Abstract
Objective
In a previous study, we investigated a sentence classification model that uses semantic features to extract clinically useful sentences from UpToDate, a synthesized clinical evidence resource. In the present study, we assess the generalizability of the sentence classifier to Medline abstracts.
Methods
We applied the classification model to an independent gold standard of high quality clinical studies from Medline. Then, the classifier trained on UpToDate sentences was optimized by re-retraining the classifier with Medline abstracts and adding a sentence location feature.
Results
The previous classifier yielded an F-measure of 58% on Medline versus 67% on UpToDate. Re-training the classifier on Medline improved F-measure to 68%; and to 76% (p<0.01) after adding the sentence location feature.
Conclusions
The classifier’s model and input features generalized to Medline abstracts, but the classifier needed to be retrained on Medline to achieve equivalent performance. Sentence location provided additional contribution to the overall classification performance.
Introduction
Most clinical questions raised by clinicians in the course of patient care can be answered by online primary literature resources, such as Medline1. However, there are critical barriers to the use of the primarily literature at the point of care. Specifically, clinicians need to search, screen, appraise, and integrate literature findings into their decision making for a particular patient. This process is labor intensive and not compatible with busy clinical workflows. Several approaches have been pursued to improve efficient consumption of the primary literature, including improvements in the search process2–6, question and answering systems7, 8, and automatic summarization of Medline abstracts and full-text articles.9 Despite substantial progress in these approaches, recent studies still show that clinicians prefer distilled recommendations in the form of guidelines and evidence summaries10, 11.
Significant effort has been dedicated to automatic biomedical text summarization.9 Yet, most previous studies aimed at generating summaries that resemble article abstracts written by study authors. However, article abstracts are written to summarize all elements of a study, such as purpose, methods, results, and conclusions. On the other hand, for patient care decision making, clinicians prefer sentences that provide patient-specific, actionable recommendations for a particular intervention as opposed to general background and study methods12–16. For example, the sentence “Apixaban 2.5 mg twice daily, starting on the morning after total knee replacement, offers a convenient and more effective orally administered alternative to 40 mg per day enoxaparin, without increased bleeding” provides an actionable treatment finding for patients who undergo total knee replacement. Specific methods are needed for extracting clinically useful sentences from clinical studies.
In a previous study we developed a feature-rich classification model for extracting clinically useful sentences from synthesized evidence resources, such as UpToDate17. The study was based on 4,824 sentences from 18 UpToDate documents on the treatment of six chronic conditions: coronary artery disease, hypertension, depression, heart failure, diabetes mellitus, and prostate cancer. In the present study, we attempt to apply the sentence classifier to the primary literature. Specifically, the study has two main goals: (1) to assess the generalizability of the feature-rich classifier on extracting clinically actionable statements from PubMed abstracts; and (2) to assess if optimization of the classifier for PubMed abstracts results in improved classification accuracy.
Background and Significance
In a previous study we designed and assessed a method for extracting clinically useful sentences from synthesized online clinical resources.17 The method’s underlying assumption is that clinically useful sentences are actionable statements that provide a specific recommendation for an intervention (e.g., medication treatment) that should be employed with a specific patient population. To capture these characteristics, the method uses three sets of semantic features from the PubMed abstracts. The method consists of a Kernel-based Bayes Network classification model with Gaussian kernel density estimators that classifies each sentence as clinically useful or not. As shown in previous research, the Kernel-based Bayesian Network is robust to highly imbalanced datasets such as the one used in this paper18, 19. This classifier is a Bayesian Network that estimates the true density of the continuous variables using kernels, which are weighting functions used to estimate random variables’ density function. The classifier is based on three domain-specific feature types extracted from UpToDate sentences: 1) treatment-related UMLS concepts and their semantic groups; 2) semantic predications; and 3) patient population. A summary of these features is provided below.
The first set of features consists of treatment-related UMLS concepts, and their corresponding semantic groups20, extracted from sentences using MedTagger, which is an extension of the cTAKES natural language processing (NLP) pipeline21. The UMLS semantic group of each concept was obtained, leading to four features according to the following semantic groups: Chemicals & Drugs (CHEM), procedures (PROC), physiology (PHYS), and disorders (DISO).
Semantic predications are relations that consist of a subject, a predicate, and an object. The sentence classifier uses treatment-related predications extracted by SemRep, a semantic NLP parser that uses underspecified syntactic analysis and structured domain knowledge from the UMLS22. The subject and object of predications are represented with UMLS concepts. Six types of predications were extracted as features: TREATS/NEG_TREATS, ADMINISTERED_TO/NEG_ADMINISTERED_TO, AFFECTS/NEG_AFFECTS, PROCESS_OF/NEG_PROCESS_OF, PREVENTS/NEG_PREVENTS, and COMPARED_WITH/HIGHER_THAN/LOWER_THAN/SAME_A. For instance, from the sentence below:
“Adding corticosteroid injection to conventional treatment in hemiplegic shoulder pain improved shoulder range of motion and decreased pain scores before treatment to the first and fourth weeks of treatment.”
SemRep produces the following output:
Shoulder Pain PROCESS_OF Hemiplegics
Injection procedure TREATS Shoulder Pain
Adrenal Cortex Hormones TREATS Shoulder Pain
which yields the following features:
Total number of predications: 3
PROCESS_OF instances: 1
TREATS instances: 2
Finally, patient population determines whether a sentence includes a description of the types of patients who are eligible to receive a certain treatment based on a pattern-based method. This produced one binary feature, which indicates whether a sentence describes the target population or not. The method uses the Stanford lexical parser23 and Tregex24. The Tregex patterns are similar to regular expressions, but more advanced in extracting patterns such as a noun phrase with two consecutive prepositional phrases, a verb phrase with two consecutive prepositional phrases, and a noun phrase preceding a subordinating conjunction. For example, in the following sentence the population extraction algorithm identifies that the sentence includes a target population (“patients with advanced NSCLC).
“The addition of vandetanib to docetaxel provides a significant improvement in PFS in patients with advanced NSCLC after progression following first-line therapy”
In the present study, we test the generalizability of the described feature-rich sentence classifier to the primary literature and whether optimization of the sentence classifier results in performance gains.
Methods
The study methods consisted of the following steps: 1) development of a gold standard of clinically useful sentences from PubMed abstracts; 2) extraction of the three feature categories (i.e., concept, predication and population features) for sentence classification; 3) optimization of the sentence classifier to identify clinically useful sentences from PubMed abstracts; and 4) assessment of classifier performance.
All features for our sentence feature-rich sentence classifier in different experiments are summarized in Table 1. Specifically, these are the inputs for the Kernel-based Bayes Network classification model with Gaussian kernel.
Table 1:
Features used to develop the classification model.
| Feature Type | Number of features | Description |
|---|---|---|
| Predication | 7 | Total number of predications with a treatment-related predicate (1 feature) and number of predication instances per treatment-related predicate (6 features). |
| Population | 1 | Whether or not a sentence includes a description of the types of patients who are eligible to receive a certain treatment. |
| Concept | 5 | Total number of concepts in the sentence (1 feature) and number of concept instances per UMLS treatment-related semantic group (4 features). |
| Location | 1 | Location of the sentences in the abstract, which can be either Conclusion, Results, or Unknown (unstructured abstracts) |
Gold Standard
The gold standard consisted of 2,146 sentences from 140 PubMed abstracts that were randomly selected from 34,913 PubMed citations of high quality clinical studies published between January 2010 and October 2014. We focused on high quality clinical studies because they are likely to be more useful for patient care decision making. Citation quality was determined using the classifier developed by Kilicoglu et al.4 Sentences from the selected citations were retrieved from the SemanticMedline database, which contains sentences, and their semantic predications, extracted from all abstracts in Medline25.
For structured PubMed abstracts, we found that the gold standard contained clinically useful sentences only in the conclusion and results sections. Thus, we excluded all the sentences that were not in these sections. This filtering was done using the NlmCategory tag of the Medline citations in XML format, which provides standard section categories (e.g., METHODS, RESULTS, CONCLUSIONS) for abstracts that are written in a structured format. For unstructured abstracts, we included all sentences. As a result, the dataset was narrowed to 954 sentences from 124 structured abstracts and all 118 sentences from 16 unstructured abstracts (i.e., total of 1,072 sentences).
Next, the sentences in the gold standard were rated by one of the study authors (GDF) according to a validated clinical usefulness scale (Table 2), which was slightly adapted from one of our previous studies26. Sentences are rated from 1 to 4, with 4 being the most useful. The core principle of this scale is that clinically useful sentences follow the PICO format, i.e. sentences that define the study patient population, the intervention under investigation, the comparison (e.g., placebo), and the study outcome. The PICO format has been recommended to clinicians for formulating well-structured clinical questions and has been applied in several biomedical information retrieval studies8, 27–29.
Table 2:
Clinical usefulness rating criteria.
| Rating | Definition | Examples |
|---|---|---|
| 1 | Sentences that, in isolation, don’t convey clear meaning. | “Lorazepam rescue was permitted after dose two.” |
| 2 | Background information, such as the epidemiology and physiopathology of a condition, mechanism of action of an intervention (e.g., a drug), justification for conducting the study, study objectives, and description of the study design (e.g., randomized controlled trial, systematic review). | “This phase III, randomised, double-blind, placebo-controlled, parallel-group study enrolled 344 individuals who received one, two or three doses of inhaled loxapine (5 or 10 mg) or a placebo.” |
| 3 | Study findings without a population, comparison, intervention and outcome (PICO); or secondary study findings. | “Death or myocardial infarction rates were reduced by fondaparinux in tertile I (age<56 years, 4.5% vs 4.8%, hazard ratio [HR] 0.94, 95% CI 0.71–1.25), in tertile II (age 56–68 years, 7.9% vs 9.7%, HR 0.80, 0.65–0.98), and in tertile III (age>=69 years, 17.2% vs 19.8%, HR 0.87, 95% CI 0.75–1.01, P for heterogeneity=0.87).” |
| 4 | Primary study findings or treatment safety finings with a population [P], intervention [I], comparison [C], and outcome [O]. | After adjustment for covariates, infants with CNS involvement [P] who had been randomly assigned to acyclovir suppression [I] had significantly higher mean Bayley mental-development scores at 12 months [O] than did infants randomly assigned to placebo [C] (88.24 vs. 68.12, P=0.046)., 4, 1, 1 |
The final dataset is available online for the research community1. The distribution of sentences according to their ratings is shown in Table 3.
Table 3:
Sentence distribution according to sentence usefulness ratings.
| Type | Rating | Total number of sentences | Average number of sentences per abstract |
|---|---|---|---|
| 1 | 102(10%) | 2.22 | |
| Not Useful | 2 | 117(11%) | 2.60 |
| 3 | 750(70%) | 5.43 | |
| Useful | 4 | 103(10%) | 1.12 |
Optimization strategies
To optimize the feature rich classifier based on PubMed abstracts, two strategies were employed. First, the feature-rich classifier was re-trained on PubMed abstracts (instead of UpToDate documents) using the same features identified in our previous study. Second, sentence location was included as an additional feature to the sentence classifier. The location feature was extracted from structured abstracts using the NlmCategory tag of Medline citations in XML format. The possible values for the location feature were Conclusions or Results for structured abstracts, and Unknown, for unstructured abstracts.
Assessment of classification performance
We conducted three experiments to test the following hypotheses:
Hypothesis 1: The feature-rich classifier trained on UpToDate sentences has comparable performance on the primary literature. To test this hypothesis, we compared the performance of the sentence classifier when applied to the original UpToDate dataset versus Medline sentences. The goal was to assess the generalizability of the sentence classifier to the primary literature.
Hypothesis 2: Re-training the feature-rich classifier on the primary literature improves performance compared to the original classifier. To test this hypothesis, we assessed the performance of the feature-rich classifier trained on Medline sentences compared to the original classifier, which was trained on UpToDate sentences. Also, the enriched feature-rich classifier was compared to a baseline classifier where all sentences in the Conclusion section of structured abstracts and the last 10% of the sentences in unstructured abstracts were labeled as clinically useful (i.e., positive class).
Hypothesis 3: Adding sentence location to the feature-rich classifier improves its performance on the primary literature. To test this hypothesis, we compared the performance of the re-trained feature-rich classifier enriched with a sentence location feature versus the re-trained classifier without sentence location.
Experiment procedures
Ordinal ratings were converted into binary values: sentences rated as “4” were considered as the positive class (i.e., clinically useful sentences) and the remaining sentences were considered as the negative class. As a result, 89% of the sentences in the gold standard were labeled as positive. This distribution is similar to the sentences in the UpToDate dataset, with 87% positive sentences.
For the first hypothesis the feature-rich classifier was trained on 4,824 UpToDate sentences from our previous study, and then tested on 1,072 Medline sentences. For the second and third hypotheses we employed a 20-fold cross-validation strategy with each fold containing 7 abstracts.
Finally, classification performance was measured according to the average precision, recall, and F-measure across the 20 folds. F-measure was defined a priori as the primary outcome for hypotheses testing. For statistical significance test of all experiments, first we applied the Friedman’s test to verify differences among multiple classifiers. If significant at an alpha of 0.05, pairwise comparisons were made with the Wilcoxon Signed-Rank test. This statistical approach is aligned with the method recommended by Demsar30.
Results
Similar to UpToDate sentences, descriptive statistics of the sentences and features in the Medline gold standard show that all feature types were correlated with useful sentences.
Hypothesis #1: The feature-rich classifier trained on UpToDate sentences has comparable performance on the primary literature
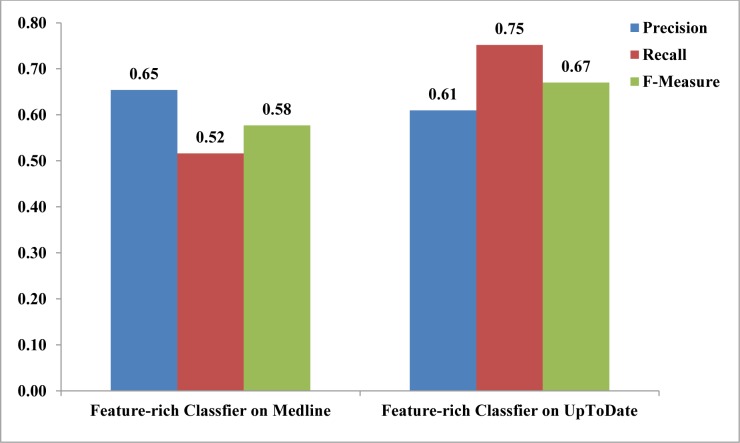
The F-measure for the feature-rich sentence classifier on the Medline dataset was 58% versus 67% on UpToDate (p<0.01) (Figure 1).
Figure 1:
Average precision, recall and F-measure of the feature-rich sentence classifier on UpToDate (from a previous study 17) and Medline sentences.
Hypothesis #2: Re-training the feature-rich classifier on the primary literature improves performance compared to the original classifier
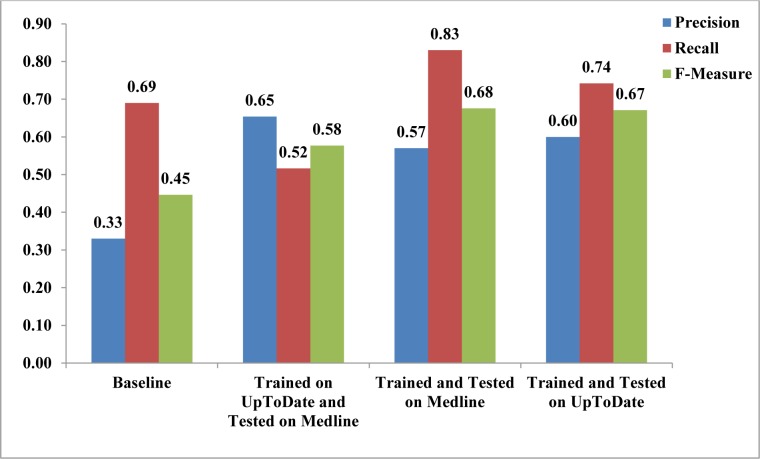
The re-trained feature-rich classifier performed significantly better than the original classifier on Medline sentences and the baseline (F-measure = 68% versus 58% and 45% respectively; p<0.001 for both comparisons) (Figure 2). Moreover, the performance of the re-trained feature-rich classifier on Medline abstracts was comparable to the performance of the feature-rich classifier on UpToDate sentences (F-measure = 68% versus 67%; p=0.53).
Figure 2:
Average precision, recall and F-measure of the baseline method compared with the feature-rich sentence classifier in different training and testing settings.
Hypothesis #3: Adding sentence location to the feature-rich classifier improves its performance on the primary literature
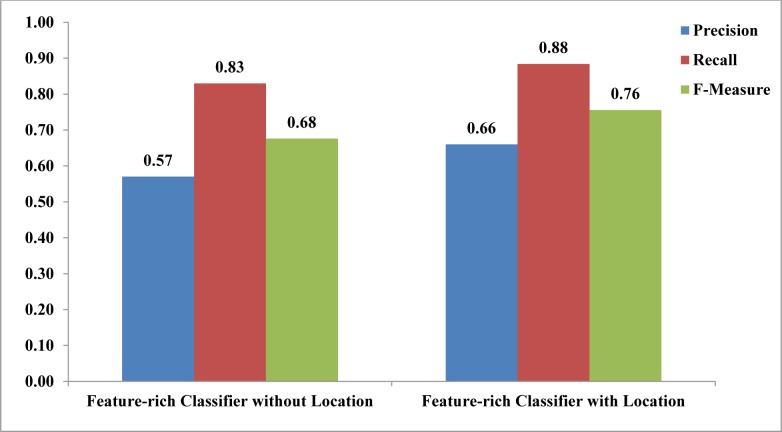
As seen in Figure 3, adding the location feature further improved the classifier performance (F-measure = 76% versus 68%, p<0.01).
Figure 3:
Average precision, recall and F-measure of the feature-rich sentence classifier, with and without location feature, and trained and tested on Medline sentences.
Discussion
This study investigated an automated method for extracting clinically useful sentences from primary literature resources such as Medline. To achieve this goal, we employed and adapted a feature-rich sentence classification model developed in a previous study. Such a method can be used in clinical decision support tools that use automatic summarization to help clinicians integrate findings from the primary literature into their decision making routine. We are currently integrating the optimized sentence classifier into one of these tools, known as the Clinical Knowledge Summary (CKS)31, 32. The CKS automatically summarizes patient-specific evidence from multiple resources and can be integrated with electronic health record (EHR) systems through the Health Level Seven (HL7) Context-Aware Knowledge Retrieval (Infobutton) Standard33, 34.
We conducted three experiments to test different hypotheses. The first experiment showed that the classifier, trained on UpToDate sentences, loses accuracy when applied to Medline sentences. Specifically, the classifier’s precision significantly decreased on Medline compared to UpToDate, although its performance in terms of recall was equivalent. A possible reason is that Medline sentences have different syntactic and semantic structure from UpToDate sentences. UpToDate provides recommendations based on synthesis of the evidence provided by multiple studies (e.g., “In patients resistant to initial therapy with hydroxychloroquine (HCQ) or sulfasalazine (SSZ), we suggest adding methotrexate (MTX) or treating with a combination of HCQ, SSZ, and MTX, rather than switching to a TNF inhibitor or to a TNF inhibitor plus MTX.”). Original studies provide a conclusion of the study findings, but in most cases there is no clear recommendation for clinical practice (e.g., “In this treatment-refractory population, tofacitinib with methotrexate had rapid and clinically meaningful improvements in signs and symptoms of rheumatoid arthritis and physical function over 6 months with manageable safety.”)
In the second experiment, re-training the classifier on Medline sentences with the exact same features resulted in improved performance, equivalent to performance on UpToDate sentences. This finding confirms that the classifier’s model and features used for UpToDate are generalizable to Medline. Also, the retrained classifier outperformed a baseline classifier, which was just based on sentence location. This shows that advanced classification methods based on NLP techniques and machine learning algorithms are worth the gained performance and classification power. The last experiment confirmed the hypothesis that sentence location in Medline abstracts further improves classification performance. This finding was expected, since study authors often summarize the main study findings and their clinical implications in the conclusion section of Medline abstracts.
Analysis of false-positives and false-negatives showed two main error categories. The first category includes recommendations that were too general, such as in “Drug therapy is recommended to stabilize and relieve symptoms in patients with preserved ventricular function.” Future studies can try to address this issue by identifying general treatment concepts using UMLS concept hierarchies. The second category was clinically useful sentences for which SemRep and MedTagger were unable to extract predications and concepts, such as in “Augment™ may represent a safe and efficacious treatment alternative to ABG during foot and ankle arthrodesis.” Fine tuning of NLP methods are needed to address this kind of problem.
Limitations
The main limitation of this study is the use of Medline abstracts as opposed to full-text articles. Medline abstracts do not report all the conclusions of a study, therefore sentence classification is limited to clinically useful sentences available in the abstract. Moreover, the gold standard consisted of high quality clinical studies published in high impact journals, which have a higher rate of structured abstracts than other studies in Medline. Since the sentence classifier benefits from standardized abstract structure, the performance of the optimized classifier applied to a dataset with a higher rate of unstructured abstracts is likely to be lower.
Future studies
We are integrating the feature-rich sentence classifier with an interactive clinical decision support tool that provides patient-specific summaries of clinical evidence from UpToDate and Medline.31 Future studies also include applying and adapting the sentence classification method to full-text articles.
Conclusion
We investigated the generalizability of a feature-rich sentence classification model, which was trained on UpToDate sentences, to Medline abstracts. The feature-rich classifier’s model and input features were generalizable to sentences from Medline abstracts, but the classifier had to be retrained on those sentences to achieve equivalent performance. Optimization of the classifier by adding a sentence location feature improved classification performance. The resulting sentence classifier can be used as a component of text summarization systems to help clinicians’ patient care decision-making.
Acknowledgments
This project was supported by grants 1R01LM011416-01 and 4R00LM011389-02 from the National Library of Medicine.
Footnotes
References
- 1.Hoogendam A, Stalenhoef AF, Robbe PF, Overbeke AJ. Analysis of queries sent to PubMed at the point of care: observation of search behaviour in a medical teaching hospital. BMC Med Inform Decis Mak. 2008;8:42. doi: 10.1186/1472-6947-8-42. Epub 2008/09/26. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aphinyanaphongs Y, Statnikov A, Aliferis CF. A comparison of citation metrics to machine learning filters for the identification of high quality MEDLINE documents. J Am Med Inform Assoc. 2006 Jul-Aug;13(4):446–55. doi: 10.1197/jamia.M2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstam EV, Herskovic JR, Aphinyanaphongs Y, Aliferis CF, Sriram MG, Hersh WR. Using citation data to improve retrieval from MEDLINE. J Am Med Inform Assoc. 2006 Jan-Feb;13(1):96–105. doi: 10.1197/jamia.M1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kilicoglu H, Demner-Fushman D, Rindflesch TC, Wilczynski NL, Haynes RB. Towards automatic recognition of scientifically rigorous clinical research evidence. J Am Med Inform Assoc. 2009 Jan-Feb;16(1):25–31. doi: 10.1197/jamia.M2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lokker C, Haynes RB, Wilczynski NL, McKibbon KA, Walter SD. Retrieval of diagnostic and treatment studies for clinical use through PubMed and PubMed’s Clinical Queries filters. J Am Med Inform Assoc. 2011 Sep-Oct;18(5):652–9. doi: 10.1136/amiajnl-2011-000233. Epub 2011/06/18. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montori VM, Wilczynski NL, Morgan D, Haynes RB, Hedges T. Optimal search strategies for retrieving systematic reviews from Medline: analytical survey. BMJ. 2005 Jan 8;330(7482):68. doi: 10.1136/bmj.38336.804167.47. Epub 2004/12/28. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Y, Liu F, Simpson P, Antieau L, Bennett A, Cimino JJ, et al. AskHERMES: An online question answering system for complex clinical questions. J Biomed Inform. 2011 Apr;44(2):277–88. doi: 10.1016/j.jbi.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang X, Lin J, Demner-Fushman D. Evaluation of PICO as a knowledge representation for clinical questions. AMIA Annu Symp Proc. 2006:359–63. Epub 2007/01/24. eng. [PMC free article] [PubMed] [Google Scholar]
- 9.Mishra R, Bian J, Fiszman M, Weir CR, Jonnalagadda S, Mostafa J, et al. Text summarization in the biomedical domain: A systematic review of recent research. J Biomed Inform. 2014 Dec;52C:457–67. doi: 10.1016/j.jbi.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook DA, Sorensen KJ, Hersh W, Berger RA, Wilkinson JM. Features of effective medical knowledge resources to support point of care learning: a focus group study. PLoS One. 2013;8(11):e80318. doi: 10.1371/journal.pone.0080318. Epub 2013/11/28. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Fiol G, Workman TE, Gorman PN. Clinical questions raised by clinicians at the point of care: a systematic review. JAMA Intern Med. 2014 May;174(5):710–8. doi: 10.1001/jamainternmed.2014.368. [DOI] [PubMed] [Google Scholar]
- 12.Hoogendam A, Stalenhoef AF, Robbe PF, Overbeke AJ. Answers to questions posed during daily patient care are more likely to be answered by UpToDate than PubMed. J Med Internet Res. 2008;10(4):e29. doi: 10.2196/jmir.1012. Epub 2008/10/18. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sayyah Ensan L, Faghankhani M, Javanbakht A, Ahmadi SF, Baradaran HR. To compare PubMed Clinical Queries and UpToDate in teaching information mastery to clinical residents: a crossover randomized controlled trial. PLoS One. 2011;6(8):e23487. doi: 10.1371/journal.pone.0023487. Epub 2011/08/23. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shariff SZ, Bejaimal SA, Sontrop JM, Iansavichus AV, Weir MA, Haynes RB, et al. Searching for medical information online: a survey of Canadian nephrologists. J Nephrol. 2011 Nov-Dec;24(6):723–32. doi: 10.5301/JN.2011.6373. Epub 2011/03/02. eng. [DOI] [PubMed] [Google Scholar]
- 15.Sheets L, Callaghan F, Gavino A, Liu F, Fontelo P. Usability of selected databases for low-resource clinical decision support. Appl Clin Inform. 2012;3(3):326–33. doi: 10.4338/ACI-2012-06-RA-0024. Epub 2012/01/01. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thiele RH, Poiro NC, Scalzo DC, Nemergut EC. Speed, accuracy, and confidence in Google, Ovid, PubMed, and UpToDate: results of a randomised trial. Postgrad Med J. 2010 Aug;86(1018):459–65. doi: 10.1136/pgmj.2010.098053. Epub 2010/08/17. eng. [DOI] [PubMed] [Google Scholar]
- 17.Morid MA, Fiszman M, Jonnalagadda S, Raja K, Del Fiol G. Classification of Clinically Useful Sentences in Clinical Evidence Resources. J Biomed Inform Under Review. doi: 10.1016/j.jbi.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Y-L, Wang R, Kwong S, Wang X-Z. Bayesian classifiers based on probability density estimation and their applications to simultaneous fault diagnosis. Information Sciences. 2014;259:252–68. [Google Scholar]
- 19.Murakami Y, Mizuguchi K. Applying the Naïve Bayes classifier with kernel density estimation to the prediction of protein–protein interaction sites. Bioinformatics. 2010;26(15):1841–8. doi: 10.1093/bioinformatics/btq302. [DOI] [PubMed] [Google Scholar]
- 20.McCray AT, Burgun A, Bodenreider O. Aggregating UMLS semantic types for reducing conceptual complexity. Studies in health technology and informatics. 2001;(1):216–20. [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H, Wagholikar K, Jonnalagadda S, Sohn S. Integrated cTAKES for concept mention detection and normalization. Proceedings of the ShARe/CLEF Evaluation Lab. 2013 [Google Scholar]
- 22.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003 Dec;36(6):462–77. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Klein D, Manning CD. Accurate unlexicalized parsing. Proceedings of the 41st Annual Meeting on Association for Computational Linguistics. 2003;1:423–30. [Google Scholar]
- 24.Levy R, Andrew G. Tregex and Tsurgeon: tools for querying and manipulating tree data structures. Proceedings of the fifth international conference on Language Resources and Evaluation. 2006:2231–4. [Google Scholar]
- 25.Kilicoglu H, Shin D, Fiszman M, Rosemblat G, Rindflesch TC. SemMedDB: a PubMed-scale repository of biomedical semantic predications. Bioinformatics. 2012 Dec 1;28(23):3158–60. doi: 10.1093/bioinformatics/bts591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishra R, Del Fiol G, Kilicoglu H, Jonnalagadda S, Fiszman M. Automatically extracting clinically useful sentences from UpToDate to support clinicians’ information needs. AMIA Annu Symp Proc. 2013;2013:987–92. [PMC free article] [PubMed] [Google Scholar]
- 27.Hoogendam A, de VriesRobbe PF, Overbeke AJ. Comparing patient characteristics, type of intervention, control, and outcome (PICO) queries with unguided searching: a randomized controlled crossover trial. J Med Libr Assoc. 2012 Apr;100(2):121–6. doi: 10.3163/1536-5050.100.2.010. Epub 2012/04/20. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schardt C, Adams MB, Owens T, Keitz S, Fontelo P. Utilization of the PICO framework to improve searching PubMed for clinical questions. BMC Med Inform Decis Mak. 2007;7:16. doi: 10.1186/1472-6947-7-16. Epub 2007/06/19. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan XF, Ni Q, Wei JP, Xu H. Evidence-based practice method of integrative Chinese and Western medicine based on literature retrieval through PICO question and complementary and alternative medicine topics. Chin J Integr Med. 2010 Dec;16(6):542–8. doi: 10.1007/s11655-010-0570-5. Epub 2010/11/27. eng. [DOI] [PubMed] [Google Scholar]
- 30.Demšar J. Statistical comparisons of classifiers over multiple data sets. The Journal of Machine Learning Research. 2006;7:1–30. [Google Scholar]
- 31.Del Fiol G, Mostafa J, Pu D, Medlin R, Slager S, Jonnalagadda S, et al. Formative evaluation of a patient-specific clinical knowledge summarization tool. Under review. doi: 10.1016/j.ijmedinf.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Del Fiol G, Pu D, Weir CR, Medlin R, Jonnalagadda S, Mishra R, et al., editors. Workshop of Interactive Systems in Healthcare. Washington, DC: 2014. Iterative design of an Interactive Clinical Evidence Summarization Tool; p. 2014. [Google Scholar]
- 33.Del Fiol G, Curtis C, Cimino JJ, Iskander A, Kalluri AS, Jing X, et al. Disseminating context-specific access to online knowledge resources within electronic health record systems. Stud Health Technol Inform. 2013;192:672–6. [PMC free article] [PubMed] [Google Scholar]
- 34.Del Fiol G, Huser V, Strasberg HR, Maviglia SM, Curtis C, Cimino JJ. Implementations of the HL7 Context-Aware Knowledge Retrieval (“Infobutton”) Standard: challenges, strengths, limitations, and uptake. J Biomed Inform. 2012 Aug;45(4):726–35. doi: 10.1016/j.jbi.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]