Abstract
Pharmacogenomics (PGx) guidelines contain drug-gene relationships, therapeutic and clinical recommendations from which clinical decision support (CDS) rules can be extracted, rendered and then delivered through clinical decision support systems (CDSS) to provide clinicians with just-in-time information at the point of care. Several tools exist that can be used to generate CDS rules that are based on computer interpretable guidelines (CIG), but none have been previously applied to the PGx domain. We utilized the Unified Modeling Language (UML), the Health Level 7 virtual medical record (HL7 vMR) model, and standard terminologies to represent the semantics and decision logic derived from a PGx guideline, which were then mapped to the Health eDecisions (HeD) schema. The modeling and extraction processes developed here demonstrate how structured knowledge representations can be used to support the creation of shareable CDS rules from PGx guidelines.
Introduction
In this work, we propose a consistent and reproducible methodology to generate platform-independent, shareable representations of PGx guidelines using existing and emerging standards. We modeled a published PGx guideline using UML activity diagrams, the HL7 vMR data model, and standard terminologies. The aim is to use these standards to build a structured, human-readable document and, ultimately, a shareable, machine-readable artifact. The motivation for developing a standards-based knowledge representation process that yields sharable policies is twofold. First, no commonly accepted, standard methodology currently exists for the extraction of CDS rules from PGx guidelines. Second, the creation of shareable CDS rules from PGx guidelines will provide clinicians and implementers with standards-based decision logic that can be used as a starting point for the design of evidence-based care delivery interventions.
Adverse drug reactions (ADR) cause more deaths than AIDS and diabetes, making it the fourth major cause of death for over a decade1. Approximately 100,000 deaths and more than 2 million acute ADRs occur annually1. Some of these ADRs are caused by variations in genes associated with drug metabolism or mechanism of action. Many of these gene-drug interactions are clinically actionable. Groups such as the Clinical Pharmacogenetics Implementation Consortium (CPIC) develop clinical guidelines that provide clinicians with information about known gene-drug interactions. These guidelines also provide clinicians with recommendations that help guide drug therapy2,3,4.
Like most clinical guidelines, pharmacogenomics (PGx) guidelines are written for clinicians and are not available in computable formats. While CPIC is beginning to include some structured text within their published guidelines, formal representations of the knowledge are not available, which hinders the extraction and implementation of CDS rules5. In addition, there are several other factors that complicate the implementation of PGx CDS. One of the main issues is the burgeoning amount of biomedical knowledge; there are currently 166 known gene drug interactions on the FDA website and 21 PGx guidelines have been published, and these numbers are growing rapidly6,7,8. Another factor is the very costly and time consuming process that is required to extract PGx knowledge from narrative guidelines, implement them as CDS rules in electronic medical record (EMR) systems, and then maintain those rules as biomedical knowledge evolves. Also, there are differences in how a clinical practice guideline can be implemented as a CDS rule, due to differences in the interpretation of the guideline and local workflows. It may be possible to reduce the cost of rule development and improve the consistency of CDS implementations by sharing CIGs, but currently there are no best practices or knowledge bases available to support the authoring of CIGs9. Therefore, a scalable approach is needed to extract, render and implement CDS rules from PGx guidelines. To accomplish this, common semantics and a platform-independent (vendor-agnostic) syntax is required.
Standardized data models and terminologies reduce variability in the representation and interpretation of clinical data by providing commons semantic meanings for concepts and terms. For example, the HL7 virtual medical record (vMR) data model was developed to represent clinical data from an EMR in a platform-neutral manner, which can be used for CDS10. Similarly, standard terminologies, such as SNOMED CT, RxNorm and LOINC, provide common definitions for clinical terms, drugs, and lab tests, respectively11,12,13. While the use of common data models and standard terminologies increases comprehension and reduces semantic ambiguity in shared data, they are not intended to represent the workflow and decision logic inherent in CDS rules. To represent these aspects, different standards are needed.
Several modeling frameworks, such as the Guideline Elements Model (GEM) and the Guideline Interchange Format (GLIF), have been developed to represent the content of clinical guidelines14,15,16. However, no existing framework has achieved broad adoption and the variety of approaches, formats and limitations of existing modeling tools prevents artifacts generated using these frameworks from being shareable in practice17,18. Despite decades of work in this area, there remains a dearth of applications that can support the authoring of platform-independent, shareable CIGs for use in CDS systems19. To address this challenge, in 2012 the Office of the National Coordinator (ONC) for Health Information Technology supported the creation of a harmonized format, known as the Health eDecisions (HeD) interchange format. The HeD standard defines a common metamodel for metadata, actions, events, and conditions as well as an expression language. HeD also recommends an XML-based serialization format and provides a schema20 for the validation of compliant documents. Together, the HeD model and its schema were designed to represent the workflow and decision logic of interventions such as CDS rules, order sets and documentation templates20.
To complement the HeD schema, in the context of the SHARPc-2B project, a multi-institution team developed a more formal representation of the HeD metamodel, expressed in the form of an OWL ontology, and a standards- and model-driven application for the authoring CDS clinical knowledge artifacts20. The HeD editor has several features that can potentially make it an ideal part of the process for modeling and testing the clinical decision logic for PGx guidelines. Specifically, it is compatible with several existing and pre-existing standards, has an intuitive user interface that can be used by knowledge engineers and non-technical staff, and uses semantic web technologies20.
In this work, we modeled a published PGx guideline using UML activity diagrams, the HL7 vMR data model, and standard terminologies. We then utilized the HeD editor to render the modeled guideline in HeD syntax. This work builds on our previous efforts to create platform-independent, standards-based, shareable representations of PGx guidelines and is another step towards the development of a generalizable approach that can be broadly applied to other PGx guidelines.
Methods
Published CPIC guidelines were reviewed and evaluated in terms of both the complexity of logic and the availability of reference implementations of corresponding CDS rules. For this project, we sought a PGx guideline that contained straightforward decision logic that was based on unambiguous PGx data (genotype and phenotype). We also prioritized guidelines that had been implemented as local, non-shareable CDS rules, which would provide additional clinical context over that which was included in the published guideline.
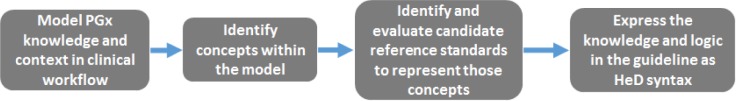
A stepwise process was developed to transform a human-readable PGx guideline into a computable HeD artifact (Figure 1). The process began by carefully analyzing the selected PGx guideline and rigorously defining all of the concepts, both explicit and implicit, that were relevant to the implementation of a PGx CDS rule. This included concepts that pertain to the clinical context of the guideline, the relevant EMR data, and terminology. The process was informed by knowledge of actual CDS implementations of the selected guideline.
Figure 1.
The workflow for modeling a PGx clinical guideline and expressing the information in HeD syntax.
Once relevant concepts were identified within the PGx guideline, candidate reference standards were reviewed and evaluated to determine which might best represent those concepts. Specifically, we evaluated the HL7 vMR, HL7 RIM, and LS DAM, to represent the clinical data for this PGx guideline21,22. We also evaluated the RxNorm, NDF-RT, SNOMED CT, and LOINC standard terminologies. The standards that were selected were used in the next step, when the PGx guideline itself was formally modeled and rendered in HeD syntax.
The parts of the PGx guideline that were targeted for modeling were the decision tree (which contained the recommended clinical workflow), the therapeutic recommendations, and the table for the genotype-informed decision logic, as the content in these sections represented the information that would comprise CDS rules. The information from these sections was augmented with knowledge of the corresponding CDS rules at Mayo Clinic, which provided additional clinical context about real-world PGx interventions. Collectively, these data were used to create a high-level model, which was expressed using UML class and activity diagrams to describe the logic contained within the PGx guideline. To reduce its complexity, the high-level model was split into logical modules that each contained a small number of decision points.
Each of the modules were analyzed in detail to identify the data elements, terminology concepts, and functions (e.g., data retrieval from the EMR, result processing) that would be necessary for implementation as a CDS rule. This information was then used to determine which entities (e.g., classes, attributes, and terms) from the selected reference standards would be needed to represent the entities within each module. Elements that represented the entities from the reference standards were added to the UML model and associated to their respective classes from the PGx guideline. Lastly, activity diagrams were generated to refine the computational steps within each workflow. Together, the UML models captured the entities, workflow, and behavior contained within the PGx guideline, but an additional method was needed to represent the logic of the CDS rules.
The logic components of the PGx guideline were expressed in pseudocode, which described the flow of data between each step and represented the functions that would be needed to implement each CDS rule. The pseudocode was based on an early version of CQL, and was used to define variables and create if-then conditional logic blocks that evaluated clinical data represented by vMR classes and attributes.
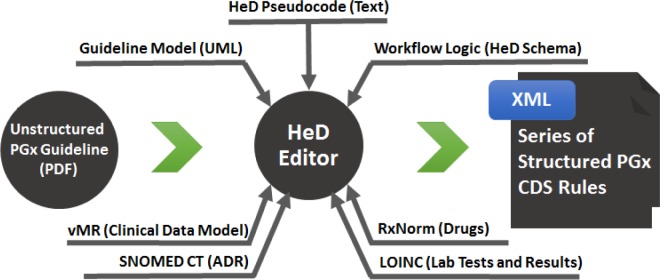
Finally, the model and pseudocode were used to render the guideline in HeD syntax, referencing the data and terminology standards (Figure 2). The HeD editor was used to ensure the resulting artifacts complied with the HeD schema.
Figure 2.
The HeD editor was used to generate structured rules from an unstructured PGx guideline, which was modeled in UML. Data and terminology standards provided common semantic meaning, and the logic of the workflow was expressed using the HeD schema.
Results
The CPIC guideline for HLA-B genotype and abacavir dosing was chosen because of its straightforward decision logic4. In particular, the HLA-B genotype is expressed as a boolean result (HLA-B*57:01 allele present/absent) that determines whether or not abacavir is contraindicated. The core recommendations from the guideline are summarized in Table 1. This guideline has been implemented as a CDS rule at Mayo Clinic23.
Table 1.
Core recommendations from the HLA-B/abacavir guideline.
| HLA-B*57:01 Genotype | Clinical Recommendation |
|---|---|
| HLA-B*57:01 alleleabsent | Abacaviris not contra indicated |
| HLA-B*57:01 allele present | Abacaviris contraindicated |
| HLA-B*57:01 status unknown | Order genotype test |
The HL7 vMR model, which was developed in part to support CDS use cases, was selected to represent clinical data elements. The vMR includes classes that represent the concepts of patient, drug order, ADR, test order and result, and communication events (e.g., CDS alerts). The RxNorm, SNOMED CT, and LOINC standard terminologies were chosen to provide coded concepts for drugs (e.g., abacavir), ADRs (e.g., adverse reaction to abacavir), and lab tests (e.g., abacavir genotype), respectively. While some of the coded concepts required for this project existed within the selected terminologies, several concepts were missing. Specifically, SNOMED CT did not contain a pre-coordinated term for an abacavir-related ADR but it would be possible to express this concept through post-coordination using “Antiviral drug adverse reaction” (292826004), “Causative agent” (246075003), and “Abacavir” (387005008). In addition, LOINC did not contain codes for the HLA-B*57:01 genotype test or results. Placeholder concept codes were used for the genotype test and results when rendering the rule in HeD syntax.
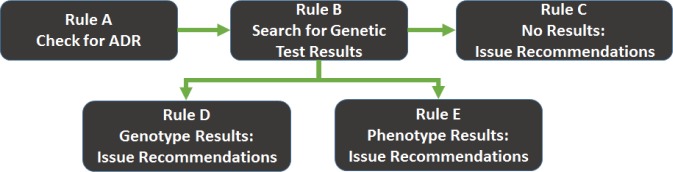
The HLA-B/abacavir guideline was modeled as five inter-related logical modules that represent the clinical workflow, therapeutic recommendations, and decision logic that would be necessary for implementation of the guideline as a CDS rule (Rules A–E, see Figure 3). These modules contain models that describe the entities, attributes, concepts, workflow relationships, and processing functions to search the patient’s record for a documented ADR (Rule A) and genetic test report (Rule B). CDS interventions are modeled in Rules C, D, and E.
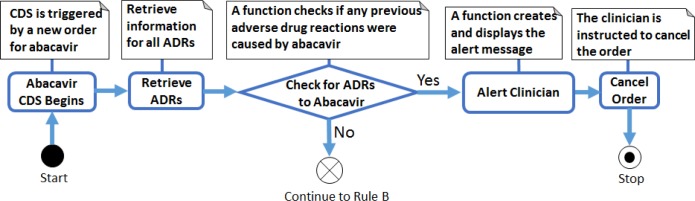
Figure 3.
Overview of the five inter-related rule modules that were generated for the HLA-B/abacavir guideline
When the CDS system detects an order for abacavir Rule A is triggered, which queries the EMR to see whether the patient has a documented adverse reaction to abacavir. If an adverse reaction is found then a CDS alert fires, instructing the clinician to cancel the drug order. If no adverse reaction exists then Rule B is triggered, which queries the patient’s record for HLA-B*57:01 genetic test results. If no results are found then Rule C is triggered, which recommends the clinician order the genetic test (if it has not already been ordered) or delay the drug order until the test results are available (if the test has been ordered and results are pending). If test results are found, Rule D is triggered, which examines the report for coded genotype results and either fires an alert to the clinician (HLA-B*57:01 allele present, abacavir is contraindicated) or allows the order to proceed (HLA-B*57:01 allele not present). If Rule D is triggered but coded genotype results are not available then Rule E is triggered, which examines the report for coded phenotype results (e.g., high/low risk of abacavir hypersensitivity). If coded phenotype results are available, the system acts accordingly (see Rule D); otherwise, the test results have not been codified and the CDS system will advise the clinician to manually review the genetic test results report.
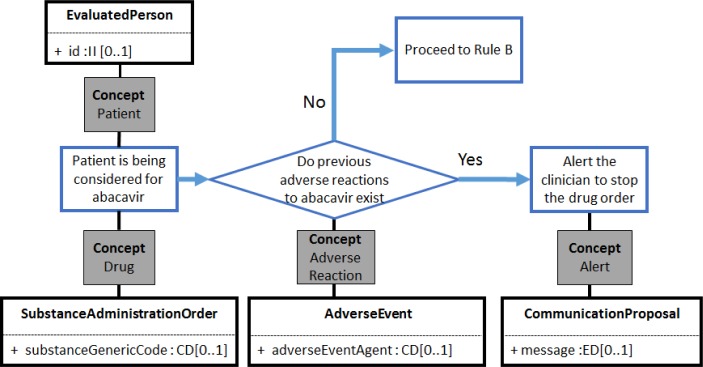
The entities in each rule were mapped to the corresponding entities from selected data and terminology standards. This step resulted in the identification of the vMR classes and attributes that would be needed for each rule. For example, the classes identified for Rule A included EvaluatedPerson, SubstanceAdministrationOrder, AdverseEvent, and CommunicationProposal (Figure 4).
Figure 4.
Clinical workflow for Rule A, with concepts identified and mapped to HL7 vMR classes. Not all attributes are shown.
Some of the attributes have a datatype of CD (concept descriptor), which reference coded concepts from terminologies that can be defined by the implementer. The standard terminologies selected for this project were used, when possible, as sources for terms that were considered to be critical for the rendering of the rule in HeD syntax. For example, “abacavir” can be represented by RxNorm RxCUI 190521, which can be used in Rule A for both SubstanceAdministrationOrder.substanceGenericCode and AdverseEvent.adverseEventAgent (Figure 4).
Each of the five workflows were refined into UML activity diagrams, which more carefully defined the actions required at each step of the workflow (Figure 5). The activity diagrams represented the final stage of UML modeling and, with the workflows described above, informed the generation of pseudocode.
Figure 5.
An activity diagram for Rule A, showing the activities that need to be performed by the CDS system. These process steps were later expressed in HeD syntax.
The penultimate step in this process involved creating pseudocode to outline the underlying processes involved in the retrieval and analysis of clinically relevant information from the EMR (see Figure 6 for an example). The pseudocode facilitated the transformation of the information specified in UML into a more pragmatically defined structured language while still retaining some aspects of human readability.
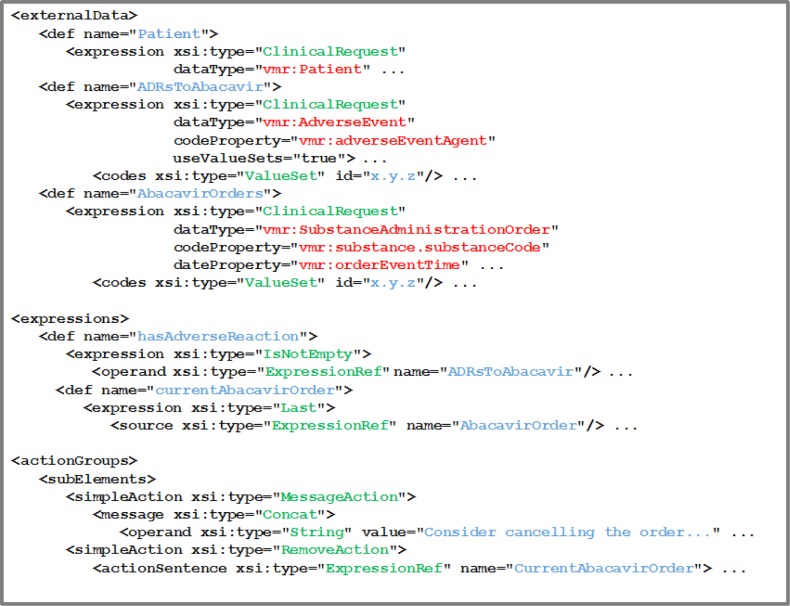
Figure 6.
An example of pseudocode corresponding to a portion of Rule A. References to the vMR are shown in red, while defined variables and expressions are shown in blue.
An initial HeD rendering of Rule A was created using the HeD editor and the knowledge artifacts previously generated (e.g., UML models, pseudocode). This rendering included references to classes and attributes from the vMR, concept codes (or placeholders) from standard terminologies, and the workflow logic expressed using the HeD schema (Figure 7).
Figure 7.
An example of the HeD syntax corresponding to a portion of Rule A. References to the vMR are shown in red; defined variables, expressions, and values are shown in blue; elements from the HeD model are shown in green.
Discussion
In this project we sought to develop a standards-based methodology to generate platform-independent, shareable representations of PGx rules. We modeled the published HLA-B/abacavir PGx guideline using UML class and activity diagrams, the HL7 vMR data model, and standard terminologies. These models were used to create pseudocode representations of the rules, which were ultimately rendered in HeD syntax. Thus, we began with an unstructured, human-readable clinical guideline and produced a shareable, machine-readable artifact.
This is the first time that PGx clinical guidelines have been formally modeled in UML with the specific aim of rendering the extracted rules in a platform-independent language, such as HeD. The use of UML to model the rules contained in the guideline facilitated the identification of data elements and coded concepts, and the subsequent mapping of those entities to standards. The UML modeling process also helped to provide a clear representation of key decision points in the rule and clinical workflow, and made explicit the computational tasks that need to be performed during the execution of the rules, such as data retrieval and processing. We anticipate this modeling approach will scalable and generalizable to other PGx guidelines. A review of other CPIC guidelines revealed variations in genetic test interpretations (e.g., phenotype) and the complexity of therapeutic recommendations, so it will be important to demonstrate this method can be used for a variety of PGx guidelines.
The HL7 vMR model, which was developed in part to support CDS use cases, was selected to represent clinical data elements. The vMR model was chosen, in part, because it provided the best coverage overall of the clinical concepts that were expressed in the PGx guideline and rules. We did not require the full expressive power and complexity of the HL7 RIM, of which vMR is a derivative, and other competing standards like FHIR were not sufficiently mature at the time this work was performed. We also did not consider logical or more detailed clinical models, such as the ones delivered as Clinical Element Models (CEMs) or OpenEHR archetypes as we decided that while we would likely find models for the more traditional concepts used in the rule, those sources did not provide PGx-specific models. For example, genetic test results were represented in the vMR as Observations, which is used for generic lab tests, but a refined domain model may be required to more fully capture the nuanced semantics of genomics data, including the concepts of allele, haplotype, copy number, and predicted phenotype. The HL7 Clinical Genomics Working Group is currently working in this area and we will evaluate the outcome of those projects when the work is complete.
The standard terminologies that were chosen for this project had variable coverage of the concepts contained in the PGx guideline. Both RxNorm and SNOMED CT contained a concept for “abacavir”; we chose to use the term from RxNorm as it is more likely to be supported by pharmacy systems. SNOMED CT did not contain a single term to represent a documented ADR for abacavir, but as noted above it would be possible to express this through post-coordination. Since the vMR model provides an attribute to specify the cause of an adverse event, though, this attribute was used with the code for “abacavir” rather than the more complex approach of using post-coordinated terms, since the class and the attribute already convey part of the semantics.
We were not able to locate an entry in LOINC that represented the HLA-B*57:01 genetic test or the results from that test; this was not a surprise, as standardized terminology is a known gap in the PGx domain. CPIC is leading a terminology harmonization effort that will provide more consistency among its guidelines and it is likely that the terms and value sets resulting from that project will be proposed for inclusion in LOINC. As those terms were not available at the time of this study, we used placeholder codes for these concepts in the HeD rendering of the rules.
The data and terminology standards provide common semantics that enable the sharing of rules derived from PGx guidelines. The actual implementation of CDS rules would require a site-specific mapping from the standard-based representation to local data models and terminologies, which reflects differences in where in the EMR data is stored and how it is coded. Similarly, while HeD is designed to represent the workflow and decision logic of interventions in a platform-independent manner, currently there is no consolidated way to execute HeD syntax or automatically transform it to an EMR-specific language beyond initial pilot implementations. These limitations represent significant opportunities for further research and tooling development.
Conclusion
In this work we demonstrated a method for modeling the HLA-B/abacavir PGx guideline in UML, which can represent the clinical workflow and decision logic as standards-based structured knowledge, and we illustrated how these models can be used to aid rendering the rules in the HeD schema. We anticipate this process will be generalizable to other PGx guidelines, although several important gaps in reference standards and tooling need to be addressed. This approach will enable the creation of sharable, platform-independent knowledge artifacts that may facilitate the implementation of and consistency among PGx CDS rules.
Acknowledgments
The authors would like to thank Pooja Raghani for reviewing the modeling protocol. This work was funded in part by the NIH/NIGMS (U19 GM61388; the Pharmacogenomics Research Network) (RRF).
References
- 1.U.S. food and drug administration Preventable adverse drug reactions: a focus on drug interactions. [Accessed 2 Feb 2015]. Available at: http://www.fda.gov/drugs/developmentapprovalprocess/developmentresources/druginteractionslabeling/ucm110632.htm.
- 2.Relling MV, Klein TE. CPIC: clinical pharmacogenetics implementation consortium of the pharmacogenomics research network. Clin Pharmacol Ther. 2011;89(3):464–67. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caudle KE, Klein TE, Hoffman JM, Muller DJ, Whirl-Carrillo M, Gong L, et al. Incorporation of pharmacogenomics into routine clinical practice: the clinical pharmacogenetics implementation consortium (cpic) guideline development process. Current Drug Metabolism. 2014;15(2):209–17. doi: 10.2174/1389200215666140130124910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin MA, Hoffman JM, Freimuth RR, Klein TE, Dong BJ, Pirmohamed M, et al. Clinical pharmacogenetics implementation consortium guidelines for HLA-B genotype and abacavir dosing: 2014 update. Clin Pharmacol Ther. 2014;95(5):499–500. doi: 10.1038/clpt.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samwald M, Gimenez Minarro JA, Boyce RD, Freimuth RR, Adlassnig KP, Dumontier M. Pharmacogenomic knowledge representation, reasoning and genome-based clinical decision support based on OWL 2 DL ontologies. BMC Med Inform Decis Mak. 2015;15:12. doi: 10.1186/s12911-015-0130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.U.S. food and drug administration Table of pharmacogenomic biomarkers in drug labeling. [Accessed 3 Mar 2015]. Available at: http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm.
- 7.PharmGKB CPIC genes/drugs. [Accessed 12 Mar 2015]. Available at: https://www.pharmgkb.org/cpic/pairs.
- 8.Eknoyan G. Why we need clinical practice guidelines in chronic kidney disease. J Ren Nutr. 2010;20 doi: 10.1053/j.jrn.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 9.AHRQ national resource center for health information technology Challenges and barriers to clinical decision support (cds) design and implementation experienced in the agency for healthcare research and quality cds demonstrations. [Accessed 12 Mar 2015]. Available at: http://healthit.ahrq.gov/sites/default/files/docs/page/CDS_challenges_and_barriers.pdf.
- 10.Lee V, Boxwala A, Shields D, Roche M, Rhodes B, McClure R, et al. Standards & interoperability (S&I) framework. [Accessed 15 Feb 2015]. [Online] Available from: http://wiki.siframework.org/file/view/HL7_vMR_Logical_Model_Release_2_for_201309_ballot.pdf.
- 11.Stearns MQ, Price C, Spackman KA, Wang AY. SNOMED clinical terms: Overview of the development process and project status. Proc AMIA Symp. 2001:662–66. [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc. 2011;18(4):441–8. doi: 10.1136/amiajnl-2011-000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forrey AW, McDonald CJ, DeMoor G, Huff SM, Leavelle D, Leland D, et al. Logical observation identifier names and codes (LOINC) database: A public use set of codes and names for electronic reporting of clinical laboratory test results. Clin Chem. 1996;42(1):81–90. [PubMed] [Google Scholar]
- 14.Peleg M. Computer-interpretable clinical guidelines: a methodological review. J Biomed Inform. 2013;46:744–63. doi: 10.1016/j.jbi.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Peleg M, Boxwala AA, Bernstam E, Tu S, Greenes RA, Shortliffe EH. Sharable representation of clinical guidelines in GLIF: relationship to the arden syntax. J Biomed Inform. 2001;34(3):170–81. doi: 10.1006/jbin.2001.1016. [DOI] [PubMed] [Google Scholar]
- 16.Shiffman RN, Karras BT, Agrawal A, Chen R, Marenco L, Nath S. GEM: a proposal for a more comprehensive guideline document model using XML. J Am Med Inform Assoc. 2000;7(5):488–98. doi: 10.1136/jamia.2000.0070488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou L, Karipineni N, Lewis J, Maviglia SM, Fairbanks A, Hongsermeier T, et al. A study of diverse clinical decision support rule authoring environments and requirements for integration. BMC Med Inform Decis Mak. 2012;12:128. doi: 10.1186/1472-6947-12-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peleg M, Tu S, Bury J, Ciccarese P, Fox J, Greenes R, et al. Comparing computer-interpretable guideline models: a case-study approach. J Am Med Inform Assoc. 2003;10(1):52–68. doi: 10.1197/jamia.M1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Latoszek-Berendse A, Tange H, van den Herik HJ, Hasman A. From clinical practice guidelines to computer-interpretable guidelines. A literature overview. Methods Inf Med. 2010;49(6):550–70. doi: 10.3414/ME10-01-0056. [DOI] [PubMed] [Google Scholar]
- 20.Sottara D, Huag PJ, Ebert M, Potrich E, Greenes R. The Health eDecisions authoring environment for shareable clinical decision support artifacts. [Accessed 16 Feb 2015]. [Online] Available from: http://ceur-ws.org/Vol-1211/paper11.pdf.
- 21.Freimuth RR, Freund ET, Schick L, Sharma MK, Stafford GA, Suzek BE, et al. Life sciences domain analysis model. J Am Med Inform Assoc. 2012;19(6):1095–102. doi: 10.1136/amiajnl-2011-000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schadow G, Russler DC, Mead CN, McDonald Integrating medical information and knowledge in the HL7 RIM. Proc AMIA Symp. 2000:764–68. [PMC free article] [PubMed] [Google Scholar]
- 23.Bielinski SJ, Olson JE, Pathak J, Weinshilboum RM, Wang L, Lyke KJ, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc. 2014;89(1):25–33. doi: 10.1016/j.mayocp.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]