Abstract
The financial incentives for data science applications leading to improved health outcomes, such as DSRIP (bit.ly/dsrip), are well-aligned with the broad adoption of Open Data by State and Federal agencies. This creates entirely novel opportunities for analytical applications that make exclusive use of the pervasive Web Computing platform. The framework described here explores this new avenue to contextualize Health data in a manner that relies exclusively on the native JavaScript interpreter and data processing resources of the ubiquitous Web Browser. The OpenHealth platform is made publicly available, and is publicly hosted with version control and open source, at https://github.com/mathbiol/openHealth. The different data/analytics workflow architectures explored are accompanied with live applications ranging from DSRIP, such as Hospital Inpatient Prevention Quality Indicators at http://bit.ly/pqiSuffolk, to The Cancer Genome Atlas (TCGA) as illustrated by http://bit.ly/tcgascopeGBM.
Introduction
Technological and policy trends are reshaping the development and deployment of Biomedical Informatics applications. The increasingly data-intensive, patient-facing nature of modern Medical Systems are translating well established architectural recommendations, such as the decoupling between the data layer and presentation layer, into specific requirements.
Furthermore, the ongoing evolution of the Web into a global data space (Heath 2011) is now a ubiquitous Big Data reality, with the Web Platform (Web Platform 2013) serving as its unifying computational environment.
As a matter of policy, public data held by government agencies in the US and many other countries must be available as Open Data (White House 2013). The technical requirements for Open Data play a foundational role in Web Technologies, defining a degree of openness along a 5 point system (Berners-Lee 2011), which ranges from being on the web (1 point) to using a linked Resource Description Framework (RDF, 5 points). Most public health data systems maintain a score of at least 3.. This is typically achieved by relying on a nonproprietary format, such as JSON, exposed through an HTTP REST API such as that provided by Socrata’s SODA data services (Socrata 2014). The availability of a wealth of health data resources satisfying this level of interoperability can be readily verified by pointing a web browser to State and Federal data resources such as health.data.ny.gov, data.medicare.gov, data.healthcare.gov, or data.cms.gov.
This report explores the feasibility of OpenHealth analytical applications that are entirely free from server-side presentation resources. The motivation for this approach derives from the unparalleled scalability promised by “beyond the data deluge” analytical solutions based on code distribution (Bell 2009). This pursuit is also informed by a number of previous results showing that client-side code distribution is no longer bound by significant algorithmic or performance limitations. Regarding the latter, browser hosted computing was shown to scale well into the realms of HPC (Wilkinson 2014). Similarly, even specialized operations, such as image (Almeida 2012a) and sequence analysis (Almeida 2012b), have, for some time, been efficiently supported by the computational engines of the Web Platform. Nevertheless, it was also abundantly clear from the onset of the study that new capabilities were needed for functionalities conventionally hosted server-side such as data caching and normalization. A third challenge to the OpenHealth route was devising an approach to the user interface assembly that would allow researchers to present prototype analytical applications to domain users with minimal overhead.
Methods
Computational application
The OpenHealth platform was developed entirely in client-side JavaScript, which is made publicly available, and is publicly hosted, with version control and open source, at https://github.com/mathbiol/openHealth. The versioned hosting feature is achieved by versioned code development in GitHub’s gh-pages branch. The architecture of the OpenHealth platform is detailed in the Results section, and includes reliance on the native NoSQL data management and storage resources of the Web Browser - IndexedDB (http://www.w3.org/TR/IndexedDB). Code development, including the use of IndexedDB, was pursued with strict adherence to W3C standards recommendations. In principle, this should render OpenHealth cross-browser, but this behavior was only tested for Google’s Chrome and Mozilla’s Firefox browsers, albeit on multiple platforms: desktop, tablet and cell phones running multiple operating systems: Mac OS, Windows, Android, and Chrome-OS.
External data and libraries
The illustrative applications make use of specialized libraries for visualization, d3.js (https://github.com/mbostock/d3), and dimensional charting (https://github.com/dc-js/dc.js). Data caching was mediated through Mozilla’s open source localForage.js library (https://github.com/mozilla/localForage). These libraries were chosen, in part, because of OpenHealth’s emphasis on public versioned hosting of open source code. Similarly, all data used in the applications described in this report are available in the public domain from a variety of State and Federal agencies, primarily hosted by health.data.ny.gov and data.medicare.gov for case study examples 1–3. In all three cases, data retrieval is performed by demand of the analysis, using the Socrata JSON formated open data services API (http://www.socrata.com/products/open-data-api) of those Open Data resources. The data normalization case study 4, which uses data from The Cancer Genome Atlas (Figure 5) illustrates a solution for patient-derived data resources made public without the complement of a data service API, or even of a Cross-Origin Resource Sharing (CORS) enabled web serving. The solution found, described in Results, is the basis for a number of recommendations in the Discussion. The TCGA data used is nevertheless in the public domain (no restricted access TCGA data sets were used), hosted by The National Cancer Institute of The National Institutes of Health, NIH/NCI, at https://tcga-data.nci.nih.gov/tcgafiles/ftp_auth/distro_ftpusers/anonymous/tumor/.
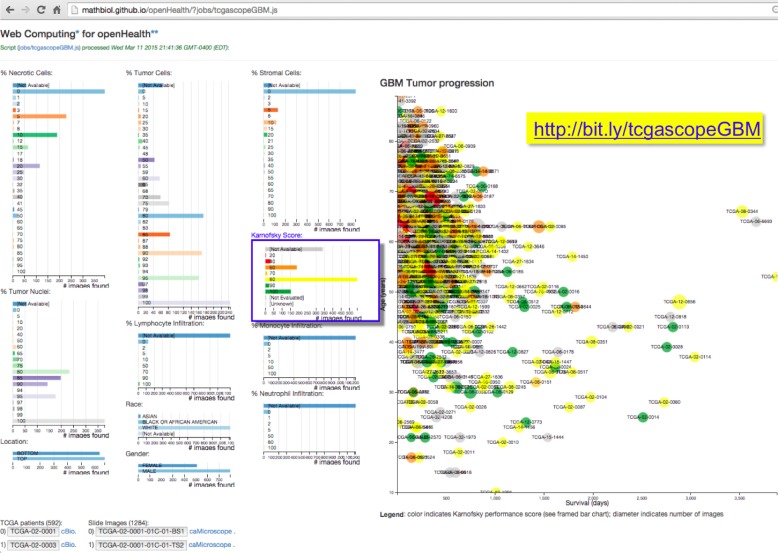
Figure 5.
Snapshot of interactive application assembled from TCGA raw text files describing TCGA patients diagnosed with Glioblastoma Multiforme, and the pathology slide images obtained from the corresponding biopsies (see text for details). The cartesian plot on the right projects, interactively, the position of each patient by age and survival (days to death), while tracking the Karnofsky score (color) and images (diameter). The effect of histopathology and demographic features can be assessed visually through interaction with the corresponding bar charts.
Results
The main goal of this project was to develop a distributable platform for data-intensive computation of OpenHealth Data entirely as a client side Web Application. This design overcomes the need for server-side components and maximizes scalability through code distribution (Bell 2009). Open Government mandates and NIH dissemination requirements have engendered a wealth of reliable, high availability, Big Data web services. These web services and their patient-resolved clinical and biomolecular data are central to the interactive systems described, which illustrate the use of the OpenHealth platform (OH).
Illustrative applications with < OpenHealth>?<analysis> URL composition
The OpenHealth platform’s data management and normalization functionalities are best visualized through illustrative examples. These illustrations will address four boundary scenarios: 1) Graphic interaction with a large data resource containing population-level data; 2) traversal of a large collection of individual patient data; 3) Cross-tabulation of multiple sources; and 4) normalization of patient-derived biomedical data available in the public domain as raw data files. In each of these examples, the same code migration URL composition pattern is invoked. OpenHealth’s core library is dereferenced with the analytical code pulled in as an additional search parameter. For example, the interactive representation in Figure 2 can be produced from http://mathbiol.github.io/openHealth/?jobs/pqiSuffolk.js.
-
Graphic interaction with a large data resource containing population level data
To illustrate the point that <openHealth>?<data analysis> is just a generative pattern, and that the analysis code can be dereferenced from both relative or absolute URLs, it is useful to note that the same result captured by the snapshot in figure 2 could be produced by http://mathbiol.github.io/openHealth/?https://rawgithub.com/SBU-BMI/openHealth/33a1b22d0e0f7fdf786bfe2ebbf024ac55523262/jobs/pqiSuffolk.js. This formulation highlights the criticality of the versioned hosting feature of OH; the analysis URL points to a specific version, hosted in a different domain. That is, for the preventable disease interactive display coded by version UID 33a1b22d0e0f7fdf786bfe2ebbf024ac55523262. The URL composition is convenient to illustrate this report, but a more useful approach may seek to engage the code migration functionality programmatically, as discussed later in this report.
traversal of a large collection of individual patient data
Cross-tabulation of care providers
-
Normalization of patient-derived biomedical data made public as raw text files.
The example application in Figure 5 tests the limits of what can be accomplished using the proposed approach when the patient-derived data is exposed without any specific provisions for web-based processing (see Discussion). This application retrieves two raw text files from https://tcga-data.nci.nih.gov/tcgafiles/ftp_auth/distro_ftpusers/anonymous/tumor/gbm/bcr/biotab/clin/, one describing the clinical data for 592 TCGA patients diagnosed with Glioblastoma Multiforme, nationwidechildrens.org_clinical_patient_gbm.txt, the other describing 1,284 pathology slide images obtained from the corresponding tumor samples, nationwidechildrens.org_biospecimen_slide_gbm.txt. While it may not lead to a noticeable delay in data retrieval and normalization, the inspection of the application code, migrated from github.com/mathbiol/openHealth/blob/gh-pages/jobs/tcgascopeGBM.js, will reveal the mediation of a cloud hosted proxy application, bit.ly/getTCGAtxt, that pass the text content across TCGA data hosting domain restrictions (see Discussion).
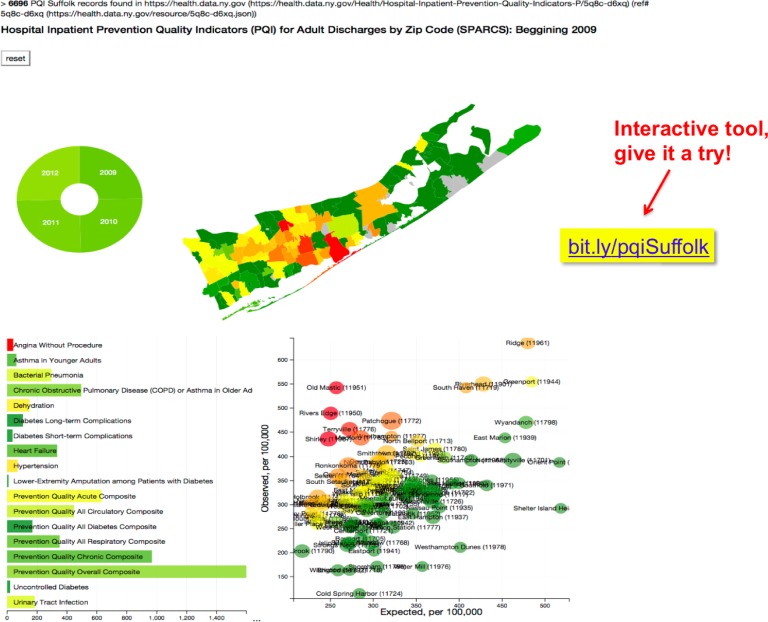
Figure 2.
Pure client-side assembly of an interactive tool for Hospital Inpatient Prevention Quality Indicators (PQI) for Suffolk County, Long Island, NY: bit.ly/pqiSuffolk. The analysis begins with OH retrieving the data for each of the relevant 107 zip codes (out of 111474 described in the reference health.data.ny.gov/resource/5q8c-d6xq source for the state of NY), and caching them in the native NoSQL browser resource, IndexedDB (see Methods). Like other native data resources, such as localStorage and WebSQL, IndexedDB will persist between sessions for the domain name. As a consequence, subsequent accesses to this interactive Web Application will not exhibit the same initial waiting period for data retrieval.
Discussion
The interactive applications and supporting data processing configuration of the OpenHealth platform is proposed to be a good fit for patient-centric, outcomes-driven health delivery programs such as Medicaid’s Delivery System Reform Incentive Payment (DSRIP, bit.ly/dsrip) and the new Patient-Centered Outcomes Research Institute (PCORI). However, the domain facing nature of modern Information and Communication Technologies (Almeida 2014) is, of course, not a reaction to these systems–quite the opposite. Healthcare is a latecomer to both the commoditized consumer-facing ICT (Mandl 2012) and the operational improvements facilitated using Big Data (Murdoch 2013, Manyika 2011). Therefore, it is reasonable to anticipate that the growing wealth of patient-resolved health data resources delivered by Open Government mandates, as articulated by the US Department of Health and Human Services (HHS 2015), will fundamentally change not only Health Information Systems, but the research and understanding of disease (Roth 2015). What data services and application development architectures will prove more effective in meeting those goals is the broader question that the OpenHealth platform described here explores.
The interactive applications in figures 2–4 provide evidence that OH’s server-less architecture, described in Figure 1B, is computationally more efficient, it is far easier to disseminate, distributing analysis that are therefore easier to reproduce. Although the applications in the Results section were assembled by a URL composition pattern, <openHealth>?<data analysis>, the core OH library can be loaded programmatically by script tag loading as in < script src="https://mathbiol.github.io/openHealth/openHealth.js"></script> or, for example, using jQuery, $.getScript(“https://mathbiol.github.io/openHealth/openHealth.js"). This is explained in detail in the project’s code development page at https://github.com/mathbiol/openHealth.
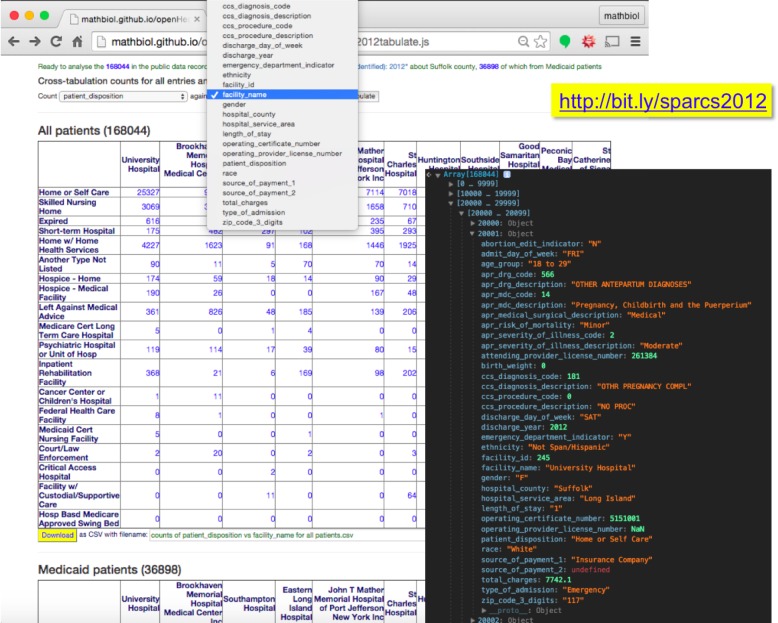
Figure 3.
Tabulation tool dereferenceable by shortcut bit.ly/sparcs2012. This example illustrates the logistics of traversing over 2.5 million de-identified hospital inpatient discharges in the state of NY in 2012 (Open Data resource health.data.ny.gov/resource/u4ud-w55t). The command line inset is a snapshot of the browser native tools, showing the values of 36 parameters for one of the 168,044 records found for Suffolk county. Note that first use of this interactive application will require pre-processing times of a few minutes, depending on the machine, but subsequent uses have OH shorten data retrieval to under half a minute, even in a moderately resourced mobile devices.
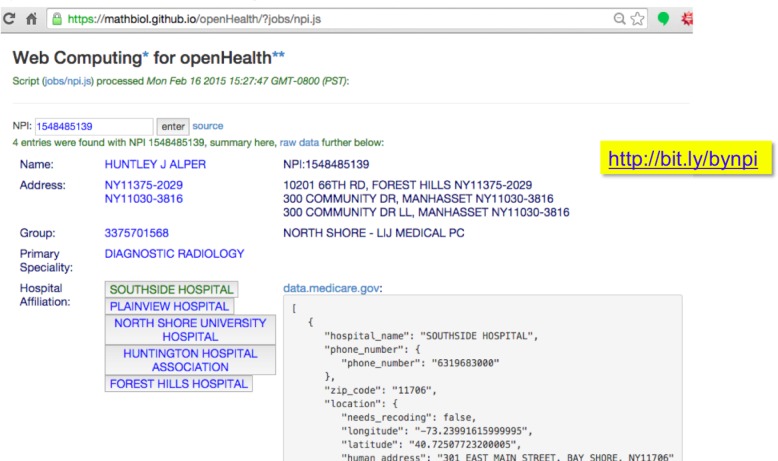
Figure 4.
Example of an interactive application that crosses two Medicare databases for an arbitrary provider identifier and the corresponding hospital affiliations. Loading times should be nearly instantaneous on any device.
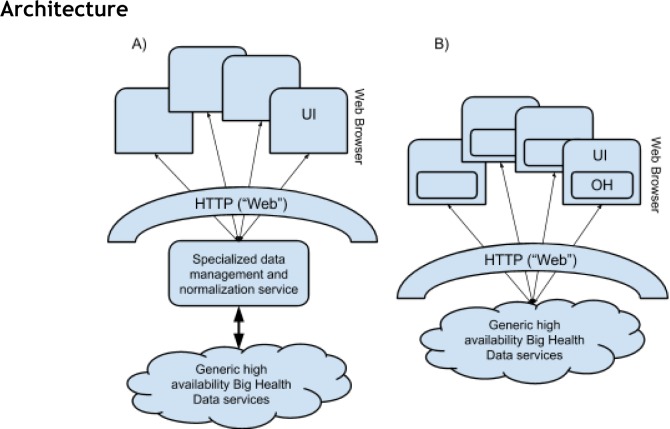
Figure 1.
Simplification of the conventional server-side data management and normalization architecture (A) by relocating that functionality (B) to the OpenHealth Platform (OH) assembled within the browser (dynamically loaded as JavaScript libraries), using native, W3C standardized, data management resources such as NoSQL IndexedDB (http://www.w3.org/TR/IndexedDB). It is critical to note that in (B), all data and computation is performed by either the data providers or by the domain consumers, not by dedicated Biomedical Informatics computation infrastructure - which is no longer needed. In other words, all that the Biomedical Informatics application layer does is provide the JS code, distributed directly to the Web Browser hosted OH component, by the standard script tag injection mechanism.
The interactive TCGA Glioblastoma (GBM) application described in figure 5, however, explores a more convoluted scenario, wherein the data is served to the public domain, but no API is available and cross-domain calling (CORS) is disabled. These obstacles have been noted by several reports over a number of years, but persist on many open repositories of public data. To overcome those barriers to interoperability with interactive web applications, we were compelled to develop a minimal proxy (server-side) mediator. To avoid a relapse to the troubled dependency on server-side resources (Fig 1A), we developed that TCGA proxy component as a Google Cloud hosted service, which can be inspected at bit.ly/getTCGAtxt. The TCGA GBM application was implemented to augment a web-based virtual microscope deployment, which hosts TCGA microscopy image data and image segmentation results. The application allows users to visualize various clinical attributes and select a subset of cases based on these attributes. Users can then interactively view these select cases using the virtual microscope platform. The need for the proxy component could have been entirely removed if the TCGA data web server included an open domain header (CORS), as noted elsewhere, namely (Robbins 2013) section 4.5 “Summary of technical recommendations for biomedical big data hosting”. On a more positive note, the normalization of the TCGA file contents did not itself present a major obstacle, as can be verified by noting the short loading/parsing times of bit.ly/tcgascopeGBM (Fig 5), and by following the cBio links (Cerami 2012) to those Web Applications.
Conclusion
OpenHealth is an in-browser JavaScript platform developed to mediate the management and normalization of Open Data in the Health Sciences domain in a manner that is scalable, secure and reproducible. Data pre-processing functionalities are conventionally associated with dedicated server-side resources. OpenHealth redirects that support to the native data resources of the modern Web Browser, which is now equipped with a computationally efficient JS interpreter, secured within a sandbox that isolates the execution of code migration from unauthorized access to additional local resources. This approach was found to be particularly effective for the development of interactive applications, and for the dissemination of reproducible analytical procedures. Mounting adoption of Open Government and Open Data mandates in Health Care suggests a key role for this architecture in measuring health outcomes and personalizing care delivery.
Acknowledgments
This work was supported in part by 1U24CA180924-01A1 from the NCI, R01LM011119-01 and R01LM009239 from the NLM. The authors also thankfully acknowledge support from Suffolk Care Collaborative Delivery System Reform Incentive Payment Program (dsrip.uhmc.sunysb.edu).
References
- Almeida JS, Dress A, Kühne T, Parida L. ICT for Bridging Biology and Medicine. Dagstuhl Manifestos. 2014;3(1):31–50. doi: 10.4230/DagMan.3.1.31. [DOI] [Google Scholar]
- Almeida JS, Grüneberg A, Maass W, Vinga S. Fractal MapReduce decomposition of sequence alignment. Algorithms for Molecular Biology. 2012b;7(1):12. doi: 10.1186/1748-7188-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida JS, Iriabho EE, Gorrepati VL, Wilkinson SR, Grüneberg A, Robbins DE, et al. ImageJS: personalized, participated, pervasive, and reproducible image bioinformatics in the web browser. Journal of pathology informatics. 2012a;3 doi: 10.4103/2153-3539.98813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G, Hey T, Szalay A. Beyond the data deluge. Science. 2009;323(5919):1297–1298. doi: 10.1126/science.1170411. [DOI] [PubMed] [Google Scholar]
- Berners-Lee T. Linked data-design issues (2006) 2011. URL http://www.w3.org/DesignIssues/LinkedData.html.
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JS, Viswanathan A, Shinavier J, Shi Y, Hendler JA. Open Government Data: A Data Analytics Approach. IEEE Intelligent Systems. 2013;28(5):0019–23. [Google Scholar]
- Heath T, Bizer C. Linked data: Evolving the web into a global data space. Synthesis lectures on the semantic web: theory and technology. 2011;1(1):1–136. [Google Scholar]
- Mandl KD, Kohane IS. Escaping the EHR trap—the future of health IT. New England Journal of Medicine. 2012;366(24):2240–2242. doi: 10.1056/NEJMp1203102. [DOI] [PubMed] [Google Scholar]
- Manyika J, Chui M, Brown B, Bughin J, Dobbs R, Roxburgh C, et al. Big data: The next frontier for innovation, competition, and productivity. 2011;5(33):222. URL http://www.mckinsey.com/Insights/MGI/Research/Technology_and_Innovation/Big_data_The_next_frontier_for_innovation. [Google Scholar]
- Robbins DE, Grüneberg A, Deus HF, Tanik MM, Almeida JS. A self-updating road map of The Cancer Genome Atlas. Bioinformatics. 2013;29(10):1333–40. doi: 10.1093/bioinformatics/btt141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth KA, Almeida JS. Computational Pathology as the New Big Data Microscope American Journal of Pathology. 2015;185(3) doi: 10.1016/j.ajpath.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Socrata Open Data API. 2014. Retrieved March 10, 2015, from http://www.socrata.com/products/open-data-api/
- The White House Introducing: Project Open Data | The White House. 2013. Retrieved March 10, 2015, from http://www.whitehouse.gov/blog/2013/05/16/introducing-project-open-data.
- US Department of Health and Human Services HHS open government plan vers. 3. 2015. http://www.hhs.gov/open/plan, http://www.hhs.gov/open/plan/open-gov-plan-v3.pdf.
- W3C Web Platform Your Web, documented · WebPlatform.org. 2013. Retrieved March 10, 2015, from https://www.webplatform.org/
- Wilkinson SR, Almeida JS. QMachine: commodity supercomputing in web browsers. BMC bioinformatics. 2014;15(1):176. doi: 10.1186/1471-2105-15-176. [DOI] [PMC free article] [PubMed] [Google Scholar]