Abstract
Targeted cancer drugs are often associated with unexpectedly high cardiovascular (CV) adverse events. Systematic approaches to studying CV events associated with targeted anticancer drugs have high potential for elucidating the complex pathways underlying targeted anti-cancer drugs. In this study, we built tcTKB, a comprehensive CV toxicity knowledge base for targeted cancer drugs, by extracting drug-CV pairs from five large-scale and complementary data sources. The data sources include FDA drug labels (44,979 labels), the FDA Adverse Event Reporting System (FAERS) (4,285,097 records), the Canada Vigilance Adverse Reaction Online Database (CVAROD) (1,107,752 records), published biomedical literature (21,354,075 records), and published full-text articles from the Journal of Oncology (JCO) (13,855 articles). tcTKB contains 14,351 drug-CV pairs for 45 targeted anticancer drugs and 1,842 CV events. We demonstrate that CV events positively correlate with drug target genes and drug metabolism genes, demonstrating that tcTKB in combination with other data resources, could facilitate our understanding of targeted anticancer drugs and their associated CV toxicities.
Introduction
Treatment outcomes in cancer patients have dramatically improved since the introduction of targeted drugs. However, targeted drugs are often associated with unexpectedly high cardiovascular (CV) toxicities in cancer patients [1–2]. The mechanisms by which targeted drugs exert their toxic effects on heart and vasculature in cancer patients are not well-understood [3–4]. To ensure safe personalized cancer treatment, research efforts are needed to understand CV toxicities associated with targeted drugs. Systematic and integrated approaches to studying CV events associated with targeted drugs have high potential for elucidating the complex pathways underlying anti-cancer drugs, identifying the on- and off-targets of undesirable CV events, and predicting unknown CV toxicities [5–7]. However, systematic study of targeted drug-induced CV toxicities has been hampered by the lack of a comprehensive and machine-understandable knowledge base of drug-CV associations. The relevant knowledge is instead buried throughout multiple disparate and complementary information sources in varying formats. It was recently demonstrated that 39% of serious events associated with targeted cancer drugs were not reported in clinical trials and 49% were not described in FDA drug labels [8]. Therefore, in order to build a comprehensive knowledge base of CV toxicities associated with targeted drugs, it is important to extract knowledge from multiple complementary data sources.
Recently, we extracted targeted anticancer drug-associated CV events from the U.S FDA Adverse Event Reporting System (FAERS) (4,285,097 records) [9]. We also developed text classification, relationship extraction, signaling filtering, and signal prioritization algorithms to extract targeted anticancer drugs associated side effects, including CV events from 13,855 full-text articles and embedded tables from the Journal of Oncology (JCO) published between 1983 and 2013 [10–11]. In this study, we built tcTKB (the CardioToxicity Knowledge Base for Targeted Cancer Drugs) by combining drug-CV pairs extracted from FAERS and JCO articles with pairs from another three large-scale and publicly available datasets: U.S. Food and Drug Administration (FDA) drug labels (44,979 drug labels), the Canada Vigilance Adverse Reaction Online Database (CVAROD) (1,107,752 records), and the vast corpus of published biomedical literature (21,354,075 MEDLINE records).
Drug toxicity knowledge contained in these five data sources is largely complementary. FDA drug labels contain known adverse events associated with commercial drugs, which are mainly gleaned from controlled clinical trials. Drug-CV pairs from FDA drug labels are highly accurate (high precision), however the recall may be limited since they are mainly obtained from pre-marketing clinical trials for well-controlled patients (i.e. patients with less comorbidities or younger patients). The two post-marketing surveillance systems (FAERS and CVAROD) contain both voluntary and mandatory reports of suspected drug adverse events from health-care professionals, consumers, and pharmaceutical companies for drugs used in less controlled ‘real-world’ patient populations. FAERS is the main spontaneous reporting system overseen by the U.S. FDA. Mining drug-side effect (drug-SE) relationships from FAERS is a highly active research area. Data mining algorithms such as disproportionality analysis, correlation analysis, and multivariate regression have been developed to detect adverse drug signals from FAERS [12–14]. Recently, we developed signaling extraction, prioritization, filtering algorithms and extracted a total of 11,173 drugCV pairs, representing 39 targeted cancer drugs and 1095 CVs, from FAERS [9]. In this study, we will extract drug-CV pairs from CVAROD, the main spontaneous reporting system overseen by Health Canada. CVAROD contains more than one million patient records, however, research effort in mining drug safety signals from CVAROD is significantly less compared to efforts in mining FAERS. Drug-CV pairs extracted from these two post-marketing surveillance systems are comprised of known true positives (those included in FDA drug labels), unknown true positives, and false positives. The main challenge is to differentiate between unknown true positives and false positives.
JCO is the official journal of the American Society of Clinical Oncology and the leading journal in oncology. JCO articles not only include pivotal clinical trials that have led to drug approval, but also trials that are still in investigational stages and even failed trials. In one of our recently studies, we downloaded a total of 13,855 full-text JCO articles published between 1983 and 2013. We combined automatic table classification and relationship extraction approaches to extract anticancer drug-associated side effects from a total of 31,255 tables embedded in these JCO articles [10]. We also developed an integrated system combining text classification, relationship extraction, signal filtering, and signal prioritization algorithms to extract targeted anticancer drug-associated side effects from the full-text part of JCO articles [11]. We demonstrated in our previous studies and in this study that full-text oncological articles contains much drug-associated side effects including CV events that are not captured in FDA drug labels, MEDLINE abstracts, or post-market drug safety surveillance systems.
Currently, more than 22 million biomedical records are publicly available on MEDLINE, making it a rich source of CV toxicity information for drugs at all clinical stages, including drugs in pre-marketing clinical trials, post-marketing clinical case reports and clinical trials. The major challenge in extracting drug-CV pairs from this rich source is that it is buried in free-text format. We recently develop approaches to extract drug-SE pairs from MEDLINE [15–16]. In this study, we will apply these approaches to extract target anticancer drug-associated CV events from MEDLINE abstracts. In this study, we show that as much as 96% and 67% targeted anticancer drug-associated CV pairs extracted from MEDLINE are not included in FDA drug labels and FAERS, respectively, demonstrating the need of building tcTKB from multiple complementary data resources. To the best of our knowledge, this is the first research effort in building a comprehensive CV toxicity knowledge base for targeted drugs from multiple complementary data sources. We demonstrated that this unique knowledge base tcTKB, in combination with other data resources such as drug target databases or drug pharmacogenetics and genomics databases, can facilitate our deeper understanding of the molecular mechanisms underlying the unexpectedly high incidence of CV toxicities associated with many targeted anticancer drugs.
Data and methods
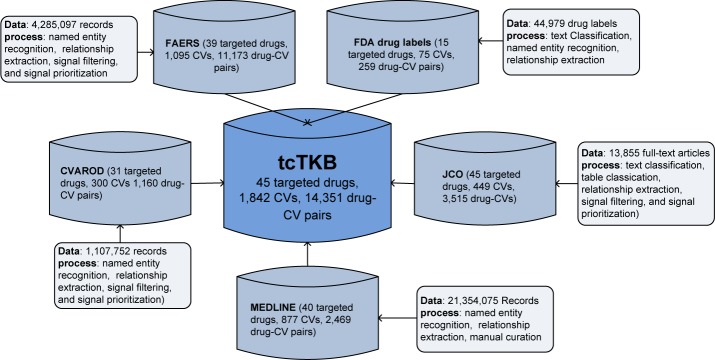
The data resources and methods that were used to construct tcTKB is depicted in Figure 1 and described later.
Figure 1:
The data resources and methods used in constructing tcTKB.
Data
The four data sources for extracting targeted anticancer drug-associated CV pairs are summarized in Table 1 and described in the sections that follow.
Table 1:
Summary of the five data sources of drug-CV associations.
| Source | Size | Type |
|---|---|---|
| FDA drug labels | 44,979 drug labels | Pre-marketing (main) and Post-marketing |
| FAERS | 4,285,097 records | Post-marketing (U.S) |
| CVAROD | 1,107,752 records | Post-marketing (Canada) |
| MEDLINE | 21,354,075 records | All stages |
| JCO | 13,855 full-text articles | pre-marketing |
FDA drug labels
We downloaded a total of 44,979 drug labels, including 21,610 human prescription labels and 23,369 human OTC labels from DailyMed1. DailyMed is maintained by the National Library of Medicine (NLM) and provides high quality FDA package inserts information about marketed drugs. The majority of drug side effect information captured on FDA drug labels is obtained from clinical trials, while some is obtained from post-marketing surveillance. We used the publicly available information retrieval library Lucene2 to create a local FDA drug label search engine with indices created on drugs, section headers such as “Indications,” “Contraindications,” and “Adverse Reactions,” and sentences. Each sentence was associated with a drug and a subsection header name.
The FDA Adverse Event Reporting System (FAERS)
FAERS is the prominent post-marketing drug safety surveillance system maintained by the U.S. FDA. In our recent study, we downloaded a total of 4,285,097 records from FAERS for the time period of 2004 to 20123. We extracted a total of 11,173 drug-CV pairs, representing 39 targeted cancer drugs and 1095 CVs, from FAERS [9].
The Canada Vigilance Adverse Reaction Online Database (CVAROD)
CVAROD is the main post-marketing drug safety surveillance system in Canada. A total of 1,107,752 patient records were downloaded4. File “reactions.txt” contains the reported adverse events. File “drug_product_ingredients.txt” contains drug information. Similar to pairs extracted from FAERS, pairs in CVAROD could contain spurious pairs as well as unknown true positives.
JCO full text articles
In our previous study, we downloaded a total of 13,855 JCO full text JCO articles published from 1983 through 2013 and extracted anticancer drug-SE pairs from both the text and the tables in the articles [10–11]. We extracted a total of 3,515 drug-CV pairs, representing 45 targeted anticancer drugs and 449 CV events.
MEDLINE data and local MEDLINE search engine
We downloaded a total of 21,354,075 MEDLINE records (119,085,682 sentences) published between 1965 and 2012 from NLM (http://mbr.nlm.nih.gov/Download/index.shtml). Each sentence was syntactically parsed with Stanford Parser [16] using the Amazon Cloud computing service (a total of 3,500 instance-hours with High-CPU Extra Large Instance were used). We created a local MEDLINE search engine with indices created on sentences, their corresponding parse trees, and abstracts.
Lexicon of targeted cancer drugs
The 45 targeted cancer drugs was obtained from the National Cancer Institute5.
Lexicon of cardiovascular event (CV) terms
We built a lexicon of CV terms based on MedDRA, a medical terminology widely used for classifying adverse events associated with drugs and other medical products6. The adverse events captured on FDA drug labels, FAERS, and CVAROD are coded with MedDRA terms. We created a lexicon of CV terms by finding all leaf nodes with the ancestor “vascular disorders” or “cardiac disorders.” This lexicon consisted of a total of 1,712 CV terms, including 1,269 vascular disorders and 527 cardiac disorders. In order to capture all the term variations in MEDLINE, we expanded the CV lexicon by including the synonyms of the terms in the lexicon. The term-synonym mappings were derived from UMLS Semantic Network [17]. After expansion, the CV lexicon consisted of 27,547 terms.
Methods
Extract drug-CV pairs from FDA drug labels
We used each of the 45 * 1,712 drug-CV combinations (45 targeted drugs and 1,712 CV terms) as search queries to the local FDA drug label search engine. Drug-CV pairs that appeared in sentences with the header “Adverse Reactions” were retrieved. We extracted a total of 259 drug-CV pairs from FDA drug labels, representing 15 targeted cancer drugs and 75 CV events. These pairs are of high quality and represent known cardiovascular events associated with targeted cancer drugs derived from both pre-marketing clinical trials (major) and post-marketing surveillance (minor).
Extract drug-CV pairs from FAERS
Drug-CV pair extraction from FAERS was done in our previous study [9]. After file linking, drug entity recognition and mapping, and CV entity recognition, we obtained a total of 11,173 drug-CV pairs, representing 39 (out of 45) targeted drugs and 1,095 (out of 1,712) CV events.
Extract drug-CV pairs from JCO articles
Drug-CV pair extractions from JCO articles (including both full-text and embedded tables) were done in our previous study [10–11]. We extracted a total of 3,515 drug-CV pairs, representing 45 (out of 45) targeted anticancer drugs and 449 (out of 1,712) CV events.
Extract and prioritize drug-CV pairs from CVAROD
We extracted drug-CV pairs from CVAROD by linking three downloaded files: (1) Report Drug.txt, which provides report drug identifiers and adverse reaction report numbers; (2) Drug_Product_Ingredient, which provides drug identifiers and active ingredient names; and (3) Reactions.txt, which provides adverse reaction report numbers and adverse reaction terms. We first linked the file “Drug_Product_Ingredient” with file “Reactions.txt” using the identifiers specified in “Report_Drug.txt”. We then extracted drug-CV pairs from the linked file. Unlike drug strings in FAERS, drugs in CVAROD were already mapped to their active ingredients, therefore no additional concept recognition and mappings were necessary. We filtered the drugs with the lexicon of 45 targeted cancer drugs. The same named entity recognition for CV terms as was completed for FAERS [9] was also performed for the data collection from CVAROD. In total, we obtained 1,160 drug-CV pairs, representing 31 targeted cancer drugs and 300 CVs.
While drug-CV pairs extracted from FDA drug labels are known true positives, pairs extracted from CVAROD contain known true positives, unknown true positives, and true negatives. In order to prioritize drug-CV pairs extracted from CVAROD, we implemented and compared six ranking algorithms in prioritizing true signals, including ranking by pairs’ frequency counts (FREQ) in FAERS, and five commonly used Disproportionality Analysis (DPA) statistical signal detection approaches: relative reporting ratio (RRR), proportional reporting ratio (PRR), reporting odds ratio (ROR), phi coefficient (PhiCorr), and information component (IC). The five DPAs are currently the most widely used approaches for automated signal detection in FAERS [11]. All these DPA methods are based on frequency analysis of 2×2 contingency tables to estimate statistical association between drugs and SEs and it intends to quantify the degree to which a drug-SE pair co-occurs disproportionally in the database. These five DPA methods differ by the statistical adjustments they apply to account for low counts.
In order to compare different ranking methods, we used 11-point interpolated average precision, a commonly used measure in evaluating information retrieval results [18]. For each ranked list, the interpolated precision was measured at the 11 recall levels of 0.0, 0.1, 0.2, …, 1.0. At each recall level, we calculated the arithmetic mean of the interpolated precision. A composite precision-recall curve showing 11 points was then graphed. In order to compare these six ranking approaches in prioritizing true signals, we used the 259 drug-CV pairs extracted from FDA drug labels as the evaluation dataset, which consisted of known true positives. If a ranking algorithm ranks known true positives highly, we can reason that it also ranks many unknown true positives highly. Note: this evaluation dataset was not used to calculate the true precisions and recalls, but to compare the six ranking approaches in prioritizing true signals.
Extraction and manual curation of drug-CV pair from MEDLINE
We used each of the 45 * 27,547 drug-CV combinations for the 45 targeted cancer drugs and 27,547 CV terms as search queries to the local MEDLINE search engine. Sentences, their associated parse trees, and abstracts that contained each pair were retrieved. Instead of simply retrieving a pairs co-occurrence counts at both sentence- and abstract-level from the search engine, we added the extra restriction that both drug and CV terms must be noun phrases in retrieved parse trees. This additional restriction was put in place to prevent the extraction of partial drug-CV pairs from sentences. For example, a CV term appeared as a substring in a noun phrase in the sentence where the noun phrase term is not included in the input CV lexicon. We extracted a total of 1,080 drug-CV pairs (38 targeted cancer drugs and 470 CVs) from MEDLINE sentences and 2,469 pairs (40 targeted cancer drugs and 877 CVs) from MEDLINE abstracts.
Unlike drug-CV pairs extracted from FAERS or CVAROD, which are hard to evaluate, pairs extracted from MEDLINE have associated abstracts which can be used for manual curation. We extracted a total of 1,080 drug-CV pairs from MEDLINE sentences and then manually curated these pairs. We used the local MEDLINE search engine to retrieve all the sentences (8,590 in total) wherein the pairs appeared. We then manually classified these 1,080 drug-CV pairs into three classes (CAUSE, TREAT, and NONE) using the sentences (and abstracts when necessary) as evidence. Three curators with graduate degrees in biomedical sciences independently performed the curation. It took an average 25 hours for each curation in annotating these sentences. Majority vote was used to decide the final classification.
Correlation analysis
We investigated whether drug-drug pairs that shared CV events also tended to share gene targets. We downloaded a total of 10,478 drug-gene pairs from DrugBank [19], a knowledge base for drugs, drug actions, and drug targets. These downloaded drug-gene pairs included a total of 24 targeted cancer drugs. For drug-drug pairs that shared different numbers of CV events, we calculated the average number of shared gene targets.
We investigated whether drug-drugs that shared CV events also tended to share drug metabolism genes. We downloaded a total of 4,399 drug-gene pairs from PharmGKB (the Pharmacogenetics and Pharmacogenomics Knowledge Base) [20], a public repository of genotype and phenotype information relevant to pharmacogenetics. The drug-gene pairs were assigned with subtypes Pharmacokinetics (“PK”) and Pharmacodynamics (“PD”). These downloaded drug-gene pairs included a total of 25 targeted cancer drugs. For drug-drug pairs that shared different numbers of CV events, we calculated the average number of shared metabolism genes.
1 Results
1.1 Description of tcTKB
tcTKB is comprised of a total of 14,351 unique drug-CV pairs extracted from five data sources, representing 45 targeted drugs and 1,842 CV events (Table 2). From the FDA drug labels, we extracted a total of 259 drug-CV pairs, representing 15 targeted drugs and 75 CV events. Unlike pairs extracted from the other three data sources, pairs extracted from FDA drug labels are mainly known true positives. From FAERS, we extracted a total of 11,173 drug-CV pairs, representing 39 targeted cancer drugs and 1,095 CV events. These CV events were reportedly associated with targeted cancer drugs in patients in real-world settings and included many CV events that have not yet been captured in the FDA drug labels. We extracted a total of 1,160 drug-CV pairs from CVAROD for 31 targeted cancer drugs and 300 CV events. This number is 10 times smaller than the number of pairs extracted from FAERS. It is unclear why significantly fewer CV events were reported in CVAROD than in FAERS although the numbers of targeted cancer drugs in both databases are comparable. From MEDLINE sentences, we extracted 1,080 drug-CV pairs for 38 targeted cancer drugs and 479 CV events. From MEDLINE abstracts, we extracted 2,469 drug-CV pairs representing 40 cancer targeted drugs and 877 CV events. The numbers of drug-CV pairs extracted from MEDLINE were smaller than that from FAERS; however, these pairs contained 40 out of the 45 targeted cancer drugs. A total of 3,515 drug-CV pairs were extracted from JCO articles. These pairs included all 45 targeted drugs, demonstrating that JCO articles is good data resource for targeted anticancer drugs.
Table 2:
Summary of tcTKB.
| Source | Drugs | CVs | Pairs |
|---|---|---|---|
| FDA drug labels | 15 | 75 | 259 |
| FAERS | 39 | 1,095 | 11,173 |
| CVAROD | 31 | 300 | 1,160 |
| MEDLINE (sentence) | 38 | 479 | 1,080 |
| MEDLINE (abstracts) | 40 | 877 | 2,469 |
| JCO | 45 | 449 | 3,515 |
| Combined | 45 | 1,842 | 14,351 |
1.2 Drug-CV pairs extracted from four data sources are largely complementary
We investigated overlaps of drug-CV pairs extracted from FDA drug labels, FAERS, CVAROD, MEDLINE, and JCO. As shown in Table 3, the information in these four sources overlaps, but is largely complementary. Column 2 in Table 3 represents the percentages of drug-CV pairs from FDA drug labels that were also included in the other data sources. Row 2 represents the percentages of drug-CV pairs from the other three data sources that were captured in the FDA drug labels. As shown in both column 2 and row 2, many of the known drug-CV pairs in FDA drug labeling appeared in the other four data sources: 95.8% in FAERS, 46.7% in CVAROD, 38.2% in MEDLINE, and 71.4% in JCO (column 2). However, the opposite is not true. Only very small percentages of drug-CV events reported in other data sources were captured in FDA drug labels (row 2). This low percentage may be due to the following reasons: First, FDA drug labels contain only 15 targeted drugs, while the other data sources contained more targeted drugs. Second, while the drug-CV pairs extracted from FDA drug labels are mostly true positives, the pairs extracted from the other sources may contain false positives and unknown true positives.
Table 3:
The overlapping matrix of drug-CV pairs in tcTKB. The number in each cell represents the percentage of pairs from one source (column) also appeared in the other source (row).
| FDA drug labels | FAERS | CVAROD | MEDLINE | JCO | |
|---|---|---|---|---|---|
| FDA drug labels | 100% | 2.2% | 10.4% | 4.0 | 0.7%% |
| FAERS | 95.8% | 100% | 96.7% | 33.1% | 48.4% |
| CVAROD | 46.7% | 10.1% | 100% | 12.2% | 13.6% |
| MEDLINE | 38.2% | 7.4% | 26.0% | 100% | 23.0% |
| JCO | 71.4% | 15.3% | 41.3% | 32.8% | 100% |
Only small percentages of pairs from FAERS were included in other data sources: 2.2% in FDA drug labels, 10.1% in CVAROD, 7.4% in MEDLINE, and 15.3% in JCO (column 3). In addition, a majority of drug-CV pairs from other data sources (except CVAROD) were not included in FAERS (row 3). This indicates that drug-CV pairs contained in FAERS are largely complementary to those in FDA drug labels or MEDLINE. Drug-CV pairs contained in CVAROD are a subset of the pairs from FAERS as 96.7% of the 1,160 pairs also appeared in FAERS. Drug-CV pairs extracted from MEDLINE are largely complementary to those in the other three data sources. For example, only 4.0% of pairs from MEDLINE appeared in FDA drug labels, 33.1% in FAERS, and 12.2% in CVAROD (Column 5). The opposite is also true. Only 38.2% drug-CV pairs from FDA labels, 7.4% from FAERS, and 26.0% from CVAROD also appeared in MEDLINE. It is also evident from the table that full-text articles contain much information not captured in MEDLINE abstracts. For example, only 23% drug-CV pairs from JCO articles also appeared in MEDLINE abstracts.
1.3 Ranking drug-CV pairs from CVAROD
Drug-CV pairs extracted from CVAROD may contain false signals. We compared six ranking algorithms in prioritizing extracted drug-CV pairs using drug-CV pairs extracted from the FDA drug labels as goldstandard. In our previous study in extracting drug-CV pairs from FAERS, we demonstrated that ranking by frequency significantly improved the precisions of top-ranked pairs as compared to other statistical ranking methods including RRR, ROR, RR, IC. Here we investigate whether the same is true in ranking drug-CV pairs extracted from CVAROD. As shown in Fig. 2, ranking based on frequency is more effective than ranking based upon the other five approaches. The precision of top-ranked pairs (at recall = 0.1) is 0.16 as measured with drug-CV pairs extracted from FDA drug labels as gold standard. This still low precision indicates that it is likely that many true positives among the top-ranked pairs from CVAROD have not yet been captured in FDA drug labeling. This is reflected by the modest overlap between pairs extracted from FDA drug labels and pairs extracted form CVAROD (Table 3).
Figure 2:
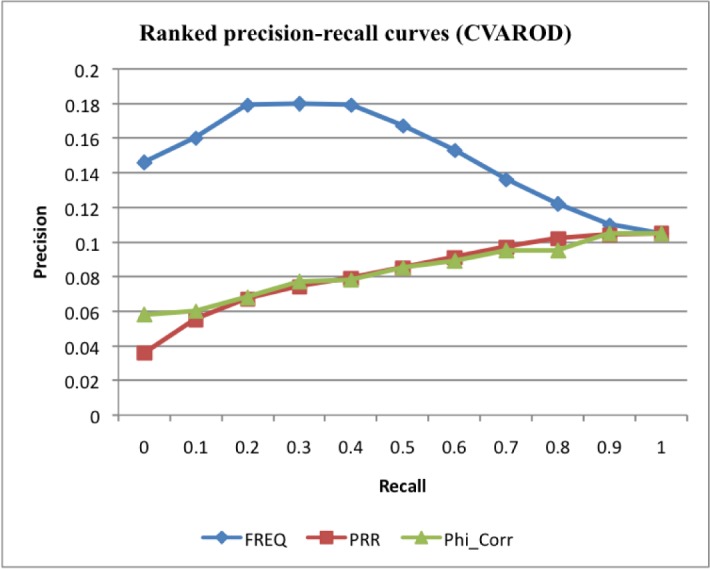
Ranked precisions at 11 recalls for drug-CV pairs from CVAROD ranked by three ranking measures: frequency (Freq), proportional reporting ratio (PRR) and phi coefficient (PhiCorr). Data for relative reporting ratio (RRR), reporting odds ratio (ROR), and information component (IC) IC are similar to that for PRR and not shown
1.4 Manual curation of drug-CV pairs extracted from MEDLINE sentences
We manually curated all 1,080 drug-CV pairs extracted from MEDLINE sentences. We did not curate the 1,389 pairs that only co-occurred in MEDLINE abstracts but not in sentences. Among the 1,080 pairs, 525 pairs (48.6%) were true positives (“drug CAUSE CV”), 356 pairs (32.9%) were “drug TREAT CV” pairs, and 199 pairs (18.5%) had no obvious semantic relationships (“drug NONE CV”) (Table 4). Moreover, among the 525 true positives, 477 pairs (90.8%) were not included in FDA drug labels.
Table 4:
Manual curation of drug-CV pairs extracted from MEDLINE sentences.
| Total | CAUSE | TREAT | NONE | Not in FDA drug labels |
|---|---|---|---|---|
| 1,080 | 48.6% | 32.9% | 18.5% | 90.8% |
1.5 Positive relationship between shared CV events and shared drug target genes
We investigated whether drug-drug pairs that shared CV events also tended to share gene targets. A positive relationship between these two entities could indicate a causal relationship between drug targets and the observed cardiovascular adverse events associated with targeted anticancer drugs. This indication could in turn open up the possibility of predicting unknown CV events by systematically studying their gene targets and discovering novel drug off-targets, such as targets related to cardiovascular systems, instead of tumor cell growth, based on observed cardiovascular events. As shown in Figure 3, there exists a positive relationship between shared CV events and shared gene targets for drug-drug pairs. For instance, the average number of shared gene targets was 1.678 for all drug-drug pairs (cut off≥0). The number significantly increased to 2.122 for drug-drug pairs that shared at least 250 CVs (cutoff≥250) and to 3.857 for drug-drug pairs that shared at least 450 CVs (cutoff≥450).
Figure 3:
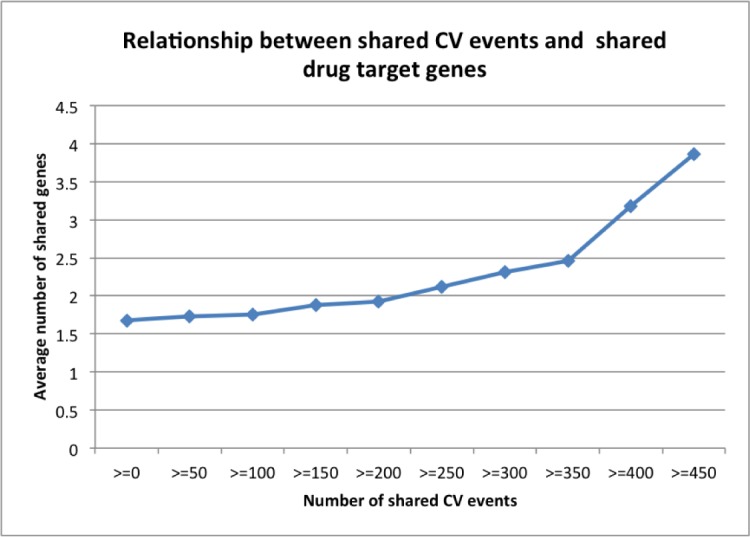
Relationship between shared CV events and shared target genes.
Since drug metabolism is responsible for many known drug-related adverse events, we then investigated whether the observed CV events were related to drug metabolism. Any positive correlations between CV events and drug metabolism genes could open up the possibility of predicting drug-associated CV events in specific cancer patients based on their metabolism genotypes (personalized cancer care). As shown in Figure 4, there is a strong positive correlation between CV events and drug metabolism genes. Drug-drug pairs that shared more CV events tended to also share more metabolism genes. The average number of shared metabolism genes for all drug-drug pairs (cutoff≥0) was 0.39. The number significantly increased to 0.583 for drug-drug pairs sharing at least 250 CV events (cutoff≥250) and 1.375 for pairs sharing at least 450 CV events (cutoff≥450). In summary, both gene targets and metabolism genes positively correlated with targeted cancer drug-associated CV events, indicating that the observed CV events may have discoverable genetic causes and that we can predict unknown cardiotoxicities and achieve personalized cancer care by systematically studying drug-associated gene targets and metabolism.
Figure 4:
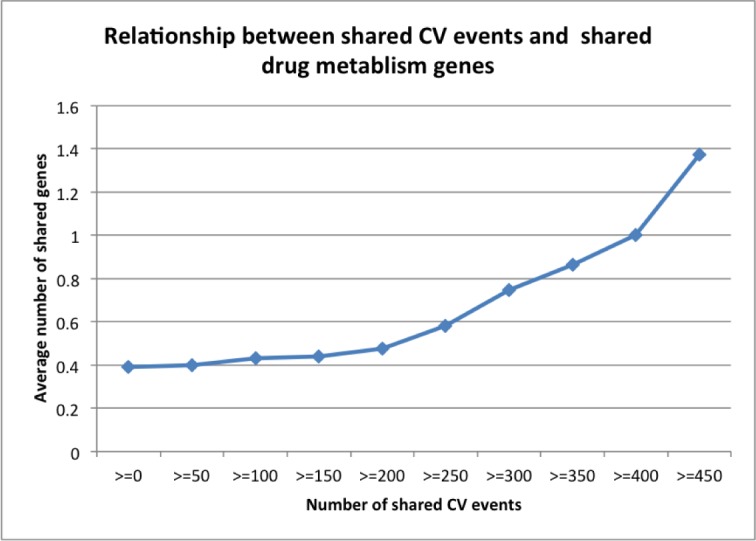
Relationship between shared CV events and shared metabolism genes.
2 Discussion
In this study, we built a comprehensive CV toxicity knowledge base for targeted cancer drugs (tcTKB) by extracting drug-CV pairs from five large scale and complementary data sources. We manually curated all drug-CV pairs that appeared in MEDLINE sentences. We systematically analyzed the correlations between the observed CV events and drug-associated gene targets and demonstrated that tcTKB, in combination with other drug-related data, represents a unique knowledge base for our understanding targeted anticancer drugs and their associated high incidence of many unexpected cardiovascular adverse events.
Nonetheless, our study has several limitations and can be further improved in the future. First, even though tcTKB includes many drug-CV pairs, it remains unknown what the actual precision and recall of these pairs are. For example, it is difficult to measure how many of the drug-CV pairs in tcTKB are true positives (including both known and unknown positives) and how many of them are true negatives. In addition, even though we used four large datasets in constructing tcTKB, we don’t know what the coverage of this knowledge base is. There may be drug-CV pairs in existence in other data sources, such as patient electronic health records (EHRs). EHRs contain rich post-marketing drug adverse event information [21]. However, obtaining EHRs requires approval from institutional review boards. tcTKB can be significantly enriched by including drug-CV associations extracted from EHRs.
Second, we observed positive relationship between CV events and drug-associated gene targets. As was previously discussed, the drug-CV pairs in tcTKB may contain noise, which can affect the observed correlations. However, we expect that the noise shall not systematically correlate with drug gene targets. Systems approach to studying drug-CV associations in tcTKB, as done for the widely used protein-protein interaction (PPI) data (which also contains noise and is largely incomplete) may generate many insightful biological hypotheses.
Third, the 45 target cancer drugs under study here have been used in treating both cancers and other, non-cancerous, conditions. For example, bevacizumab is an FDA-approved drug for the treatment of colorectal cancer and lung cancer. Bevacizumab is also used to treat eye diseases such as age-related macular degeneration and diabetic retinopathy. In constructing tcTKB, we did not take patient disease characteristics into account. It is possible that certain cardiovascular events only appear when a targeted drug is taken by cancer patients. In order to further stratify drug-CV pairs in tcTKB by patient characteristics, we need detailed information about patient disease characteristics and demographics. Patient demographics information can be extracted from MEDLINE abstracts [22], full-text JCO articles as well as FAERS. In the future, we will enrich tcTKB with patient demographics information.
Last but not least, we only extracted cardiovascular events associated with targeted cancer drugs in this study. In the future, we will extract richer sets of toxicities that are associated with targeted cancer drugs, including neurotoxicities, nephrotoxicities, heptotoxicities, and hematotoxicities, and develop systems approaches to studying these observed drug phenotypes in order to understand the molecular mechanisms underlying these toxicities.
Acknowledgments
Xu and Wang have jointly conceived the idea, designed and implemented the algorithms and prepared the manuscript. We would like to thank the three curators from ThinTek for the manual curation.
Funding
RX was supported by the Eunice Kennedy Shriver National Institute Of Child Health & Human Development of the National Institutes of Health under the NIH Directorś New Innovator Award number DP2HD084068, the Training grant in Computational Genomic Epidemiology of Cancer (CoGEC) (R25 CA094186-06), and the Grant #IRG-91-022-18 to the Case Comprehensive Cancer Center from the American Cancer Society.
Footnotes
References
- 1.Cleeland CS, Allen JD, Roberts SA, Brell JM, Giralt SA, Khakoo AY, Skillings J. Reducing the toxicity of cancer therapy: recognizing needs, taking action. Nature reviews Clinical oncology. 2012;9(8):471–478. doi: 10.1038/nrclinonc.2012.99. [DOI] [PubMed] [Google Scholar]
- 2.Keefe DM, Bateman EH. Tumor control versus adverse events with targeted anticancer therapies. Nature Reviews Clinical Oncology. 2012;9(2):98–109. doi: 10.1038/nrclinonc.2011.192. [DOI] [PubMed] [Google Scholar]
- 3.Ewer MS, Ewer SM. Cardiotoxicity of anticancer treatments: what the cardiologist needs to know. Nature Reviews Cardiology. 2010;7(10) doi: 10.1038/nrcardio.2010.121. [DOI] [PubMed] [Google Scholar]
- 4.Mellor HR, Bell AR, Valentin JP, Roberts RR. Cardiotoxicity associated with targeting kinase pathways in cancer. Toxicological Sciences. 2010:kfq378. doi: 10.1093/toxsci/kfq378. [DOI] [PubMed] [Google Scholar]
- 5.Gibb S. Toxicity testing in the 21st century: a vision and a strategy. Reproductive Toxicology. 2008;25(1):136–138. doi: 10.1016/j.reprotox.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 6.Pujol A, Mosca R, Farrs J, Aloy P. Unveiling the role of network and systems biology in drug discovery. Trends in pharmacological sciences. 2010;31(3):115–123. doi: 10.1016/j.tips.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Raschi E, De Ponti F. Cardiovascular toxicity of anticancer-targeted therapy: emerging issues in the era of cardio-oncology. Internal and emergency medicine. 2012;7(2):113–131. doi: 10.1007/s11739-011-0744-y. [DOI] [PubMed] [Google Scholar]
- 8.Seruga B, Sterling L, Wang L, Tannock IF. Reporting of serious adverse drug reactions of targeted anticancer agents in pivotal phase III clinical trials. Journal of Clinical Oncology. 2010 doi: 10.1200/JCO.2010.31.9624. JCO-2010. [DOI] [PubMed] [Google Scholar]
- 9.Xu R, Wang Q. Automatic signal prioritizing and filtering approaches in detecting post-marketing cardiovascular events associated with targeted cancer drugs from the FDA Adverse Event Reporting System (FAERS) Journal of Biomedical Informatics. 2014 2014 Feb;47:171–7. doi: 10.1016/j.jbi.201310.008. Epub 2013 Oct 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu R, Wang Q. Combining automatic table classification and relationship extraction in extracting anticancer drug-side effect pairs from full-text articles. Journal of Biomedical Informatics. 2015 2015 Feb;53:128135. doi: 10.1016/j.jbi.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu R, Wang Q. Large-scale automatic extraction of side effects-associated with targeted anticancer drugs from full-text oncological articles. Journal of Biomedical Informatics. 2015 Jun;55:64–72. doi: 10.1016/j.jbi.2015.03.009. Epub 2015 Mar 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bate A, Evans SJW. Quantitative signal detection using spontaneous ADR reporting. Pharmacoepidemiology and drug safety. 2009;18(6):427–436. doi: 10.1002/pds.1742. [DOI] [PubMed] [Google Scholar]
- 13.Harpaz R, DuMouchel W, Shah NH, Madigan D, Ryan P, Friedman C. Novel Data? Mining Methodologies for Adverse Drug Event Discovery and Analysis. Clinical Pharmacology & Therapeutics. 2012;91(6):1010–1021. doi: 10.1038/clpt.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu R, Wang Q. Large-scale combining signals from both biomedical literature and the FDA Adverse Event Reporting System (FAERS) to improve post-marketing drug safety signal detection. BMC Bioinformatics. 2014 2014 Jan 15;15:17. doi: 10.1186/1471-2105-15-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu R, Wang Q. Toward creation of a cancer drug toxicity knowledge base: automatically extracting cancer drug-side effect relationships from the literature. Journal of the American Medical Informatics Association. 2013 doi: 10.1136/amiajnl-2012-001584. amiajnl-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu R, Wang Q. Automatic construction of a large-scale and accurate drug-side-effect association knowledge base from biomedical literature. Journal of Biomedical Informatics, J Biomed Inform. 2014 2014 Oct;51:191–9. doi: 10.1016/j.jbi.201405.013. Epub 2014 Jun 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein D, Manning CD. Accurate unlexicalized parsing. Proceedings of the 41st Annual Meeting on Association for Computational Linguistics; Association for Computational Linguistics; 2003. Jul, pp. 423–430. [Google Scholar]
- 17.Humphreys BL, Lindberg DA, Schoolman HM, Barnett GO. The Unified Medical Language System An Informatics Research Collaboration. Journal of the American Medical Informatics Association. 1998;5(1):1–11. doi: 10.1136/jamia.1998.0050001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manning CD, Raghavan P, Schtze H. Introduction to information retrieval. Vol. 1. Cambridge: Cambridge university press; 2008. p. 496. [Google Scholar]
- 19.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic acids research. 2008;36(suppl 1):D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDonagh EM, Whirl-Carrillo M, Garten Y, Altman RB, Klein TE. From pharmacogenomic knowledge acquisition to clinical applications: the PharmGKB as a clinical pharmacogenomic biomarker resource. Biomarkers in medicine. 2011;5(6):795–806. doi: 10.2217/bmm.11.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu R, Garten Y, Supekar K, Altman RB, Garber AM. (2007) Extracting Subject Demographics From Abstracts of Randomized Clinical Trials; World Congress on Medical and Health Informatics(MEDINFO); 2007. pp. 824–828. [PubMed] [Google Scholar]