Abstract
Cancer Registries record cancer data by reading and interpreting pathology cancer specimen reports. For some Registries this can be a manual process, which is labour and time intensive and subject to errors. A system for automatic extraction of cancer data from HL7 electronic free-text pathology reports has been proposed to improve the workflow efficiency of the Cancer Registry. The system is currently processing an incoming trickle feed of HL7 electronic pathology reports from across the state of Queensland in Australia to produce an electronic cancer notification. Natural language processing and symbolic reasoning using SNOMED CT were adopted in the system; Queensland Cancer Registry business rules were also incorporated. A set of 220 unseen pathology reports selected from patients with a range of cancers was used to evaluate the performance of the system. The system achieved overall recall of 0.78, precision of 0.83 and F-measure of 0.80 over seven categories, namely, basis of diagnosis (3 classes), primary site (66 classes), laterality (5 classes), histological type (94 classes), histological grade (7 classes), metastasis site (19 classes) and metastatic status (2 classes). These results are encouraging given the large cross-section of cancers. The system allows for the provision of clinical coding support as well as indicative statistics on the current state of cancer, which is not otherwise available.
Introduction
Cancer notified from pathology is the primary method of identifying population based cancer incidence and is an important and fundamental tool for cancer monitoring, service planning and research. The Cancer Registry receives cancer specimen reports from pathology laboratories, which are subsequently abstracted by expert clinical coders for key cancer characteristics. The information is often trapped in the language of these reports, which are in the form of unstructured, ungrammatical and often fragmented free-text. The effort required for information abstraction can therefore be an extremely labour and time intensive exercise. Furthermore, the abstraction is also subject to errors and inconsistent interpretations due to the need for repeated interpretation of the results by coders with differing levels of experience and training potentially leading to differing conclusions, repeated data entry into collection systems, and when cases are misinterpreted or keywords are missed.
An approach whereby reports are electronically received and automatically processed, abstracted and analysed has the potential to support expert clinical coders in their decision-making and assist with improving accuracy in data recording. Improving the cancer notifications process would provide significant benefits to oncology service providers, health administrators, clinicians and patients.
An automated medical text analysis system that extracts cancer notifications data from any notifiable electronic cancer pathology report is proposed. A rule-based approach utilising natural language processing (NLP) and symbolic reasoning using SNOMED CT* were adopted in the system. Selected Queensland Cancer Registry business rules were also incorporated to mimic the interpretations and coding standards that expert clinical coders would adopt. The system was deployed to process pathology HL7 feeds from across the state of Queensland in Australia. The utility of the system was assessed and showed promising results on a set of reports containing a large cross-section of cancers.
Background
There has been a number of clinical language processing systems or studies relating to the extraction of key cancer characteristics from pathology free-text. Most research has focused on data extraction tasks for specific cancers such as colorectal, breast, prostate and lung.
The medical text analysis system/pathology (MedTAS/P) proposed by Coden et al.1 uses NLP, machine learning and rules to automatically extract or classify cancer characteristics. Selected cancer characteristics were evaluated and showed promise with F-measures ranging from 0.9–1.0 for most extraction tasks including histological type, primary site, and grade on a corpus of colon cancer pathology reports.
Martinez and Li2, similarly, used a colorectal cancer database to automatically predict cancer characteristics using machine learning (and in some cases complemented with rules) with 5 of the 6 multiclass problems achieving an F-measure above 74.9% using simple feature representations. Primary site, however, proved difficult to predict with an F-measure of 0.58.
Ou and Patrick3 extracted pertinent colorectal cancer information from narrative pathology reports using supervised machine learning and automatically populated the cancer structured reporting template using rule-based methods. They achieved an overall F-measure of 81.84% over a large range of structured reporting data fields.
Currie et al.4 presented a method of automated text extraction using specific rules and language patterns to extract over 80 data fields from breast and prostate cancer pathology reports with 90–95% accuracy for most fields.
Buckley et al.5 studied the feasibility of using natural language processing to extract clinical information from over 76,000 breast pathology reports from 3 institutions. They reported that there was widespread variation in how pathologists reported common pathologic diagnoses. For example, 124 ways of saying ‘invasive ductal carcinoma’, 95 ways of saying ‘invasive lobular carcinoma’ and over 4000 ways of saying ‘invasive ductal carcinoma was not present’. Reported sensitivity and specificity of the system were 99.1% and 96.5% when compared to expert human coders.
The Medical Text Extraction (Medtex) pipeline proposed by Nguyen et al.6 used a symbolic rule-based approach to parse pathology reports using NLP to identify SNOMED CT concepts of relevance, and tested whether these concepts were subsumed by concepts relating to cancer staging factors. Lung cancer staging and synoptic reporting were used to illustrate the symbolic rule-based approach7,8. The symbolic rule-based system performed within the bounds of human staging accuracy as observed in studies of registry data8.
As these studies are cancer (or tumour stream) specific, more generalized approaches are needed to extract cancer characteristics for all cancers. More recent research using Medtex has been applied to the extraction and coding of cancer characteristics such as basis of diagnosis, histological type and grade, cancer site and laterality from pathology free-text for all cancers9. Preliminary results on a small evaluation set of 61 cancer notifiable reports comprised of a range of cancers have shown that cancer characteristics can be extracted with an overall accuracy of 80%.
In this paper, we present the architecture and deployment of Medtex on streaming pathology HL7 feeds from public pathology laboratories across the state of Queensland, Australia. Challenges here included the vast individual pathologists and institutional variations in the textual contents of the reports. A subset of 220 pathology reports from the deployment was selected from patients with a range of cancers to evaluate and analyse the performance of the system over seven cancer characteristic categories, each potentially containing a large number of possible classes, namely, basis of diagnosis (4 classes), primary site (330 classes), laterality (5 classes), histological type (1036 classes), histological grade (9 classes), metastasis site (330 classes) and metastatic status (2 classes). In contrast to previous work9, the work presented here evaluated the utility of a Medtex deployment using a different and larger evaluation dataset and, unlike all previous tumour stream specific studies, this paper presents cancer characteristic extraction results on a wide range of cancer sites and types. The robustness of the system is also presented by comparing the evaluation results against those obtained from the development set and from a majority class classifier. An error analysis of the poorer performing cancer characteristic categories was also performed to determine the underlying limitations of the system.
Method
System Description
The medical text analysis system, Medtex, is a Java-based NLP software platform created for the development of clinical language engineering analysis engines to support data-driven analytic tasks6. Medtex incorporates a (1) free-text to SNOMED CT mapping engine to normalize the free text (i.e. unify the language of the reports) by identifying medical concepts, abbreviations and acronyms, short-hand terms, dimensions and relevant legacy codes, (2) relate key medical concepts, terms and codes using contextual information and report substructure, and (3) use formal semantics, via a SNOMED CT ontology server, for medical text inference and reasoning. Additional analysis engines can be incorporated to infer or classify complex clinical notions relevant to a particular health application using handcrafted algorithms and rules and/or machine learning techniques. Medtex has been applied to small scale datasets for research purposes; however its utility on real-time data streams and larger datasets may be inadequate if the computational time for the analysis of reports cannot keep up with the demands of the incoming data stream.
To address this issue, the Java messaging service (JMS) was chosen as the messaging broker for providing an intermediary to allow Java applications to be loosely coupled and reliably create, send, receive and read messages10. This messaging service is built on the concept of message queues, producers (senders), and consumers (receivers). A message producer is used for sending messages to a specific queue. The message consumer is then used for receiving messages from a specified queue. Multiple message consumers can be set up in parallel to receive messages from the same queue such that only one message is received by only one of the consumers. Furthermore, consumers acting on data can publish their results to another queue called a message topic, whereby other consumers wishing to register and subscribe to the topic can receive messages from the topic. This scenario allows for multiple consumer applications to act on the same messages published from a given consumer.
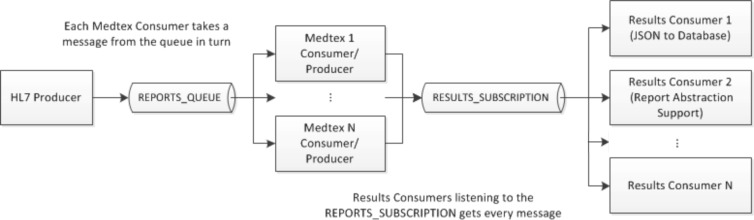
The proposed Medtex service for analysing HL7 messages from a statewide pathology information system is illustrated in Figure 1. It aims to automate a number of Cancer Registry tasks such as the notification of cancer reports and the coding of notifications data. Apache ActiveMQ†, an open source message broker, which fully implements JMS, was used to implement the messaging service. The message producer (HL7 Producer) accesses pathology HL7 messages and through the selection of report types that are relevant for subsequent processing, messages are sent to a specified queue (REPORTS_QUEUE). Multiple Medtex consumers can be set-up such that each consumer will take a message from the queue in turn. The results from the Medtex analysis are encoded in JSON‡ format and published to a message topic (RESULTS_SUBSCRIPTION) where the topics can be subscribed to by end-user applications (Results Consumer), for example, to consolidate patient results and store them in a database and/or provide support for clinical coders to abstract clinical information from medical reports.
Figure 1.
Messaging architecture for analysing pathology HL7 messages.
Within Medtex itself, the system would automatically process and analyse free-text HL7 pathology reports. The system selects from the pathology feed, histology and cytology reports, and filters non-notifiable cancer reports12. For notifiable cancer reports, cancer characteristics extracted for a Cancer Registry notification consist of basis of diagnosis, primary site, laterality, histological type, histological grade, metastasis site, metastatic status, among others. Those relevant for the evaluation in the current study include:
Basis of diagnosis encodes the method by which the cancer was diagnosed. For cytology and histology reports, the basis of diagnosis can either be encoded as cytology or haematology (06), histology of metastasis (07), histology of primary (08), or autopsy and histology (09). The basis of diagnosis is often used to assess the reliability of the cancer diagnosis, where the most conclusive information is found from histological reports.
Histological type records the characteristics of the tumour. It is encoded using an ICD-O morphology code, which consist of a prefix M followed by a five-digit code ranging from M-80000 to M-99893. The cell type and behaviour make up the morphology code. The first 4 digits refer to the histology, while the last digit is the behaviour code and identifies whether the neoplasm is benign (0), uncertain and unknown behaviour (1), in situ (2), malignant (3), secondary or metastatic (6), or malignant but unknown whether it’s a primary or metastatic site (9). Behaviour codes 6 and 9 are not used by Cancer Registries11 but are instead flagged or derived from other cancer characteristic data elements such as basis of diagnosis, metastatic site or metastatic status.
Histological grade, differentiation or phenotype describes how much or how little a tumour resembles normal tissue. It is encoded using a one-digit code. Only malignant tumours are graded and are represented by code numbers 1 to 4, designating grades I (well differentiated) to IV (undifferentiated), respectively. For a lymphoma or leukaemia, separate code numbers 5 to 8 are used to identify immunophenotype differentiation such as T-cell, B-cell, Null cell, and NK cell origin, respectively. If grading is unknown, not applicable or cannot be determined, then a code number of 9 would be assigned.
Primary site describes the origin of the cancer in the body and is represented by an ICD-O topology code ranging from C00.0 to C80.9. It is encoded using a four-character code using a prefix C to identify topography codes. The first two digits represent the site (e.g. C34 for Lung), while the last digit defines the sub-site (e.g. C34.1 for Upper lobe of lung).
Laterality indicates the side affected by the tumour for cancers of paired organs (e.g. breast, lung, kidney, etc.). It is encoded using a one-digit code: right (1), left (2), bilateral (3), not applicable (8) and unknown (9).
Metastasis site describes the site of spread from which the cancer originated. In this study, it was proposed to assign an equivalent ICD-O topography code to represent the metastatic site, although ICD-O is not usually used for this purpose.
Metastatic status is a flag to reflect whether the behaviour of a tumour indicates a metastasis (including lymph node metastasis).
The cancer characteristic codes were defined from ICD-O Third Edition13 for primary site, histological type and histological grade; and other notifications data according to classification codes recorded in the Queensland Cancer Registry11. Table 1 summarises the list of the cancer characteristics along with their codes (or classes). In general, the cancer characteristic categories are multiclass where there are more than 2 classes within the category. The vast number of histological types and primary sites for classification show the complexity of the extraction tasks involved.
Table 1.
Cancer characteristic categories and code description.
| Category | Code | Number of Classes |
|---|---|---|
| Basis of Diagnosis | 06 – Cytology or Haematology | 4 |
| 07 – Histology of metastasis | ||
| 08 – Histology of primary | ||
| 09 – Autopsy and histology | ||
| Histological Type | ICD-O morphology code – M-xxxxx | 1036 |
| Histological Grade | 1 – Grade 1 – well differentiated | 9 |
| 2 – Grade II – moderately differentiated | ||
| 3 – Grade III – poorly differentiated | ||
| 4 – Grade IV –undifferentiated or anaplastic | ||
| 5 – T-cell | ||
| 6 – B-cell, Pre-B, B-precursor | ||
| 7 – Null cell, Non T, Non-B (For leukaemias only) | ||
| 8 – NK Cell | ||
| 9 – Grade or differentiation not determined, not stated or not applicable. | ||
| Primary Site | ICD-O topography code – Cxx.x | 330 |
| Laterality | 1 – Right | 5 |
| 2 – Left | ||
| 3 – Bilateral | ||
| 8 – Not applicable | ||
| 9 – Unknown | ||
| Metastatic Site | Equivalent ICD-O topology code – Cxx.x | See primary site |
| Metastatic Status | Not applicable or 2 (metastasis) | 2 |
An expert clinical coder analysed the development set (see Corpus Description) to help build the ground truth and extraction modules for each of the cancer characteristic categories. A combination of NLP, domain knowledge and rules, and in particular SNOMED CT14 manipulation and querying were used to classify cancer characteristics. The algorithm and rules were iteratively refined based on measuring and analysing the performance of the system on the development data set. Examples of the cancer characteristic classification methods are tabulated in Table 2.
Table 2.
Example of methods used for the extraction of cancer notifications data.
| Method | Notifications Data | Example |
|---|---|---|
| Queensland Cancer Registry coding rules (including special casings) | Histological Type | Select the highest morphology if more than one morphology is stated. |
| Histological Grade | Assign the highest grade or differentiation code. | |
| Primary Site | Code all leukaemia except myeloid sarcoma (M-99303) to C42.1 (bone marrow). | |
| Domain knowledge | Primary Site | List of one-to-one only ICD-O morphology to site mappings. |
| SNOMED CT property access | Histological Type | Restrict SNOMED CT concepts to those with a ‘morphologic abnormality’ semantic category and those that have alternate terms with the following regular expression “M-[0–9]{5}”. |
| Primary Site | Restrict SNOMED CT concepts to those with a ‘body structure’ semantic category. | |
| SNOMED CT to ICD-O topography cross-maps | Primary Site | Map SNOMED CT ‘body structure’ concepts to ICD-O topography codes. |
| SNOMED CT Subsumption querying | Histological Type | Candidate ‘leukaemia’ concepts are found by testing subsumption by the ‘128931003 | Leukemia – category’ concept. |
| SNOMED CT Concept relationship querying | Primary Site | “Procedure site – Direct” and “Finding site” relationship values from concepts are used as candidate sites. |
| SNOMED CT querying using ad-hoc term expansion | Histological Type | The histological type and grade’s preferred terms were used to search for a more specific concept. For example, the query for “Follicular Lymphoma” + “Grade 3” would return the histological type M-96983, which is “Follicular Lymphoma, Grade 3”. |
| Relation extraction | Basis of Diagnosis | Identification of multiple concepts or terms within a search scope such as metastasis and lymph nodes within a sentence. |
| Keyword/phrase spotting | Histological Grade | Detect keywords or phrases that were unable to be (or unreliably) mapped to SNOMED CT. For example “poorly to moderately differentiated”. |
Corpus Description
Access to the Queensland statewide pathology data was obtained from the Queensland Oncology Repository with research ethics approval from the Queensland Health Research Ethics Committee. The data covers HL7 pathology feeds from public pathology laboratories in the state of Queensland. A corpus consisting of 500 pathology reports was used for system development of which 201 of them were notifiable cancers (and thus relevant for the current cancer characteristic extraction task). Non-notifiable cancers such as non-malignant cancers, and squamous cell carcinoma (SCC) and basal cell carcinoma’s (BCC) of the skin were identified, but removed and flagged by the system; a separate study addressed these issues by filtering notifiable reports from non-notifiable reports12. For system evaluation, a separate 220 pathology reports from the deployment of the system for processing the backlog of pathology feeds was selected from patients with a range of cancers (i.e. tumour-stream stratified sampling) to evaluate and analyse the performance of the system. The ground truth used for system evaluation was based on the reference data set annotated by the same expert clinical coder who helped develop the system. Table 3 shows the cancer characteristic statistics from the development and evaluation corpus.
Table 3.
Notifiable cancer characteristic corpus statistics.
| Category | Number of Classes | Majority Class | Frequency Range (Mean ± Std Dev) | Number of Unseen Classes in Eval. | ||
|---|---|---|---|---|---|---|
|
| ||||||
| Dev./Eval. | Dev. | Eval. | Dev. | Eval. | ||
| Basis of Diagnosis | 3 | 08 | 08 | 19–146 (67±69) | 21–175 (73±88) | – |
| Histological Type | 64/94 | M-81403 | M-81403 | 1–35 (3.1±5.4) | 1–21 (2.3±3.1) | 65 (69%) |
| Histological Grade | 7 | 9 | 9 | 3–110 (28.7±38.1) | 1–129 (31.4±44.6) | – |
| Primary Site | 58/66 | C50.9 | C42.1 | 1–21 (3.4±4.1) | 1–39 (3.3±5.9) | 30 (45%) |
| Laterality | 4/5 | 8 | 8 | 20–129 (50.3±52.6) | 1–134 (44.0±52.4) | 1 (20%) |
| Metastatic Site | 17/19 | NA | NA | 1–170 (11.2±39.7) | 1–192 (11.0±42.6) | 10 (53%) |
| Metastatic Status | 2 | NA | NA | 31–170 (101±98) | 28–192 (110±116) | – |
Dev., development set (N=201); Eval., evaluation set (N=220); Std Dev, standard deviation; NA, ‘Not Applicable’ class
The distributions between the development and evaluation set for each cancer characteristic category were quite similar. However, within the categories the class frequencies can vary quite significantly, with large variation in ranges and standard deviations, e.g. basis of diagnosis, histological grade, laterality, etc. On the other hand, in some categories, there are a large number of small frequency classes resulting in low means and standard deviations, e.g. histological type and primary site, due to the large and diverse number of cancer types and sites, respectively. Furthermore, there is a large portion of classes contained in the evaluation set that were not found in the development set (e.g. histological type, primary site and metastatic site). The corpus and cancer characteristic category statistics show the challenges and complexity of the extraction tasks. In addition, the varied writing style and language contained within the reports from different pathologists and laboratories creates additional challenges that will test the robustness and generalizability of the extraction modules when measuring the performance of the system on the evaluation set.
Performance Measures
The measures used to evaluate results are based on the counts of true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN) resulting from the classification decisions. The multiclass classification performance for a given cancer characteristic category, C, is measured using the micro-average recall (R or sensitivity), precision (P or positive predictive value), and balanced F-measure (F).
Overall system performance is reported as the macro-average measure across all the categories, with equal weight to every category.
A majority class classifier was used as a baseline to determine whether the system’s results were significantly better than one that naively classifies results simply based on the majority class. In addition, to assess the generalizability of the system, the system’s results from the evaluation set were compared with that from the development set. Other cancer notification extraction systems are tumour specific and thus were not suitable for use as a benchmark.
Results
The Medtex messaging service was applied to a statewide pathology HL7 message feed. The backlog and new incoming HL7 messages from pathology laboratories in Queensland, Australia were used to analyse the pathology reports as well as test the load on the service. Using 3 instances of Medtex, the system’s average processing rate was 3.6 seconds per message and achieved the processing of a year’s worth of messages within just under 5 days.
An increase in report analysis throughput was achieved by using the messaging framework and multiple instances of Medtex consumers in parallel. The use of 3 Medtex instances in parallel resulted in a 2.5 times speed-up over the sequential single instance of Medtex in operation. Depending on system resources, further speed-ups are possible if additional instances of Medtex and/or multiple instances of Medtex’s shared resources such as the SNOMED CT ontology and concept-mapping servers were made available.
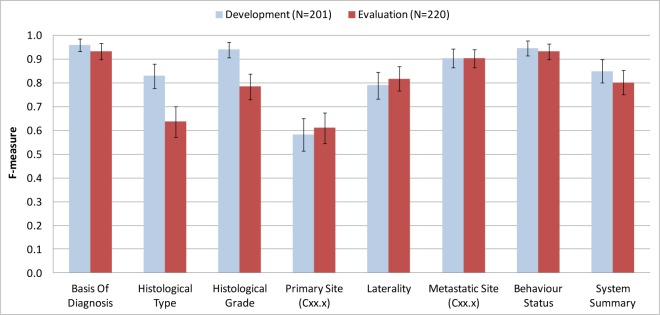
The classification performances of the system with respect to the cancer characteristic categories are shown in Table 4. Figure 2 summarises the F-measure comparison of cancer characteristic categories between the development and evaluation set.
Table 4.
Cancer characteristic classification performances.
| Recall | Precision | F-measure | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Dev. | Eval. | Dev. | Eval. | Dev. | Eval. | |
| Basis of Diagnosis | 0.955 | 0.918 | 0.965 | 0.948 | 0.960 | 0.933 |
| Histological Type | 0.796 | 0.577 | 0.865 | 0.710 | 0.829 | 0.637 |
| Histological Grade | 0.935 | 0.773 | 0.945 | 0.798 | 0.940 | 0.785 |
| Primary Site | 0.522 | 0.546 | 0.656 | 0.694 | 0.582 | 0.611 |
| Laterality | 0.786 | 0.805 | 0.794 | 0.831 | 0.790 | 0.818 |
| Metastatic Site | 0.891 | 0.886 | 0.918 | 0.920 | 0.904 | 0.903 |
| Metastatic Status | 0.945 | 0.932 | 0.945 | 0.932 | 0.945 | 0.932 |
| Macro-average | 0.833 | 0.777 | 0.870 | 0.833 | 0.850 | 0.803 |
Figure 2.
Cancer characteristic classification performances (F-measure) shown with 95% confidence intervals.
The system’s results were compared to a majority class classifier to determine whether the system was significantly better than one that naively classifies results simply based on the majority class. Table 5 shows the F-measure results for both the system and the majority class classifier.
Table 5.
Comparison of F-measure results between the system (Medtex) and majority class classifier.
| Development (N=201) | Evaluation (N=220) | |||
|---|---|---|---|---|
|
| ||||
| Majority Classifier (95% CI) | Medtex (95% CI) | Majority Classifier (95% CI) | Medtex (95% CI) | |
| Basis of Diagnosis | 0.73 (0.66–0.79) | 0.96 (0.93–0.99) | 0.80 (0.74–0.85) | 0.93 (0.90–0.97) |
| Histological Type | 0.17 (0.12–0.23) | 0.83 (0.78–0.88) | 0.10 (0.06–0.14) | 0.64 (0. 57–0.70) |
| Histological Grade | 0.55 (0.48–0.62) | 0.94 (0.91–0.97) | 0.59 (0.52–0.65) | 0.79 (0.73–0.84) |
| Primary Site | 0.11 (0.06–0.15) | 0.58 (0.51–0.65) | 0.18 (0.13–0.23) | 0.61 (0.55–0.68) |
| Laterality | 0.64 (0.58–0.71) | 0.79 (0.73–0.85) | 0.61 (0.55–0.67) | 0.82 (0.77–0.87) |
| Metastatic Site | 0.85 (0.80–0.90) | 0.90 (0.86–0.95) | 0.87 (0.83–0.92) | 0.90 (0.86–0.94) |
| Metastatic Status | 0.85 (0.80–0.90) | 0.95 (0.91–0.98) | 0.87 (0.83–0.92) | 0.93 (0.90–0.97) |
| Macro-average | 0.56 (0.49–0.62) | 0.85 (0.80–0.90) | 0.57 (0.51–0.64) | 0.80 (0.75–0.86) |
CI, confidence interval
Discussion
Overall system performance on the evaluation set reported as F-measure was 0.80. At a cancer characteristic level, the F-measure performances within each category were 0.93 for basis of diagnosis, 0.64 for histological type, 0.79 for histological grade, 0.61 for primary site, 0.82 for laterality, 0.90 for metastatic site and 0.93 for metastatic status. The results are promising given the challenges previously discussed regarding the large number of classes from certain categories, varied and skewed distributions within each cancer characteristic category, and the large number of unseen classes being classified in the evaluation set. The results show the system’s robustness and generalizability by achieving extraction performances on the evaluation set that is comparable with that obtained from fine-tuning the system using the development set, and also its superiority to that obtained when using a majority class classifier. When compared to previous studies that focused on certain tumour streams1,3,5, the results show that generalizing the extraction algorithms to accommodate for all tumour streams has its challenges and therefore is sub-optimal to the tumour stream specific results. However, it would be a very costly exercise to build specific tumour stream cancer characteristic classifiers for each and every possible cancer. The trend in extraction performances across the categories are also consistent with previous works2 whereby primary site was found to be the most challenging.
The results between the development and evaluation set were in general not significantly different. Only the histological type and histological grade generated results that exhibited non-overlapping 95% confidence intervals suggesting that the algorithms and rules adopted in these extraction modules likely over-fitted the development data. That said, there was a large number of histological types in the evaluation set that were not seen in the development set; despite having almost 70% of the histological types unseen by the system during development, the system was able to classify the category with a recall of 0.58, precision of 0.71 and an overall F-measure of 0.64. In terms of the free-text to SNOMED CT mapping engine, it was observed that 83.2% of the histological types from the development set could have been found within the mapped concepts. This provides an upper bound to the classification performance by the system, unless other methods are introduced to infer the histological type.
For histological grade, error analysis revealed that at a per-class level, all classes had non-overlapping 95% confidence intervals, except for the histological grade of 3 (poorly differentiated). The errors were more pronounced for histological grades 4 through to 6, from which 5 and 6 relate to lymphoma or leukaemia histological types.
The extraction of primary site was also a challenge for the system as evident from its performance. Despite having 45% of the cases in the evaluation set unseen by the system during development, the results between the development and evaluation set had overlapping 95% confidence intervals. One source of error was due to the lack of co-referencing of specimens between the macroscopic and microscopic sections of the pathology report. As a result, the relation between different evidences in the free-text could not be classified correctly. Again in terms of the free-text to SNOMED CT mapping engine, 70.3% of the primary sites (excluding unknown primary C80.9 sites) from the development set were found in the mapped concepts. This suggests that the use of the current concept-mapping algorithm has its limitations in giving the system the ability to correctly identify primary sites. Further inspection of the errors also revealed that many of the cases actually classified the ICD-O site (Cxx) correctly (e.g. site = C34 for Lung) but not the sub-site (Cxx.x; e.g. sub-site = C34.1 for Upper lobe of lung). Figure 3 illustrates this effect where results at a site level (Cxx) were significantly better than that classified at a sub-site level (Cxx.x) with non-overlapping 95% confidence intervals.
Figure 3.
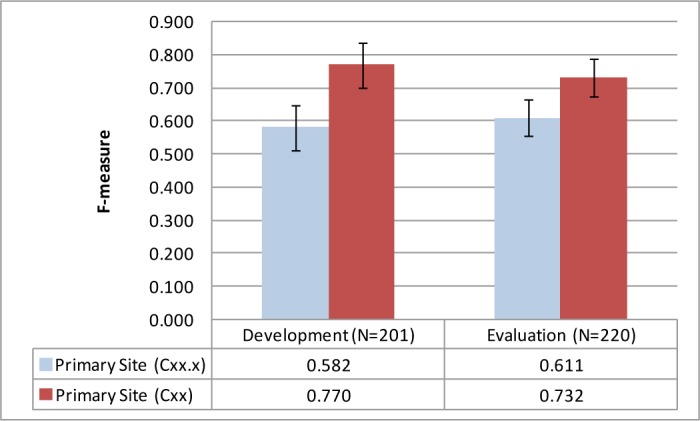
Primary site classification performances at a site (Cxx) and sub-site (Cxx.x) level along with 95% confidence intervals.
The metastatic site and metastatic status data items are also worth deeper analysis. Although, performing well with high F-measures, the large bias towards the majority class did not make the system perform much better than the majority class classifier. As a result, it is likely that there is poor recall and precision for the minority classes.
Future research will need to focus on iteratively improving the system’s performance, especially for critical cancer notifications data such as primary site and histological type. Preliminary error analysis have revealed areas of focus for further system improvement such as histological grade for lymphoma and leukaemia cancers, including the need to investigate other concept-mapping algorithms for improving both histological type and primary site categories. These error analyses facilitate the identification of the relevant Queensland Cancer Registry business rules to be incorporated, abstraction errors by human experts, and also feedback the type of errors generated for further system development.
Other limitations of the system include limited number of development reports given the large number of possible histological types and primary sites. The evaluation corpus is also biased towards uncommon cancers due to the tumour-stream stratified sampling that was applied to ensure that a range of cancers were represented in the evaluation set. In addition, the system at present cannot distinguish between multiple cancers reported within a single report.
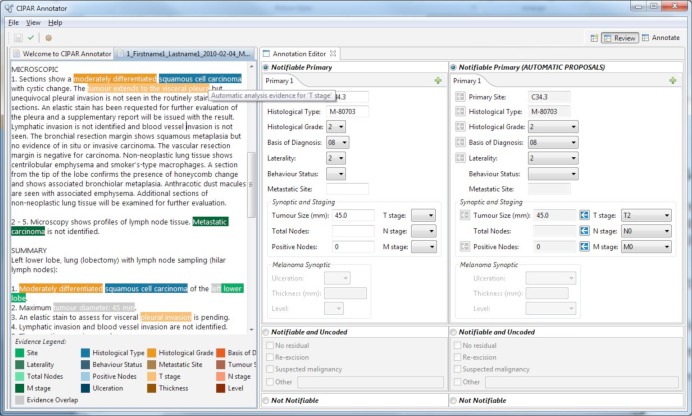
In a clinical coding workflow setting, the system could be used to support clinical coders and hence improve data collection capture at Cancer Registries by highlighting and pre-populating cancer notification items for validation (for example, see Figure 4). Here, the free-text report is shown in the leftmost panel. The highlights shown over the free-text report correspond to the evidence used to generate the system’s suggested coding, which is shown in the rightmost panel. Clinical coders can then use the system’s suggestions to either accept (using the arrows in the rightmost panel) or enter in an alternate code to populate the fields within the clinical coder’s coding panel (as depicted by the middle panel). Investigations into the utility of a guided and interactive annotation process on cancer characteristic abstraction tasks is reserved for future work.
Figure 4.
The Medtex software can process narrative pathology reports and generates structured data to aid clinical coders in cancer abstraction tasks.
The proposed architecture for the processing and analysis of streaming pathology reports has potential to overcome the multi-year delay in the reporting of cancers by providing indicative population-level statistics on the current incidence of cancer. The system supports future data extension requirements such as cancer stage, and also can be applied on other sources of cancer data with free text, e.g. death data and radiology reports.
Conclusion
Analysis of the contents of electronic pathology reports will have a profound impact on cancer care. As a result, a number of automatic cancer data extraction systems have been developed, however their utility in automating Cancer Registry tasks has to be adequately assessed. In this study, the Medtex system was assessed and showed promise in terms of stream processing at a statewide level and also in terms of cancer characteristic extraction performances and its ability to track and identify specific system limitations for future improvements.
The use of Medtex’s messaging technologies that take advantage of the parallelism of consumers/producers can be an effective real-time processing solution for data streams. These technologies greatly increase the throughput of medical text analytics for clinical decision support and/or research activities involving real-time data streams or large datasets.
The cancer notifications data extraction results from Medtex show promise with an overall F-measure performance of 0.80 on a broad range of cancers and cancer characteristic categories. Despite some cancer characteristic categories performing well, with an F-measure score of above 0.90, the histological type and primary site extraction results proved more challenging due to its large number of possible classes. The system is extensible and cancer stage including other synoptic data can also be incorporated. Future work will analyse errors between the system and the reference standard to feedback into the iterative development process.
The system is proposed to streamline and support the clinical coding workflow at Cancer Registries by identifying cancer notifiable reports and then highlighting and pre-populating cancer notification items for clinical coder validation. It is hoped that such automation would help overcome the multi-year delay in the reporting of cancer statistics with Cancer Registries able to have access to up-to-date population-level statistics on the current state of cancer.
Footnotes
Systematized nomenclature of medicine - clinical terms
References
- 1.Coden A, Savova G, Sominsky I, et al. Automatically extracting cancer disease characteristics from pathology reports into a Disease Knowledge Representation Model. J Biomed Inform. 2009;42(5):937–949. doi: 10.1016/j.jbi.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Martinez D, Li Y. Information extraction from pathology reports in a hospital setting; ACM International Conference on Information and Knowledge Management; Glasgow, Scotland, UK: 2011. pp. 1877–1882. [Google Scholar]
- 3.Ou Y, Patrick J. Automatic Structured Reporting from Narrative Cancer Pathology Reports. electronic Journal of Health Informatics. 2014;8(2):e20. [Google Scholar]
- 4.Currie A-M, Fricke T, Gawne A, Johnston R, Liu J, Stein B. Automated Extraction of Free-Text from Pathology Reports; AMIA Annual Symposium Proceedings; 2006. p. 899. [PMC free article] [PubMed] [Google Scholar]
- 5.Buckley J, Coopey S, Sharko J, et al. The feasibility of using natural language processing to extract clinical information from breast pathology reports. J Pathol Inform. 2012;3(1):23. doi: 10.4103/2153-3539.97788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen A, Lawley M, Hansen D, Colquist S. A simple pipeline application for identifying and negating SNOMED clinical terminology in free text. Health Informatics Conference. 2009:188–193. [Google Scholar]
- 7.Nguyen A, Lawley M, Hansen D, Colquist S. Structured pathology reporting for cancer from free text: Lung cancer case study. electronic Journal of Health Informatics. 2012;7(1):e8. [Google Scholar]
- 8.Nguyen A, Lawley M, Hansen D, et al. Symbolic rule-based classification of lung cancer stages from free-text pathology reports. Journal of the American Medical Informatics Association. 2010;17(4):440–445. doi: 10.1136/jamia.2010.003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen A, Moore J, Lawley M, Hansen D, Colquist S. Automatic Extraction of Cancer Characteristics from Free-Text Pathology Reports for Cancer Notifications. Studies in health technology and informatics. 2011:117–124. [PubMed] [Google Scholar]
- 10.Richards M, Monson-Haefel R, Chappell DA. Java Message Service. 2nd ed. O’Reilly Media, Inc; 2009. [Google Scholar]
- 11.Queensland Cancer Registry. Clinical Coding Manual Version 3.
- 12.Nguyen A, Moore J, Zuccon G, Lawley M, Colquist S. Classification of Pathology Reports for Cancer Notifications. Studies in health technology and informatics. 2012:150–156. [PubMed] [Google Scholar]
- 13.World Health Organization . International Classification of Diseases for Oncology. 3 ed. Geneva: World Health Organization; 2007. [Google Scholar]
- 14.International Health Terminology Standards Development Organisation . SNOMED Clinical Terms User Guide. 2008. [Google Scholar]