Abstract
Recently, in response to the rising costs of healthcare services, employers that are financially responsible for the healthcare costs of their workforce have been investing in health improvement programs for their employees. A main objective of these so called “wellness programs” is to reduce the incidence of chronic illnesses such as cardiovascular disease, cancer, diabetes, and obesity, with the goal of reducing future medical costs. The majority of these wellness programs include an annual screening to detect individuals with the highest risk of developing chronic disease. Once these individuals are identified, the company can invest in interventions to reduce the risk of those individuals. However, capturing many biomarkers per employee creates a costly screening procedure. We propose a statistical data-driven method to address this challenge by minimizing the number of biomarkers in the screening procedure while maximizing the predictive power over a broad spectrum of diseases. Our solution uses multi-task learning and group dimensionality reduction from machine learning and statistics. We provide empirical validation of the proposed solution using data from two different electronic medical records systems, with comparisons to a statistical benchmark.
Introduction
Early identification of individuals who are at a higher risk of developing a chronic disease is of significant clinical value, as it creates opportunities for slowing down or even reversing the pace of the disease [1]. In particular, 80% of the cases of cardiovascular disease and diabetes, and 40% of the cancer cases can be treated successfully at an early stage [2], preventing the need for expensive medical procedures due to complications occurring during the standard care of chronically ill patients. This early stage intervention is also of significant cost value, as avoiding these expensive procedures and associated costs of care leads to a reduction in healthcare spending. In 2012, nearly 76% of Medicare spending was on patients, comprising just 17% of the Medicare population, with five or more chronic diseases: Alzheimer’s disease, cardiovascular disease, diabetes, renal disease, and lung disease [3].
Recently, one solution to this rising cost burden has been the introduction of incentive programs by an increasing number of self-insured companies, i.e., companies who pay for healthcare costs of their workforce. In 2013, 86% of all employers, which include Johnson & Johnson, Caterpillar, and Safeway, offered such incentivized programs. These programs involve, among many other aspects, incentivizing employees to undergo a health risk assessment (HRA) in order to identify employees with risk factors for major chronic diseases. The HRA typically involves collecting basic lab results like lipid panels, or other relevant biomarkers such as blood pressure, age, gender, height and weight. This data about the employee is then used to calculate risk factors based on simple clinical rules (see [4] for a list such rules). However, these simple clinical rules suffer from poor prediction accuracy since they rely on few biomarkers and do not account for risk variations in different populations, resulting in reduced efficacy of the program and undermining its objectives, suggesting the need for more effective methods for multiple disease screening and early identification. In particular, a false positive (mistakenly assigning high risk to a healthy patient) leads to unnecessary intervention costs while a false negative (mistakenly assigning low risk to a high risk patient) would be a lost opportunity to avert a large future healthcare bill to the employer, and it has been shown that higher predictive accuracy directly improves the cost-effectiveness of such programs [5].
Statistical data-driven methods present one principled way to obtain a set of biomarkers for such an assessment which provides high disease prediction accuracy. The use of such methods for disease prediction has grown rapidly over the past few years. An admittedly incomplete list of studies include: stroke prediction via Cox proportional hazard models [6], relational functional gradient boosting in myocardial infarction prediction [7], support vector machines and naïve Bayes classifiers for cancer prediction [8], neural networks for mortality prediction [9], time series in infant mortality [10], cardiac syndromes [11] or infectious disease prediction [12], and dimensionality reduction for unstructured clinical text [13]. However, these past works have focused on individual disease prediction, rather than determining a set of biomarkers which are jointly predictive of several diseases. While it is possible to combine the relevant biomarkers selected by each individual disease prediction model, this leads to a larger set of biomarkers than is necessary, and we will later show this is less cost effective than our proposed method.
In this paper, we present a method which relies on multi-task learning [14] model and group regularization [15]. The multitask learning model we consider allows for determining a small set of biomarkers which are optimal for prediction of multiple diseases, and is hence low cost. The model acts by learning the relevant biomarkers for multiple diseases simultaneously, while forcing these relevant biomarkers to be sparse [14, 16, 17], resulting in a lower cost model. Furthermore, it is known to be highly generalizeable and thus to adapt well to new data; we refer the reader to [18] for a detailed survey.
While these aforementioned works share methodology with our proposed method, to the best of our knowledge none have not yet addressed a setting where one desires to build a small, and thus low cost, universal set of features (biomarkers in our case) that can be used to predict multiple diseases. The work of [19] also considers multitask learning in the context of logistic regression and multiple disease prediction, however, their model does not enforce the disease prediction tasks to share the same set of biomarkers. Their model constructs a shared set of latent predictors as linear combinations of groups of features (diagnosis history) and a ℓ1 regularizer is used to ensure that each individual disease prediction relies on a small set of these predictors. Hence, their approach does not enforce a shared group of biomarkers to be used as predictors for all diseases, which makes their method less suitable for our purpose. Our model also uses the raw biomarkers as the predictors, which is more interpretable and enables our model to directly ensure that the set of biomarkers is small. Thus we present our proposed method as a solution to obtaining an accurate, low cost disease screening methodology. We evaluate our method on two patient populations, and compare the cost of our method to the current statistical benchmark, individual disease prediction. We find that our method has comparable, and in some cases improved, accuracy at much lower cost.
1 Methods
1.1 Patient Data
We considered two patient populations for developing our prediction tool, the Kaggle Practice Fusion dataset [20], which is publicly available, and the patient records from Stanford Hospital and Clinics, which will be referred to as the SHC dataset throughout1. The SHC dataset consisted of 73,842 patients, and 1,313 possible laboratory exams; while the Kaggle dataset consisted of 1,096 patients with 285 possible exams; a comparison summary is given in Table 2.
Table 2:
Comparison of SHC and Kaggle dataset
| SHC | Kaggle | |
|---|---|---|
| Number of patients | 75,619 | 1,096 |
| Number of available lab exams | 1,313 | 258 |
| Median number of patients taking a lab | 19 | 5 |
| Median number of labs taken per patient | 24 | 12 |
| Minimum patient age | 0 | 19 |
| Maximum patient age | 110 | 90 |
The patient data from the SHC dataset was taken as follows. For each disease j (j = 1, …,K, with K = 9 for the SHC dataset), we collected the biomarkers of patients during a one-month period, which occurred during the year 2010 to 2011. These biomarkers included quantities such as age, gender, and the results of laboratory exams such as hemoglobin A1C, cholesterol, as well as blood cell counts2. If there were multiple laboratory exam results during this month, they were averaged. If patients had results over multiple months, we took the first month for which results occurred. If the patient was diagnosed with disease j during this month, we removed the patient from the dataset, i.e., we only considered patients who had never had disease j before the end of the month in which their results were taken. We collected these biomarkers into a vector Xi, i = 1, …,n, where Xij took the value of the result of biomarker j for patient i, and where n was the total number of patients collected. We assigned each patient i a label for each disease j, which we defined to be +1 if the patient has a positive diagnosis of disease j within one to thirteen months, beginning the month after biomarkers were collected, and −1 otherwise. Lastly, we took the intersection over all diseases for which we collected patients. For the SHC dataset, we noticed that the set of patient-months for all diseases had a large intersection containing 75,619 patients, as shown in Table 2. Two of these diseases had an extremely low incidence in the Kaggle dataset, and were not studied when evaluating on the Kaggle dataset. These diseases are highlighted in Table 1 with an asterisk (*). The diseases we studied were obtained from [21, 22] and are shown in Table 1, with their formal names and abbreviations which will be used throughout this paper.
Table 1:
List of diseases under study.
| Disease | Abbrev. |
|---|---|
| Coronary artery disease | CAD |
| Malignant cancer of any type | CANCER |
| Congestive heart failure | CHF |
| Chronic obstructive pulmonary disorder | COPD |
| Diabetes* | DB |
| Dementia | DEM |
| Peripheral vascular disease | PVD |
| Renal failure* | RF |
| Severe chronic liver disease* | SCLD |
Diseases marked with were only studied on SHC data due to sparsity or missing labels in Kaggle.
In the Kaggle dataset, due to lack of access to the exact time stamp for the lab results or diagnosis codes, the feature vectors Xi were formed for each patient from the laboratory results and age information, without any use of temporal information. The label was determined by checking whether the patient had a positive diagnosis for disease j in the system. Due to the small size of the Kaggle dataset, the bootstrap method was used to estimate the average and standard error of the AUC values.
For both datasets, the patients did not have results for all of the possible laboratory exams. In particular, the matrix X has 98%, and 93% missing values in SHC and Kaggle data respectively. To deal with the missing data entries, we used the standard method of mean imputation. We gave each the average value of the laboratory exam, where the average was taken over all patients who had taken the exam. We also repeated our analysis on Kaggle data set using multiple imputation (MI) with 100 imputations and the results were consistent. In particular, the relative accuracy of all algorithms and the sparsity of the models did not change3. Finally, we standardized the observations so that the values of each biomarker have zero mean and variance one across the observations, which is a standard preprocessing step for many statistical methods.
1.2 Cost Data
Data for the cost analysis was taken from [23], maintained by Health One, Incorporated. Health One, Incorporated offers laboratory exams and has physical locations in all states in United States. Health One Lab’s website provides competitive prices of various laboratory exam packages, which are comprised of several laboratory exams. These prices were used to determine the cost of the set of laboratory exams which were selected by our methods to be the most important for disease prediction.
We emphasize that most laboratory exams are not sold individually but in packages. Thus, in order to find the cost of our screening procedure, we solved a set-covering problem, formulated as a mixed-integer optimization problem where we minimized the cost over the possible sets of packages but enforced the choice of packages to include all of the laboratory exams sought. This problem can also be formulated as a convex linear programming problem, but since the size of the problem did not lead to a computational impasse, the mixed integer program was suitable for our needs. More specifically, we solved the optimization problem given by
| (1) |
where is the set of all packages, is the set of laboratory exams for which we wish to obtain the total minimum cost, cS is the cost of a package, and qS is an indicator variable which indicates whether a particular package S ∈ is chosen for inclusion. We solved this optimization problem using the software of [24]. We then calculated the cost of the laboratory exams by adding the prices of the set of packages S ∈ for which qS = 1 at the solution of (1).
1.3 Statistical Methods
We denote the different disease prediction models in this section as follows. We will refer to our main model as the multitask learning model (MTL). For each value of a tuning parameter, λ, this model selects M biomarkers as relevant to the disease prediction. We derive a model from this, the ordinary logistic regression model (OLR-M), which takes as input data the results of the M biomarkers which have been selected by the multitask model at each value of the tuning parameter, λ. We compare both of these models, the multitask model and the OLR-M model, to the results of solving one individual disease prediction at a time, for each disease. We will henceforth refer to this model as single task learning (STL), since this type of model only involves a single disease prediction. First, we begin with logistic regression (LR), a preliminary for our models.
Logistic Regression (LR)
We briefly describe logistic regression, which we will denote by LR, before describing our methods. Given n p-dimensional data vectors, Xi, and labels yi ∈ {0,1}, i = 1, …,n, logistic regression finds the optimal values of a p-dimensional parameter vector θ and a scalar b such that the conditional probability of the observing yi given the data Xi for all data vectors i = 1,…,n, is maximized. This optimization problem is given by: = arg maxθ, b where P(yi|Xi;θ,b) denotes the conditional probability that one observed the label yi given the data observation Xi, which is parameterized by (θ,b). We will refer to this term as the likelihood. This probability is commonly given the form P(yi|Xi;θ,b) = 1/[1 + exp(−yi(θ⊤Xi + b))]. This form of P(yi|Xi;θ, b) is used in this paper.
Single Task Learning Model (STL)
The single task learning model formulates the goal of maximizing the log-likelihood while keeping the set of nonzero elements in the parameter vector θ to be small. This model consists of a logistic regression and a regularization term, commonly known as the LASSO penalty [25]. In the context of disease prediction, for each disease j = 1, …, K, where K denotes the total number of diseases considered, the model finds the set of parameters that maximizes the conditional probability that a patient will be diagnosed with disease given his or her vector of biomarkers. More specifically, for each disease j, we solve the problem
| (2) |
where as defined previously, n is the number of patients, is an indicator of whether patient i had disease j, and Xi is the vector of biomarkers for patient i, of length p. The vector θj and scalar bj are the learned parameters for the model for disease j, with θjm representing the mth element of θj. If θjm is nonzero, then the model has selected this element as relevant for prediction. The magnitude of the parameter θjm represents the importance of biomarker m to the prediction of disease j, and the larger this value the greater its importance for achieving higher prediction accuracy. The first term in (2) is the logistic regression, and the second term is the regularization term with λ as the regularization parameter. The value of λ controls the effect of enforcing, for a particular value of j, the number of values of m for which θjm are nonzero to be small. A larger value of λ encourages the effect more strongly. Note that a different model is learned for each value of λ.
Multitask Learning Model (MTL)
The multi-task learning model formulates the goal of maximizing the log-likelihood and enforcing that the set of biomarkers which is jointly predictive of all diseases of interest to be small. This model consists of a logistic regression and a regularization term known as the GLASSO [15], or group lasso, penalty. In contrast to the STL model, we jointly learn the parameters for all disease predictions, by solving the problem given by
| (3) |
where all variables are as defined previously. As with the single task learning model, the first term is a logistic regression, and the second term is the group regularization term, which encourages biomarker m to be either nonzero or zero across all diseases together, i.e. that for each m, θjm should be nonzero or zero for all j. Larger values of λ enforce this more strongly. Intuitively, such co-variation of weights through this group regularization can be interpreted as transfer of information between different disease predictions during the training of the model and can further lead to greater generalization ability of the model. When K = 1, the penalty term then reduces to the penalty term in the STL model, and the parameters are learned for prediction of only one disease; thus, this model reduces to the STL model. As with the STL model, a different model is learned for each value of λ.
OLR-M Model
The OLR-M model is derived from the MTL model. After solving for the optimal parameters for the MTL model with parameter λ, we retrained a truncated model, as follows. For each value of λ, we determined the nonzero biomarkers, i.e., the values of m for which the quantity was above a threshold. We will denote this number of nonzero biomarkers by M, and give further details on how the number of nonzero biomarkers was determined, in the section Learning the Model Parameters. We then retrained the model over only these M biomarkers for each disease prediction, j = 1, …,K, formulated as an ordinary logistic regression.
Learning the Model Parameters
The optimization package MinFunc [26], which utilizes spectral projected gradient, was used for solving the associated optimization problems of the considered approaches, given for the STL, MTL, and OLG-M model described below. For each value of λ, all models were trained and tested using five-fold cross validation. During training, due to the lack of positive examples, in each cross-validation fold (or bootstrap sample, in the case of the Kaggle dataset) the training data was sampled so that, for the SHC dataset, the ratio of positive to negative examples in the training set was 1:1, and for the Kaggle dataset it was 1:3. The main performance metric for all prediction tasks is area under ROC curve (AUC). All demonstrated AUC values in the figures are cross-validated, thus averaged, AUCs.
For each value of the regularization parameter λ, we considered biomarker i to have been selected by the model if for any value of j, it had a nonzero value of θij. We determined this as follows. For the MTL model, for a particular value of λ, the number of nonzero features was determined by counting the number of biomarkers m for which the quantity was greater than a threshold of τ = 10−6 for at least one value of j = 1, …,K. For the STL model, we considered a biomarker as nonzero if for some value of was greater than τ. Thus each value of λ corresponds to a set of nonzero biomarkers, and a cost can be derived via solving the optimization problem (1), as given in the section Cost Data.
2 Results
2.1 Biomarker Selection for Multiple Disease Prediction
We compare the results of the MTL, OLR-M, and STL models for disease prediction, number of biomarkers necessary to achieve the given results for prediction, and cost. As our evaluation metric, we will use area under the receiver-operating characteristic (ROC) curve, henceforth abbreviated as AUC. The AUC values shown in the succeeding figures are averaged over the five cross-validation folds, as well as averaged over the nine different diseases of interest.
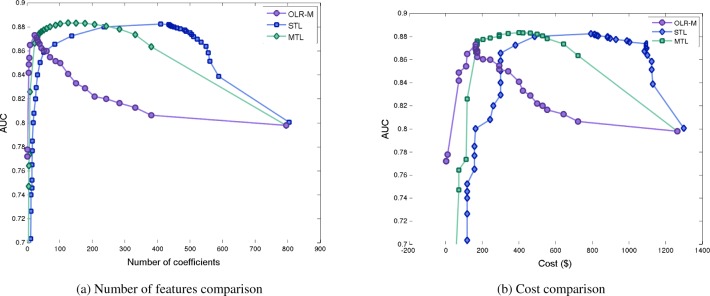
The MTL and OLR-M models achieve comparable accuracy to the STL model with a smaller number of biomarkers. Figure 1a presents this result visually, in which the given curves show the effect on the value of the AUC is when the number of biomarkers selected by the model is varied, for each of the different modeling approaches. This results also translates when considering the cost of the biomarkers needed to achieve each AUC value on the curve. Figure 1b presents the same result as Figure 1a, but calculates the cost of the biomarkers at each point on the curve in Figure 1b. Thus the effect is amplified when comparing cost of the MTL and OLR-M models to the STL model, as the MTL and OLR-M models are much lower in cost. The OLR-M was the most succinct of all the considered approaches, and in particular, the value M = 30 was found to be the optimal value for M using cross validation to have the highest AUC, as presented in Figures 1a and 1b.
Figure 1:
Comparison of number of biomarkers selected and associated cost by the MTL, STL, and OLR-M models. Each point on each curve is obtained by changing the regularization parameter.
2.2 Sensitivity Analysis
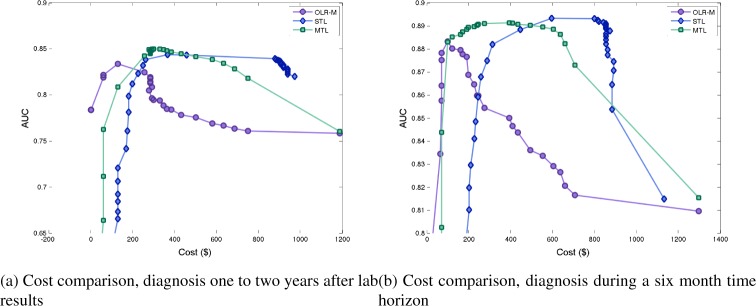
We study the effect of varying the length of, as well as a shift in, the time interval over which we identify a patient as having a positive diagnosis. In the previously presented results, the time horizon considered for diagnosis was one to thirteen months after biomarkers were taken. We consider a shift of a year, i.e. a time horizon of twelve to twenty-four months after the biomarkers were taken. We also study the performance when the time horizon considered is varied, one to six months, and one to eighteen months, after the biomarkers were taken. Due to space limitations, in Figure 2 we only demonstrate the results for twelve to twenty-four months interval and one to six months interval. In all cases, we observe the same pattern as in previous results.
Figure 2:
Comparison of costs and accuracy of models, for a shift and varied lengths of time horizon.
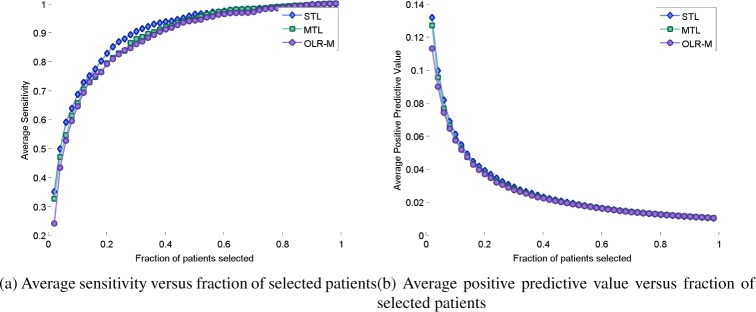
We also compare predictive accuracy of the three models using other metrics than AUC. In particular, Figure 3 shows that the three models have comparable accuracy with respect to sensitivity (or true positive rate). However, with respect to positive predictive value, the MTL and OLR-M models are indistinguishable and closely follow the STL model.
Figure 3:
Comparison of predictive accuracy of models based on sensitivity and positive predictive value.
2.3 Biomarkers Selected for OLR-M Model
As aforementioned, the value of M which achieved the highest AUC, when cross-validated, was M = 30. We have given a list of the top M = 30 labs selected by the OLR-30 model in Table 3. All of these labs are associated with the diseases of interest in this paper, and as expected from the design of our method, many of them are associated with several diseases. For example, abnormal glucose levels are associated with signs of diabetes, cancer, renal disease, liver disease, and heart disease. Similarly, abnormal alkaline phosphatase levels can be associated with liver disease, cancer, as well as heart disease. A vitamin B12 test can be given to those experiencing symptoms of anemia, as well as dementia and memory problems, but higher levels can also be associated with diabetes, liver disease, and heart problems. Additionally, the considered model also has the ability to discover associations between these biomarkers and the diseases which may not be explicitly known; hence, the associations go beyond known ones, some of which have just been mentioned.
Table 3:
List of the top features with their rank determined by OLR-30. Smaller rank means the feature group received a larger value of in multi-task learning.
| Rank | Lab Name |
|---|---|
| 1 | Creatinine |
| 2 | Age, Gender |
| 3 | Hemoglobin A1C |
| 4 | Platelet Count |
| 5 | Glucose |
| 6 | Alkaline Phosphatase |
| 7 | QRSD Interval |
| 8 | Prothrombin Time |
| 9 | Urea Nitrogen |
| 10 | EGFR |
| 11 | Vitamin B12 |
| 12 | HDL Cholesterol |
| 13 | T-Wave Axis |
| 14 | Triglyceride |
| 15 | Globulin |
| 16 | RDW |
| 17 | Alpha-1 Antitrypsin |
| 18 | INR |
| 20 | Bilirubin |
2.4 Generalizability of OLR-M to Different Patient Populations
We investigate the performance of the OLR-M model on a different patient population, the Kaggle dataset. As aforementioned, the Kaggle dataset is a less ideal dataset, with fewer biomarker values available per patient. As M = 30 achieved the best predictive accuracy and lowest cost when validating on the SHC dataset, we will use this model, and refer to it as OLR-30. The OLR-M model is able to achieve comparable accuracy to the STL model, with far fewer biomarkers, and thus has similar performance on this dataset as on the SHC dataset. The OLR-30 model consists of 30 biomarkers, as aforementioned, much fewer than the STL model, which requires the results of 63 biomarkers, in addition to age and gender information. Table 4 provides the confidence intervals at the 5% level for the AUC values for both models, for each disease under study. The results were obtained using 5-fold cross validation and the method of DeLong [27] for the confidence intervals.
Table 4:
Confidence intervals for the AUC values of OLR-30 and STL approaches
| Disease | OLR-30 | STL |
|---|---|---|
| CAD | 0.7535 ± 0.0468 | 0.7412 ± 0.0483 |
| CHF | 0.6003 ± 0.0489 | 0.5961 ± 0.0476 |
| COPD | 0.6492 ± 0.1067 | 0.6604 ± 0.1043 |
| DEM | 0.6359 ± 0.1192 | 0.6447 ± 0.1131 |
| PVD | 0.9184 ± 0.0716 | 0.9183 ± 0.0676 |
| RF | 0.7957 ± 0.0684 | 0.7925 ± 0.0656 |
Remark
The Kaggle dataset, as aforementioned, is a lower quality dataset than SHC, which required us to process the data differently than the SHC dataset as discussed in the section Patient Data. However, we observe that the results follow the same pattern as the results for the SHC dataset, as presented in Table 4.
3 Discussion
We have proposed two approaches, the MTL and OLR-M models, for developing a clinical model for multiple disease prediction. Our study has focused, in particular, on how to design a health-risk assessment (HRA) for prediction of multiple diseases, using statistical methods and patient data. In contrast to the STL model, which is one appealing and commonly used statistical approach in the literature, we propose a solution which relies on group regularization methods to jointly learn a small set of biomarkers are the most relevant for prediction, for multiple diseases.
Although our results are encouraging, we note that our study has limitations. Although we would like to also compare the prediction accuracy of our method against common scoring rules, such as the Framingham score, used for individual diseases, we are unable to do so at this time because of the unavailability of necessary biomarkers in our data for these scores, such as blood pressure, whether the patient is a smoker, or whether they are on dialysis. Hence, we limit our comparison to the state-of-the-art statistical method for disease prediction, STL. The lack of temporal information regarding disease diagnosis in the Kaggle dataset is surely a limitation. Validation on this dataset has allowed us to compare the OLR-M and STL methods, however, we cautiously use these results to guide our conclusion that the OLR-M model is a viable option for a low cost multiple disease prediction model.
The results of our investigation show that the MTL and OLR-M models provide one solution to designing a lower cost HRA. We have presented experiments to validate that our models achieve comparable accuracy to the STL model, but achieve this accuracy with the use of fewer biomarkers, which directly translates to cost. Our MTL model uses group regularization to select a small set of biomarkers, which can be used to jointly predict all of the diseases at once. By learning which biomarkers are jointly successful for prediction, the MTL model is able to achieve comparable accuracy to the STL model, but with fewer biomarkers. The OLR-M model enhances these benefits by producing a model with fewer biomarkers than either of the other two considered approaches, and keeping a high accuracy by refining over the MTL model. Since the MTL model is a biased model, the OLR-M acts to remove the bias from this model, increasing accuracy of the MTL model at the points for which it has the highest AUCs.
The same pattern of results is observed when we consider different time horizons for considering a positive diagnosis of a patient, i.e, that the MTL and OLR-M models produce lower cost models with comparable accuracy. We have studied the performance of the approaches under a one to six month time horizon, as well as twelve to twenty-four months, when there is a lag of a year between when the patients’ biomarkers were taken and when the positive diagnosis occurred. We observed similar accuracy between the models as well when we studied the performance under other metrics, other than AUC.
Lastly, we have validated the OLR-M model on a new patient population, that of the Kaggle dataset. This dataset is a significantly smaller dataset, with fewer patients, fewer biomarkers per patient. However, we observe the same result to hold for this new dataset, as the OLR-M model is still as accurate as the STL model but much less costly. Thus we believe the the performance of the OLR-30 model is not restricted to the SHC dataset from which it was derived, but can be extended to new populations.
Footnotes
Due to HIPAA privacy restrictions, this dataset cannot be made publicly available.
However, we did not have access to biomarkers such as blood pressure, height, and weight.
We only restricted MI to Kaggle data set due to its high memory and computation requirement on SHC data. For mean imputation we exploited the fact that most entries of each column are equal and therefore X is sparse. MI leads to a large non-sparse matrix that significantly increases memory and speed requirements. Results have been omitted due to space constraints.
References
- [1].Bates DW, Saria S, Ohno-Machado L, Shah A, Escobar G. Big Data In Health Care: Using Analytics To Identify And Manage High-Risk And High-Cost Patients. Health Aff. 2014;33:1123–1131. doi: 10.1377/hlthaff.2014.0041. [DOI] [PubMed] [Google Scholar]
- [2].WHO. Chronic diseases and health promotion World Health Organization. 2014. report available at http://www.who.int/chp/chronic_disease_report/part1/en/index11.html.
- [3].PFCD Almanac of Chronic Disease. 2012. Available from: http://almanac.fightchronicdisease.org/Home.
- [4].MDCalc MDCalc Online. 2014. Available from: http://www.mdcalc.com/
- [5].Bayati M, Braverman M, Gillam M, Mack K, Ruiz G, Smith M, et al. Data-Driven Decisions for Reducing Readmissions for Heart Failure: General Methodology and Case Study. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0109264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Khosla A, Chiu HK, Lin CC, Chiu HK, Hu J, Lee H. An Integrated Machine Learning Approach to Stroke Prediction. KDD. 2010 [Google Scholar]
- [7].Weiss J, Natarajan S, Peissig P, McCarty C, Page D. Machine Learning for Personalized Medicine: Predicting Primary Myocardial Infarction from Electronic Health Records. AI Magazine. 2012 [PMC free article] [PubMed] [Google Scholar]
- [8].Cruz J, Wishard D. Applications of Machine Learning in Cancer Prediction and Prognosis. Cancer Informatics. 2006;2:59–77. [PMC free article] [PubMed] [Google Scholar]
- [9].Song X, Mitnitski A, Cox J, Rockwood K. Comparison of machine learning techniques with classical statistical models in predicting health outcomes. Stud Health Technol Inform. 2004;107:736–40. [PubMed] [Google Scholar]
- [10].Saria S, Rajani AK, Gould J, Koller D, Penn AA. Integration of early physiological responses predicts later illness severity in preterm infants. Science Translational Medicine. 2010 Sep;2(48) doi: 10.1126/scitranslmed.3001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Syed Z, Leeds D, Curtis D, Nesta F, Levine RA, Guttag JV. A Framework for the Analysis of Acoustical Cardiac Signals. IEEE Trans Biomed Engineering. 2007;54(4):651–662. doi: 10.1109/TBME.2006.889189. [DOI] [PubMed] [Google Scholar]
- [12].Wiens J, Guttag J, Horvitz E. Patient Risk Stratification for Hospital-Associated C diff as a Time-Series Classification Task. NIPS. 2012 [Google Scholar]
- [13].Halpern Y, Horng S, Nathanson LA, Shapiro NI, Sontag D. A Comparison of Dimensionality Reduction Techniques for Unstructured Clinical Text. ICML 2012 Workshop on Clinical Data Analysis. 2012 [Google Scholar]
- [14].Caruana R. Multitask learning. Machine learning. 1997;28(1):41–75. [Google Scholar]
- [15].L Meier PB, van de Geer S. The group lasso for logistic regression. J R Statist Soc B. 2008;70:53–71. [Google Scholar]
- [16].Ando RK, Zhang T. A framework for learning predictive structures from multiple tasks and unlabeled data. The Journal of Machine Learning Research. 2005;6:1817–1853. [Google Scholar]
- [17].Evgeniou A, Pontil M. Advances in neural information processing systems: Proceedings of the 2006 conference. Vol. 19. The MIT Press; 2007. Multi-task feature learning; p. 41. [Google Scholar]
- [18].Pan SJ, Yang Q. A Survey on Transfer Learning. IEEE Transactions on Knowledge and Data Engineering. 2010;22(10):1345–1359. [Google Scholar]
- [19].Wang X, Wang F, Hu J, Sorrentino R. AMIA Annual Symposium Proceedings. Vol. 2014. American Medical Informatics Association; 2014. Exploring Joint Disease Risk Prediction; p. 1180. [PMC free article] [PubMed] [Google Scholar]
- [20].Practice Fusion Diabetes Classification Kaggle competition dataset. 2012. Available from: http://www.kaggle.com/c/pf2012-diabetes.
- [21].Iezzoni L, Heeren T, Foley S, Daley J, Hughes J, Coffman G. Chronic conditions and risk of in-hospital death. IEEE Trans Biomed Engineering. 1994;29(4):435–460. [PMC free article] [PubMed] [Google Scholar]
- [22].Atlass D. The Dartmouth Atlas of Health Care. 2011. Available online at http://www.dartmouthatlas.org/downloads/methods/Chronic_Disease_Codes.pdf.
- [23].One H. Tests Offered at Health One. 2014. Available from: http://www.healthonelabs.com/
- [24].CVX Research I CVX: Matlab Software for Disciplined Convex Programming, version 2.0. 2012. http://cvxr.com/cvx.
- [25].Tibshirani R. Regression shrinkage and selection with the Lasso. J Royal Statist Soc B. 1996;58:267–288. [Google Scholar]
- [26].Schmidt M.Function for unconstrained optimization of differentiable real-valued multivariate functions; 2012MinFunc Package Available from: http://www.di.ens.fr/~mschmidt/Software/code.html
- [27].DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988:837–845. [PubMed] [Google Scholar]