Abstract
Clinical laboratory results are stored in electronic health records (EHRs) as structured data coded with local or standard terms. However, laboratory tests that are performed at outside laboratories are often simply labeled “outside test” or something similar, with the actual test name in a free-text result or comment field. After being aggregated into clinical data repositories, these ambiguous labels impede the retrieval of specific test results. We present a general multi-step solution that can facilitate the identification, standardization, reconciliation, and transformation of such test results. We applied our approach to data in the NIH Biomedical Translational Research Information System (BTRIS) to identify laboratory tests, map comment values to the LOINC codes that will be incorporated into our Research Entities Dictionary (RED), and develop a reference table that can be used in the EHR data extract-transform-load (ETL) process.
Introduction
Laboratory test results are stored in electronic health records (EHRs) as structured data coded with local or standard terms. These formally coded laboratory tests can facilitate retrieval and reuse of EHR data. However, laboratory tests conducted at an outside laboratory are often simply labeled or coded as “outside test” or something similar, with the actual name included in the free text result, comment, or note (e.g. “Test requested: Lyme Disease Serology Test performed at: Mayo Medical Laboratories Rochester, MN Test result: Lyme Disease Serology, S - Negative Expected values – Negative”). As a result, these outside laboratory tests with nonspecific names cannot be differentiated during retrieval, impeding tasks such as patient care, data sharing, integration, analysis, and decision support. Manual clarification of such data is tedious and redundant.
Our goal is to develop a generalized method to make outside unspecific laboratory data available for secondary use. Our approach seeks to code nonspecific tests with appropriate codes and standard terms from the Logical Observation Identifiers Names and Codes1 (LOINC) using a fuzzy matching approach2 that comprises four steps: 1) identify outside laboratory tests results, 2) based on the text fields, map the test to a specific standard LOINC code, 3) develop local codes, 4) recode outside unspecific laboratory test results proactively when loading new data into the EHR. We demonstrate this approach with the Biomedical Translational Research Information System3 (BTRIS), a repository of EHR data at the National Institutes of Health (NIH).
Background
Logical Observation Identifiers, Names and Codes (LOINC)
LOINC, a standard for reporting clinical observations (including laboratory test results) used in EHR systems, includes names and identifiers for more than 68,000 medical terms. Approximately 300 LOINC codes cover more than 95% of laboratory test orders in the U.S.4 Based upon available laboratory information (e.g. name, unit of measure), we can use the LOINC Mapping Assistant (RELMA5) to match LOINC codes with local codes. Over the past two decades, the benefits of LOINC mapping for EHR interoperability have been well discussed, as have the challenges in mapping practice6, 7, 8. Basic LOINC mapping guidelines are derived from an accumulated experience with MIMIC-II lab codes7.
The Biomedical Translational Research Information System (BTRIS)
BTRIS is a clinical research data repository at the National Institutes of Health (NIH) that collects EHR data from over 50 NIH sources3. Laboratory data are obtained from multiple systems including the current hospital laboratory system, archived data from previous systems, and institutional clinical trials management systems. All of these sources include results from outside laboratories (e.g. Mayo Medical Laboratories).
All data in BTRIS are coded with the NIH’s Research Entities Dictionary (RED), a terminology resource that includes the concepts related to laboratory tests or panels in five categories (chemistry, hematology, immunology/flow cytometry, microbiology, and transfusion medicine). Like other RED concepts, laboratory test concepts have properties, roles, and associations to represent comprehensive knowledge about the tests they represent.
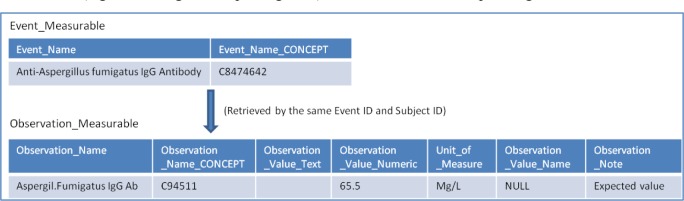
BTRIS uses a hybrid relational and Entity–Attribute–Value (EAV) database model3, in which most laboratory data are represented with columns in two tables: Event_Measurable and Observation_Measurable. The Event_Measurable table contains laboratory order information with orderable laboratory tests and panels stored in the Event_Name column along with their RED concepts in the Event_Name_CONCEPT column. Laboratory test results are recorded in the Observation_Measurable table. Laboratory finding values (results) are in Observation_Value_Text or Observation_Value_Numeric columns. Additional textual information is stored in Observation_Note and Observation_Value_Name columns. Figure 1 displays a sample of the two tables. Additional information (e.g. status flags and reporting time) are stored in the corresponding EVA tables.
Figure 1.
Sample data on the Event_Measurable and Observation_Measurable tables (selected columns only).
Fuzzy Lookup Transformation
Fuzzy matching or lookup uses mathematical processes to determine the similarities between strings and to find similar (non-exact) matches. We expect this method to be sufficient for matching newly received results with similar previously encountered results, since they typically share similar patterns and formats. Fuzzy matching can be expected to overcome minor differences such as unique numeric results, misspellings, or inconsistent abbreviation usage. For example “Aspergillus fumigatus Ab IgE 78.4 ku/l laboratory” and “Aspergillus Fumigatus Aby Ige 36 Ku/L” will be readily recognized as matching.
The development of fuzzy technology has a wide range of potential uses in health care (e.g. diagnosis, decision making); its role in EHR data cleaning and standardization is relevant to the current project9–12. EHR data extract-transform-load (ETL) processes consolidate disparate clinical data into a data repository3, 13. Microsoft SQL Server Integration Services (SSIS) features a fuzzy lookup transformation in the ETL package for data cleaning and standardization2, 14. We plan to use the SSIS platform to perform fuzzy matching to recode unspecific laboratory data and incorporate it into the ETL processes to check new data during real-time data loading.
Methods
As previously described, the nonspecific laboratory tests are those conducted at an outside laboratory and the results are stored in clinical databases without specific test names. Our goal is to develop a generalized method that makes outside unspecific laboratory data available for secondary use. The following illustrates our four-step approach.
Step 1: Identifying nonspecific outside laboratory tests
To find unspecified outside (or reference) laboratory tests, we looked in the RED for suspicious local terms referring to outside laboratory tests (e.g. “ref lab”). We then obtained actual laboratory results from BTRIS that were coded with these terms.
Step 2: LOINC mapping
We selected samples from these laboratory test results and designed mapping strategies according to laboratory test types. For microbiology tests, our initial focus in the study, we extracted the laboratory test information from Observation_Value_Text or Observation_Note. After a simple normalization (e.g. spacing and capitalization), we used regular expression functions to parse comment strings into four parts: 1) Test requested; 2) Test results; 3) Laboratory location; and 4) Additional information. After removing duplicate “test requested” strings, we used test names, laboratory locations, and test results for LOINC mapping.
For tests performed at a known laboratory (e.g., Mayo Medical Laboratories), we reviewed the laboratory’s Web site to obtain test names, synonyms and LOINC codes where they were available. We used RELMA to map additional test terms to LOINC codes by matching main parts (e.g. components measured, the unit of amount of substance, timing and sample type) with available information found in test results 1, 7. For example, for the test name “B. burgdorferi ELISA serum,” we obtained information from the result “IgM-Negative and IgG-Negative” and matched 4 LOINC parts: component (B. burgdorferi Ab.IgG & IgM), method (ELISA), system (serum), and Scale (Ord). We classified coding results as either “mapped”, “likely mapped (ambiguous match)” or “not mapped”.
Step 3: Developing local codes
We checked the RED for existing test terms with the corresponding LOINC codes; where they did not exist, we created new local test terms and annotated them with information about the outside laboratory tests.
Step 4: Developing a reference table for fuzzy lookup transformation
Based on the results of Steps 2 and 3, we created a reference table to associate the text results of outside laboratory tests to the mapped LOINC codes and local codes, which will be used for fuzzy lookup transformation in an ETL process to code new data.
Results
Identified unspecific outside laboratory tests in BTRIS
We found that the majority of outside laboratory tests had names that conveyed what the tests were being performed (e.g. “Anti-Influenza Virus B IgG Antibody Serum Test by Mayo”). We found that nonspecific tests most often included the word “other” in their names. Table 1 shows the summary statistics of distinct RED concepts and patient rows for all the records associated with RED concepts for outside laboratory tests in the Event_Measurable table.
Table 1.
Counts of distinct RED concepts and incidences in the event table.
| RED Concepts | Patient Rows | |
|---|---|---|
| All lab orders | 8,549 | 89,105,930 |
| Identified outside lab | 881 | 252,361 |
| Outside unspecific lab | 27 | 32,590 |
We identified a total of 27 RED concepts used to code outside unspecific laboratory tests. Among them, 4 RED concepts are associated with microbiology tests: “other microbiology test,” “other mayo clinic microbiology Test,” “other micro mayo contract laboratory test,” and “other micro AML Test.”
We extracted patient rows from the joined tables of Event_Measurable and Observation_Measurable where the value of Event_Name_CONCEPT was one of these 27 concepts. As a result, we obtained 14,082 unspecified laboratory test results that are stored in Observation_Note and Observation_Value_Text columns. We found each of the microbiology tests had a specific, consistent report pattern, including units, and reference ranges, although naturally the values and interpretations varied.
LOINC mapping
From the “other microbiology test”, we selected 1,000 patient data rows as a convenience sample data set and extracted 342 unique laboratory test names from the comment fields. Of these, 298 test names were from 16 known laboratories (Table 2). We mapped 329 of 343 (95.9%) laboratory tests to 102 unique LOINC codes through laboratories’ Web sites and RELMA. No match was found for 14 tests because of the lack of minimal information, or a new LOINC code required (see examples in Table 3).
Table 2.
LOINC mapping results.
| Laboratory | Test Names | LOINC Codes | |||
|---|---|---|---|---|---|
| Website | RELMA | Unmapped | Unique Code | ||
| Mayo Medical Laboratories | 106 | 100 | 1 | 5 | 48 |
| Beacon Diagnostics Laboratory | 61 | 61 | 1 | ||
| Unknown | 45 | 42 | 3 | 31 | |
| Quest/Focus Diagnostics | 40 | 39 | 1 | 28 | |
| MiraVista Diagnostics | 28 | 28 | 6 | ||
| Fungus ing Lab, University of Texas | 11 | 11 | 4 | ||
| Johns Hopkins University | 11 | 11 | 1 | ||
| ViroMed Laboratories | 11 | 11 | 3 | ||
| Centers for Disease Control and Prevention | 9 | 8 | 1 | 6 | |
| Maryland State Lab | 6 | 4 | 2 | 3 | |
| National Jewish Health | 5 | 3 | 2 | 1 | |
| Specialty Laboratories | 3 | 3 | 3 | ||
| Center for Anti-Infective Research, Hartford, CT | 2 | 2 | 2 | ||
| University Of Minnesota | 2 | 2 | 1 | ||
| Immunetics | 1 | 1 | 1 | ||
| Palo Alto Medical Foundation | 1 | 1 | 1 | ||
| American Medical Laboratories | 1 | 1 | 1 | ||
Table 3.
Examples of unmapped test terms.
| Test Requested | Test Results | Comments |
|---|---|---|
| Beta-D-Glucan | No Content | Lack information to determine the test |
| Blastomyces Ab, EIA, S | Negative | In process of code application (or mapping) |
| Anti-mycobacterial Drug Panel | Ethambutol Level 0.97 Micrograms/Ml Isoniazid Level 0.94 Micrograms/Ml Pyrazinamide Level 14.46 Micrograms/Ml |
In process. No panel for antimycobacterial susceptibility testing |
| Ova and Parasite Identification | Microsporidia Detected By PCR | The result indicates a test ‘Microsporidia Molecular Identification,’ that can not be inferred from ‘Q&P’. No match in LOINC using ‘PCR.’ Also, this example shows the inconsistency between lab test and result. |
| West Nile PCR | West Nile Virus, Eastern Equine Encephalitis Virus Saint Louis Encephalitis Virus, Lacrosse Virus Not detected by RT-PCR. |
A full arboviral panel conducts when a West Nile is ordered. |
| Bordetella Pertussis Antibodies, Igg | B. pertussis Ab, Igg W/Reflex 2.0 U/Ml B. pertussis Ab, Igm W/Reflex 1.2 U/Ml, B. pertussis Ab, Igm Immunoblot: No Igm Antibodies A\against Bordetella FHA and PT detected. |
This is a reflex test. The naming of panels with reflex components in LOINC has not been addressed.1 |
Developing local codes
We added 102 new terms to the RED when no laboratory test name had been represented in the RED, based on mapping and information from the outside laboratory.
Creating a reference table for fuzzy lookup transformation
Multiple columns are included on the reference table for fuzzy lookup transformation, including local code, local term, full text string, LOINC code and name for laboratory results. Table 4 shows the structure of the reference table with sample data.
Table 4.
Transformation reference table.
| Local Code | Local Term | FULL TEXT Sample | Text Requested | Ref. Lab | LOINC Code | LOINC Common Name | LOINC Short Name |
|---|---|---|---|---|---|---|---|
| C92484111 | RED | Test requested: Aspergillus fumigatus AB IgE (Mayo) Test result:36.2 kU/L Class 4 (Strongly Positive 17.6–50.0) Test | Aspergillus Fumigatus Ab Ige | Mayo | 6025-1 | Aspergillus fumigatus IgE Ab [Units/volume] in SerumA fumigatus IgE | A fumigatus IgE Qn |
| C92484121 | RED | Test requested: Azithromycin Level (National Jewish) Test result: 1.68 mcG/mL For full | Azithromycin Level | NJH | 25233-8 | Azithromycin [Mass/volume] in Unspecified specimen | Azithromycin XXX-mCnc |
| C92484133 | RED | TEST REQUEST: NEISSERIA GONORRHOEA DNA PROBE TEST RESULT: NEGATIVE TEST PERFORMED AT AMERICAN | Neisseria Gonorrhoea Dna Probe | AML | 24111-7 | Neisseria gonorrhoeae DNA [Presence] in Unspecified specimen by Probe and target method | N gonorrhoea DNA XXX Ql PCR |
| C92484212 | RED | TEST REQUEST: COCCIDIOIDES ANTIBODY TEST RESULTS: 1:2(4+), 1:4(4+), 1:8(4+), 1:16(3+), 1:32(0), 1:64(0), 1:128(0), | Coccidioides Antibody | unknown | 31048-2 | Coccidioides immitis Ab [Presence] in Body fluid by Complement fixation | C immitis Ab Fld Ql CF |
| C92484123 | RED | TEST REQUEST:HHSV-8 PCR-SPECIALTY LABORATORIES TEST RESULT: NOT DETECTED TEST PRFORMED BY SPECIALIY | Hhsv-8 Pcr | Specialty | 49403-9 | Herpes virus 8 DNA [#/volume] (viral load) in Unspecified specimen by Probe and target method | HHV8 DNA # XXX PCR |
| C92484549 | RED | TEST REQUEST:MYCOBACTERIUM TUBERCULOSIS AMPLIFIED DIRECT TEST TEST RESULY: NO MYCOBACTERIUM | Mycobacterium Tuberculosis Amplified Direct | Mayo | 17296-5 | Mycobacterium tuberculosis complex rRNA [Presence] in Unspecified specimen by DNA probe | MTB Cmplx rRNA XXX Ql Prb |
Discussion
Data in EHRs are often uncoded or coded to a level that is inadequate for re-use. Outside laboratory test results, such as those in our study, are commonly found in EHRs and are particularly troublesome, especially when attempting to merge such data with data that are more explicitly coded. In this study, we demonstrated that such tests do actually have appropriate LOINC codes and automated methods can be used to achieve mapping where neither the outside laboratory nor the receiving laboratory system are able to do so.
LOINC mapping
The LOINC code is an intermediate between local and outside laboratory tests. Our mapping is based on textual comments reported by the outside laboratory as results that actually include test names and results, which are different from mapping local laboratory tests based on the names from data dictionary7. Our mapping strategy is a specific-general approach. We choose a specific LOINC code for the laboratory result with multi-part matching. Considering test results with less information, we prefer a relatively general code that can cover laboratory results with similar reporting patterns. The laboratory results from well-known laboratories use similar report formats for specific tests, so that the mapping results can be shareable within any laboratory system that obtains results from these laboratories. Our general mapping strategy produces a high success rate; however, we do not consider the possibilities of multiple LOINC codes and the influences of specific methods used in the laboratory tests. Although our mappings appear correct, we have not conducted a formal evaluation of their accuracy.
Local coding
We need to add new terms or codes into the list of laboratory tests, if these terms are not in an EHR system. In clinical data repositories within the i2b2 model, the i2b2 ontology management cell manages new test names under accurate concept paths15. The RED concept structure allows the representation of outside laboratory test information in “synonym” properties and their associated attributes as local data sources. However, when we add as many new terms as we can identify from unspecific outside laboratory test results, we bear the burden of the need for additional concepts and an efficient method to manage the concepts.
Fuzzy lookup transformation
Microsoft SQL server integration services (SSIS)2, 14 provides a platform for ETL processes that supports routines for outside laboratory test results transformation into the daily work flow. In BTRIS, we will apply the featured fuzzy lookup function in the ETL processes to identify unspecified laboratory results to match them with previously encountered text result (perhaps differing only by numeric result or punctuation). The input data source is the file with original outside laboratory results. The program will produce similarity scores for each sample in the reference table. The input data will be added to the output file with the specific laboratory test name with the highest similarity score above a given threshold. So far, we have worked on sample data retrieved from BTRIS. Additional experiments on new incoming laboratory data are needed.
Conclusion
We identified the meanings of outside unspecified laboratory tests results using medical standards and developed an outside laboratory test reference table for use in fuzzy lookup transformation processes. This study suggests that a modest effort can lead to improved coding of these outside nonspecific laboratory data, such that we do not have to settle for having a portion of the patient’s records be unusable but rather can bring these additional data to bear on data re-use tasks. The heterogeneous and dynamic nature of EHR data reminds us of the challenges in the implementation of standards.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM) and Lister Hill National Center for Biomedical Communications (LHNCBC). This research was also supported in part by an appointment to the NLM Research Participation Program, administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the US Department of Energy (DoE) and the NLM.
Footnotes
Disclaimer
The views and opinions of the authors expressed herein do not necessarily state or reflect those of the National Library of Medicine, National Institutes of health or the US Department of Health and Human Services.
Competing Interests
None
References
- 1.McDonald C, Huff S, Deckard J, et al. Logical Observation Identifiers Names and Codes (LOINC®) users’ guide. 2014. Available from: http://loinc.org/downloads/files/LOINCManual.pdf.
- 2.Microsoft Corporation Fuzzy Lookups and groupings provide powerful data cleansing capabilities. Available from: https://msdn.microsoft.com/en-us/magazine/cc163731.aspx.
- 3.Cimino JJ, Ayres EJ, Remennik L, Rath S, Freedman R, Beri A, Chen Y, Huser V. The National Institutes of Health’s Biomedical Translational Research Information System (BTRIS): design, contents, functionality and experience to date. J Biomed Inform. 2014 Dec;52:11–27. doi: 10.1016/j.jbi.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Regenstrief Institute A universal code system for tests, measurements, and observations. Available from. www.loinc.org.
- 5.Regenstrief Institute RELMA® Regenstrief LOINC® Mapping Assistant user manual. Available from: http://loinc.org/downloads/files/RELMAManual.pdf.
- 6.Baorto DM, Cimino JJ, Parvin CA, Kahn MG. Using Logical Observation Identifier Names and Codes (LOINC) to exchange laboratory data among three academic hospitals. Proc AMIA Annu Fall Symp. 1997:96–100. [PMC free article] [PubMed] [Google Scholar]
- 7.Abhyankar S, Demner-Fushman D, McDonald CJ. Standardizing clinical laboratory data for secondary use. J Biomed Inform. 2012 Aug;45(4):642–50. doi: 10.1016/j.jbi.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee LH, Groß A, Hartung M, Liou DM, Rahm E. A multi-part matching strategy for mapping LOINC with laboratory terminologies. J Am Med Inform Assoc. 2014 Sep-Oct;21(5):792–800. doi: 10.1136/amiajnl-2013-002139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres A, Nieto JJ. Fuzzy logic in medicine and bioinformatics. J Biomed Biotechnol. 2006;2:91908. doi: 10.1155/JBB/2006/91908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joffe E, Byrne MJ, Reeder P, Herskovic JR, Johnson CW, McCoy AB, Sittig DF, Bernstam EV. A benchmark comparison of deterministic and probabilistic methods for defining manual review datasets in duplicate records reconciliation. J Am Med Inform Assoc. 2014 Jan-Feb;21(1):97–104. doi: 10.1136/amiajnl-2013-001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adamusiak T, Shimoyama N, Shimoyama M. Next generation phenotyping using the unified medical language system. JMIR Med Inform. 2014 Mar 18;2(1):e5. doi: 10.2196/medinform.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skeppstedt M, Kvist M, Nilsson GH, Dalianis H. Automatic recognition of disorders, findings, pharmaceuticals and body structures from clinical text: an annotation and machine learning study. J Biomed Inform. 2014 Jun;49:148–58. doi: 10.1016/j.jbi.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 13.Post AR, Krc T, Rathod H, Agravat S, Mansour M, Torian W, Saltz JH. Semantic ETL into i2b2 with Eureka! AMIA Jt Summits Transl Sci Proc. 2013:203–207. [PMC free article] [PubMed] [Google Scholar]
- 14.Microsoft Corporation Fuzzy lookup transformation. Available from: https://msdn.microsoft.com/en-us/library/ms137786.aspx.
- 15.i2b2 design document ontology management (ONT) cell. Available from: https://www.i2b2.org/software/projects/ontologymgmt/Ontology_Design_15.pdf.