVISU is flexible. The core REST API can be easily connected to any CDW. In the current architecture, the API queries a relational database (using the i2b2 star-schema). The API could be easily modified to query other types of storage system, including NoSQL databases. In the current version, we used a regex approach know to have a good sensitivity with a potentially low specificity. Thanks to a flexible architecture, adding new “sources” of concept could be easily done (i.e. using external NLP processes to identify concepts in the documents). Our goal is to provide several ways to feed the system (for different purpose).
Clinical significance
FASTVISU has been developed at HEGP only recently, but is already used in several studies at the Department of Medical Informatics. It can cover many different cases for a wild range of applications (including patient selection, visit selection, phenotype annotation at patient or visit level).
Limitation
The system is at an early stage of development and is not completely integrated in the workflow. We plan to store the results of patient selections in the CDW for later reuse. The system today does not allow voting for single concept occurrences, therefor does not yet allow validating an automatic annotation. Voting on a set of mixed typed entities (text, lab result, care procedure, drug prescription) is not implemented yet, and validating a billing system is not yet feasible. We plan to add these new functionalities in the next version of the software.
Perspective
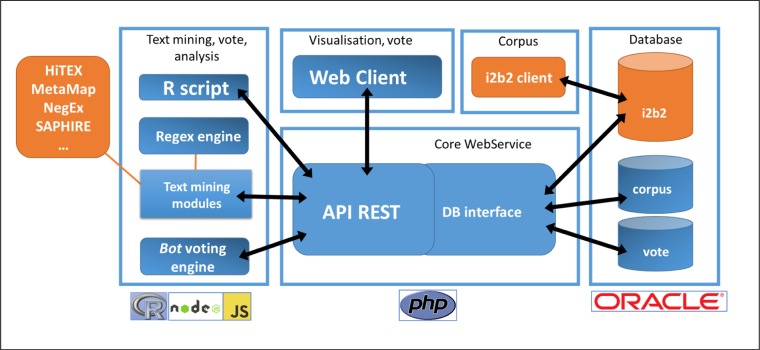
Today, results collected using FASTVISU are not recorded in our CDW. We plan to “close the loop” and to export the results of extractions or patient selection in i2b2. This would allow leveraging concept manually validated in for other usage. We also plan to provide the user with information not only from the text reports but also from other sources, such as structured data, directly in the interface. The combination of complementary sources of information would give a broader and more complete perspective to the reviewers. Our short term perspective is to connect community-issued NLP tools to FASTVISU (as text processing module in FASTVISU, see Figure 1). FASTVISU modular architecture should enable the creation of such connectors easily. Additional automated modules (bots) could augment the scope of the software. For example, FASTVISU could be used as a workbench to evaluate or to use machine learning algorithms on a set of selected documents.
Figure 1.
Modular architecture of FASTVISU.
Conclusion
Cohort selection, phenotypic annotations, validation of concept detected through NLP methods are common tasks in clinical research. FASTVISU allow to easily review set of patient documents, and to vote at different levels of granularity (patient selection in a cohort, encounter selection, phenotype presence or absence). FASTVISU was built upon a REST web-service API, which allows connecting a web-client with several components: a web client, an entity recognition module and a voting module. In this study, we showed that FASTVISU can be used to efficiently detect the presence of auto-immune diseases from a large cohort of patients.
Acknowledgments
This work was supported in part by the SIRIC CARPEM (Cancer Research and Personalized Medicine) program.
References
- 1.Murphy SN, Mendis ME, Berkowitz DA, Kohane I, Chueh HC. Integration of clinical and genetic data in the i2b2 architecture. AMIA Annu Symp Proc AMIA Symp AMIA Symp. 2006;1040 [PMC free article] [PubMed] [Google Scholar]
- 2.Lowe HJ, Ferris TA, Hernandez PM, Weber SC. STRIDE--An integrated standards-based translational research informatics platform. AMIA Annu Symp Proc AMIA Symp AMIA Symp. 2009;2009:391–395. [PMC free article] [PubMed] [Google Scholar]
- 3.Danciu I, et al. Secondary use of clinical data: the Vanderbilt approach. J Biomed Inform. 2014;52:28–35. doi: 10.1016/j.jbi.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cimino JJ, Ayres EJ. The clinical research data repository of the US National Institutes of Health. Stud Health Technol Inform. 2010;160:1299–1303. [PMC free article] [PubMed] [Google Scholar]
- 5.Zapletal E, Rodon N, Grabar N, Degoulet P. Methodology of integration of a clinical data warehouse with a clinical information system: the HEGP case. Stud Health Technol Inform. 2010;160:193–197. [PubMed] [Google Scholar]
- 6.Li L, Chase HS, Patel CO, Friedman C, Weng C. Comparing ICD9-encoded diagnoses and NLP-processed discharge summaries for clinical trials pre-screening: a case study. AMIA Annu Symp Proc AMIA Symp AMIA Symp. 2008:404–408. [PMC free article] [PubMed] [Google Scholar]
- 7.Bertaud V, Lasbleiz J, Mougin F, Burgun A, Duvauferrier R. A unified representation of findings in clinical radiology using the UMLS and DICOM. Int J Med Inf. 2008;77:621–629. doi: 10.1016/j.ijmedinf.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Fiszman M, Chapman WW, Aronsky D, Evans RS, Haug PJ. Automatic detection of acute bacterial pneumonia from chest X-ray reports. J Am Med Inform Assoc JAMIA. 2000;7:593–604. doi: 10.1136/jamia.2000.0070593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn U, Romacker M, Schulz S. MEDSYNDIKATE--a natural language system for the extraction of medical information from findings reports. Int J Med Inf. 2002;67:63–74. doi: 10.1016/s1386-5056(02)00053-9. [DOI] [PubMed] [Google Scholar]
- 10.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc JAMIA. 2004;11:392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bakken S, Hyun S, Friedman C, Johnson SB. ISO reference terminology models for nursing: applicability for natural language processing of nursing narratives. Int J Med Inf. 2005;74:615–622. doi: 10.1016/j.ijmedinf.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Denny JC, Smithers JD, Miller RA, Spickard A., 3rd ‘Understanding’ medical school curriculum content using KnowledgeMap. J Am Med Inform Assoc JAMIA. 2003;10:351–362. doi: 10.1197/jamia.M1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stenetorp P, et al. BRAT: A Web-based Tool for NLP-assisted Text Annotation. Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics; Association for Computational Linguistics; 2012. pp. 102–107. at < http://dl.acm.org/citation.cfm?id=2380921.2380942>. [Google Scholar]
- 14.Pham A-D, et al. Natural language processing of radiology reports for the detection of thromboembolic diseases and clinically relevant incidental findings. BMC Bioinformatics. 2014;15:266. doi: 10.1186/1471-2105-15-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bejan CA, Xia F, Vanderwende L, Wurfel MM, Yetisgen-Yildiz M. Pneumonia identification using statistical feature selection. J Am Med Inform Assoc JAMIA. 2012;19:817–823. doi: 10.1136/amiajnl-2011-000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao KP, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res. 2010;62:1120–1127. doi: 10.1002/acr.20184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carroll RJ, Eyler AE, Denny JC. Naïve Electronic Health Record phenotype identification for Rheumatoid arthritis. AMIA Annu Symp Proc AMIA Symp AMIA Symp. 2011;2011:189–196. [PMC free article] [PubMed] [Google Scholar]
- 18.Aronson AR, Lang F-M. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc JAMIA. 2010;17:229–236. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34:301–310. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 20.Shivade C, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc JAMIA. 2014;21:221–230. doi: 10.1136/amiajnl-2013-001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harkema H, Dowling JN, Thornblade T, Chapman WW. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform. 2009;42:839–851. doi: 10.1016/j.jbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng QT, et al. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak. 2006;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cuggia M, et al. Roogle: an information retrieval engine for clinical data warehouse. Stud Health Technol Inform. 2011;169:584–588. [PubMed] [Google Scholar]
- 24.Zapletal E, Le Bozec C, Degoulet P, Jaulent M-C. A collaborative platform for consensus sessions in pathology over Internet. Stud Health Technol Inform. 2003;95:224–229. [PubMed] [Google Scholar]