Abstract
Assigning labels from a hierarchical vocabulary is a well known special case of multi-label classification, often modeled to maximize micro F1-score. However, building accurate binary classifiers for poorly performing labels in the hierarchy can improve both micro and macro F1-scores. In this paper, we propose and evaluate classification strategies involving descendant node instances to build better binary classifiers for non-leaf labels with the use-case of assigning Medical Subject Headings (MeSH) to biomedical articles. Librarians at the National Library of Medicine tag each biomedical article to be indexed by their PubMed information system with terms from the MeSH terminology, a biomedical conceptual hierarchy with over 27,000 terms. Human indexers look at each article’s full text to assign a set of most suitable MeSH terms for indexing it. Several recent automated attempts focused on using the article title and abstract text to identify MeSH terms for the corresponding article. Despite these attempts, it is observed that assigning MeSH terms corresponding to certain non-leaf nodes of the MeSH hierarchy is particularly challenging. Non-leaf nodes are very important as they constitute one third of the total number of MeSH terms. Here, we demonstrate the effectiveness of exploiting training examples of descendant terms of non-leaf nodes in improving the performance of conventional classifiers for the corresponding non-leaf MeSH terms. Specifically, we focus on reducing the false positives (FPs) caused due to descendant instances in traditional classifiers. Our methods are able to achieve a relative improvement of 7.5% in macro-F1 score while also increasing the micro-F1 score by 1.6% for a set of 500 non-leaf terms in the MeSH hierarchy. These results strongly indicate the critical role of incorporating hierarchical information in MeSH term prediction. To our knowledge, our effort is the first to demonstrate the role of hierarchical information in improving binary classifiers for non-leaf MeSH terms.
1. Introduction
Indexing biomedical articles is an important task that has significant impact on how researchers search and retrieve relevant information. This is especially essential given the exponential growth of biomedical articles indexed by PubMed®, the main search system developed by the National Center for Biotechnology Information (NCBI). PubMed lets users search over 22 million biomedical citations available in the MEDLINE bibliographic database curated by the National Library of Medicine (NLM) from over 5000 leading biomedical journals in the world. To keep up with the explosion of information on various topics, users depend on search tasks involving Medical Subject Headings (MeSH®) that are assigned to each biomedical article. MeSH is a controlled hierarchical vocabulary created by the NLM and consists of medical subjects that form a directed acyclic graph (DAG). Once articles are indexed with MeSH terms, users can quickly search for articles that pertain to a specific subject of interest instead of relying solely on keyword based searches.
Since MeSH terms are assigned by librarians who look at the full text of an article, they capture the semantic content of an article that cannot easily be captured by keyword or phrase searches. Thus assigning MeSH terms to articles is a routine task for the indexing staff at NLM. This is empirically shown to be a complex task with 48% consistency because it heavily relies on indexers’ understanding of the article and their familiarity with the MeSH vocabulary [1]. As such, the manual indexing task takes a significant amount of time leading to delays in the availability of indexed articles. It is observed that it takes about 90 days to complete 75% of the citation assignment for new articles [2]. Moreover, manual indexing is also a fiscally expensive initiative [3]. Due to these reasons, there have been many recent efforts to develop automatic ways of assigning MeSH terms for indexing biomedical articles including an on-going indexing challenge (http://www.bioasq.org/). However, automated efforts (including ours) mostly focused on predicting MeSH terms for indexing based solely on the abstract and title text of the articles. This is because most full text articles are only available based on paid licenses not subscribed by many researchers.
Many efforts in MeSH term prediction generally rely on two different methods. The first method is the k-nearest neighbor (k-NN) approach where k articles that are already tagged with MeSH terms and whose content is found to be “close” to the new abstract to be indexed are obtained. The MeSH terms from these k articles constitute candidate terms for the new abstract [2]. A second method is based on applying machine learning algorithms to learn and index the best binary classifier models for each MeSH term. A new candidate abstract would then be put through all these classifiers and the corresponding MeSH terms of classifiers that return a positive prediction are chosen as candidate terms for the abstract. This approach has been termed as meta-learning and researchers at the NLM [4] have demonstrated that depending on the individual terms different classifiers might yield different results thus justifying the approach.
In one of their recent results, Jimeno-Yepes et al. [5] identify six MeSH terms that have been found to be difficult to predict using existing indexing approaches at the NLM. Out of these, two terms, hormones and infection, were shown to have unusually high numbers of false positive instances (with existing classification approaches) that are actually tagged with the descendants of the corresponding terms in the MeSH hierarchy. That is, there were many citations (abstract and title text) that were originally assigned a more specific descendant of an ancestor term but where the ancestor term classifier has classified each of them as a positive instance. Using this as motivation, in this paper, we explore classification methods that utilize training examples from the descendants as a way of improving non-leaf MeSH term prediction.
The rest of the paper is organized as follows. In the rest of this section, we first discuss the essential background on the size and structure of the MeSH terminology and discuss some related efforts. We also discuss a motivating scenario for incorporating descendants of non-leaf terms in building non-leaf term binary classifiers. In Section 2, we discuss our main methods that exploit descendant training instances to reduce false positive errors. We present our experiments using a large dataset and the corresponding results obtained with associated discussion in Section 3.
1.1. Background and Related Work
MeSH’s main purpose is to index biomedical articles and hence strict notions of meronymy were not used in its design; the hierarchical relationships are actually guided by “aboutness” of a child to its parent1. Hence a term could be a descendant of multiple other terms whose least common consumer is not one of them. That is, a term could have multiple paths from the root. In the 2013 version of MeSH, there are 26578 main subject headings. The scatter plot of number of descendants and depth of the term is shown in Figure 1. Note that there are 16 individual hierarchies in MeSH but the machine processable UMLS Metathesaurus2 files combine these hierarchies with additional place holder nodes to have a unified root. In the figure, the depth of the root is taken as 0 and the first valid MeSH nodes for each of the 16 hierarchies start at depth 1. From the figure, we can see that the number of descendants is in hundreds for many nodes at depths ≤ 4. By analyzing the MeSH hierarchy (Figure 2), we also found that there are at least 15,000 MeSH nodes with depth ≤ 4.
Figure 1:
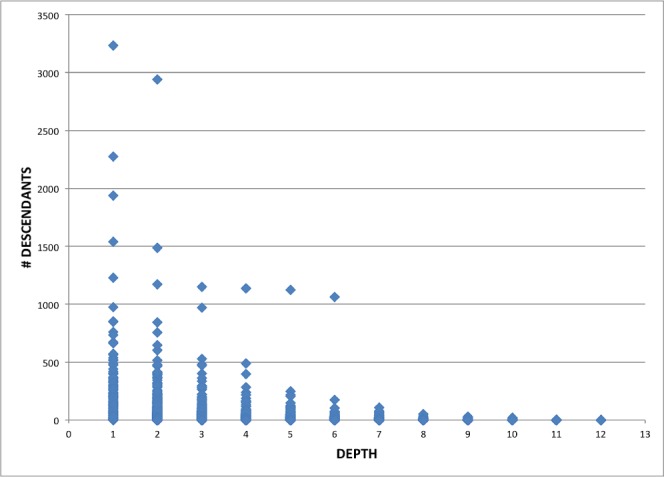
Scatter Plot of Term Depth and Number of Descendants
Figure 2:
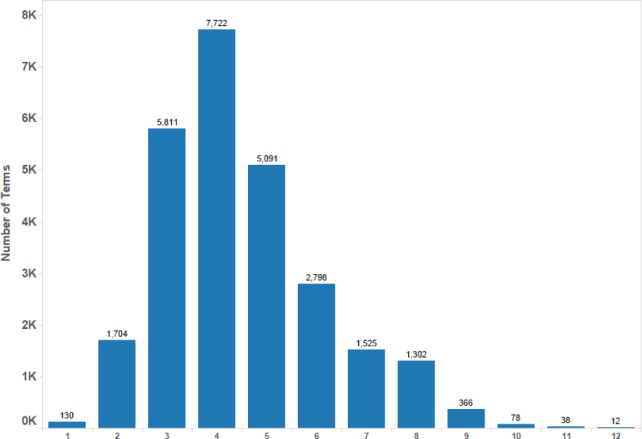
Number of MeSH Terms at a Given Depth on X-axis
NLM initiated efforts in automatic MeSH term extraction with their Medical Text Indexer (MTI) program that uses a combination of k-NN based approach and named entity recognition (NER) based approaches with other unsupervised clustering and ranking heuristics in a pipeline [6]. MTI recommends MeSH terms for NLM indexers to assist in their efforts to expedite the indexing process3. Another recent approach by Huang et al. [2] uses k-NN approach to obtain MeSH terms from a set of k already tagged abstracts and use the learning to rank approach to carefully rank the MeSH terms. They use two different gold standard datasets one with 200 abstracts and the other with 1000 abstracts and achieve an F-score of 0.5 and recall 0.7 on the smaller dataset compared to MTI’s F-score of 0.4 and recall 0.57. Vasuki and Cohen [7] also use the k-NN approach but employ reflective random indexing to find the nearest neighbors in the training dataset and use the indexing based similarity scores to rank the terms from the neighboring citations.
Several other attempts incorporated different machine learning approaches with novel feature selection [8] and training data sampling [9] techniques. In our earlier effort, we also explored purely unsupervised approaches that rely on term co-occurrence counts and named entity recognition [10], which we subsequently extended [11] to a supervised framework exploiting latent associations between MeSH headings based on reflective random indexing. A recent effort by Jimeno-Yepes et al. [4] uses a large dataset and uses meta-learning to train custom binary classifiers for each MeSH term and indexes the best performing model for each label to be applied on new abstracts; we request the reader to refer to their work for a recent review of machine learning methods used for MeSH term assignment. In a related effort [5] they identify six MeSH terms that have been found to be difficult to predict using existing indexing approaches at the NLM. In this work, we look at all non-leaf nodes (8756 ≈ 33% of all MeSH terms) in the MeSH hierarchy with varying number of descendants For these terms we develop and evaluate binary classifiers that utilize examples of their descendants. Given recent efforts [4, 12] that heavily rely on accurate binary classifiers for each term, we believe our work is very relevant to further the state-of-the-art in automated indexing of biomedical articles.
1.2. Motivating Scenario & Baseline Approach
In this section we demonstrate the presence of false positives arising from descendant nodes using the following six terms with varying descendant counts.
Membrane Proteins: 971 descendants
Neoplasms: 663 descendants
Hormones: 221 descendants
Infection: 154 descendants
Mutation: 47 descendants
Plasmodium: 10 descendants
Here Hormones and infection are from the challenging terms used in experiments in [5]; among the terms considered in these related efforts, these two had many descendants and high descendant attributable FPs. In addition to these, we chose four other terms arbitrarily with varying numbers of descendants. For our motivating scenario, we built training and testing datasets for each of these terms by extracting positive and negative examples from over 20 million Medline citations that are already fully tagged with MeSH terms by trained coders at the NLM. We made sure that our training and testing examples both had a non-empty abstract field as books and other artifacts indexed might not have an abstract. The number of positive and negative examples for each of the terms in the training and testing data are shown in Table 1.
Table 1:
Training and Testing Instance Counts
| MeSH Term | Training
|
Testing
|
||
|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |
| Membrane Proteins | 98401 | 340780 | 5674 | 255535 |
| Neoplasms | 134082 | 338308 | 4138 | 257071 |
| Hormones | 15679 | 340416 | 1026 | 260183 |
| Infection | 12930 | 340327 | 1060 | 260149 |
| Mutation | 230905 | 336020 | 5451 | 255758 |
| Plasmodium | 3206 | 343440 | 253 | 260956 |
In Table 1, the sum of positive examples in both training and testing sets for each term is equal to the total number of citations (with a non-empty abstract field) tagged with that term in the Medline citation database (2014 Baseline). We randomly selected approximately 5% of the available citations for each term to be included as the corresponding positive examples in the testing data. Most MeSH terms are tagged for very few citations and hence, we chose a very high number of negative examples as shown in the negative testing instance count column; the positive examples constitute a maximum of around 2% of the total testing dataset for each term.
For our experiments, we chose as baseline the LIBLINEAR [13] logistic regression implementation through the scikit-learn [14] framework. We used unigrams and bigrams, with a minimum frequency of five citations per n-gram, as our binary features. After applying the classifiers on the corresponding testing sets for each of the six terms, we looked at the number of FPs that can be attributed to descendants of the six terms under consideration. These observations are shown in Table 2 from which we notice that a significant number of FPs (more than 40% for four terms) are due to misclassification of descendants. Based on these observations, we set out to design and evaluate classification approaches that exploit descendant instances.
Table 2:
False Positives due to Descendants
| MeSH Term | # FPs | # Desc. FPs | % Desc. FPs |
|---|---|---|---|
| Membrane Proteins | 7547 | 3126 | 41% |
| Neoplasms | 8133 | 3533 | 44% |
| Hormones | 1349 | 790 | 56% |
| Infection | 1283 | 356 | 28% |
| Mutation | 13268 | 1733 | 13% |
| Plasmodium | 227 | 157 | 69% |
2. Methods: Hierarchical Strategies to Handle Descendant FPs
Motivated by observations in Table 2, we proceed to exploit instances of descendants terms in building better binary classifiers for non-leaf MeSH terms. Here, the descendant set of a given MeSH term is defined recursively as the union of the set of its children and all their descendants in the MeSH hierarchy. The baseline method is simply building binary classifiers with unigram and bigram features using support vector machines (SVMs). Next, we outline the five different methods that incorporate descendant instances to be compared with the baseline approach. We eventually use a dataset of one million biomedical citations and all non-leaf terms that are assigned to at least 100 citations in this dataset. Details of dataset and results achieved using our methods will be elaborated in Section 3.
Before we proceed, we first note that it is a valid indexing approach to assign a term and one or more of its descendants to the same biomedical article. NLM’s multi-label assignment process allows this when the content of the article warrants such an assignment. As such, the task of assigning MeSH terms is a relaxation of the non-mandatory leaf node prediction [15, Section 4.4] scenario encountered in hierarchical classification. Given this, to best differentiate a term’s instances from its descendants’ instances, we curate training data of ancestor terms that are not tagged with any of their descendants. Similarly, we curate training instances of descendant terms that are not tagged with the corresponding ancestors. If T is the size of the training set of a given ancestor term selected as explained above, we randomly pick T/d training instances for each of its d descendant terms to approximately create equal sized datasets. While we allow appropriate data imbalance to persist in our main experiments, we selected balanced datasets to distinguish between main terms and their descendants, with equal contribution from each descendant4. This is due to the presence of extremely large number of instances when summed over all descendants of a given term. Because of the way we incorporate these descendant instances into our methods (the rest of the section), we believe this descendant specific instance selection is appropriate given the testing data set is chosen randomly while preserving realistic imbalance. Next, we describe each of the five methods that employ descendant training instances.
2.1. Multiclass Prediction with a Third Descendant Class
Unlike the binary classification approach used in our baseline method, in this approach, for each term we model the prediction as a multiclass text classification problem with three classes: 1. main non-leaf term class of interest to us, 2. its descendants class, and finally 3. the ‘other’ class that has negative examples of all other non-descendant instances. The non-descendant ‘other’ instances are citations in our dataset that are not tagged with the non-leaf term of interest or its descendants. The positive class instances are those that are tagged with the non-leaf term but none of its descendants. The descendants’ training instances are curated as explained in the beginning of this section with equal contributions from each descendant. We used the built-in one-vs-all approach in LIBLINEAR with the same set of features used for our baseline method in Section 1.2.
2.2. Two Stage Hierarchical Prediction
Here we follow the conventional hierarchical approach and build two different classifiers. The top level binary classifier combines instances of either the main term or any of the descendants as one positive class, the negative class being the ‘other’ class from Section 2.1. Intuitively, this classifier is expected to identify testing instances of a candidate term or its descendants. Once an instance is predicted as positive with this classifier, a second binary classifier that distinguishes between the main term and its descendants’ instances is applied. This second classifier is built using the main non-leaf term class and the descendants class from Section 2.1.
2.3. Baseline Result Filtering using Descendant Classifier
Instead of applying the second stage classifier in Section 2.2 to the positive instances of the top level classifier in the hierarchical approach, we simply use it to filter the positive instances output by the baseline classifier from Section 1.2. Given the high proportion of FPs attributable to descendant instances (Table 2) and the reasonably high performance we observed in the 2nd stage classifier in the two stage hierarchical approach, we chose to do this filtering.
2.4. Baseline Filtering with Descendant Classifier and FN recovery
Our primary focus thus far has been on minimizing FPs. In this context, clearly, the percentages of total FP reduction should be looked at in the context of the recall loss due to new false negatives (FNs) induced. Just as many FPs can be attributed to descendant instances, it is also possible that several FNs can be attributed to descendants. This is because of the multi-label indexing nature where both a term and any of its descendants can be assigned to a given article as discussed in the beginning of this section. That is, many negative predictions that end up in the descendant class in Section 2.1 or of the second stage classifier in Section 2.3 could actually be positives. To reduce these descendant attributable FNs, we propose to move several of these positives from the incorrect descendant predictions to their correct ancestor predictions using a binary FN recovery classifier that distinguishes between instances that are tagged with “only descendants” (negative class) and instances that are tagged with “both the ancestor term and at least one descendant” (positive class). We apply this binary classifier to the negative predictions from the second stage classifier of the baseline filter approach (Section 2.3). Those that are classified as belonging to the class “both the ancestor term and at least one descendant” are considered positive instances of the non-leaf ancestor term in the end. To build this binary classifier, we obtain the positive instances by considering all citations tagged with the non-leaf term and any of its descendants. The negative instances dataset is again a subset of the descendant-only examples as explained at the beginning of this section.
2.5. Combining NER and Supervised Classification
Our final approach involves combining named entity recognition with supervised classification. It is also essentially a filter on positive classifications of the baseline classifier from Section 1.2. In this method, we first use a state-of-the-art biomedical named entity recognition tool, MetaMap [16], on the input citation of each positive instance output by the baseline classifier to identify MeSH terms mentioned in its title and abstract. If any of these MeSH terms is actually a descendant of the candidate term, we subject it to a corresponding new binary classifier that distinguishes between instances that are tagged with only descendants and those tagged with both a main term and at least one of its descendants (from Section 2.4). Those that are classified into the “only descendants” are treated as negatives in this filter. Intuitively, since the input instances to this filtering approach are already deemed positive for the main term (since they are the positives from the baseline classifier), we only need to identify those instances that should be exclusively tagged with descendant terms only. However, just because a citation contains a descendant MeSH term does not necessarily mean it can only take a descendant MeSH term as a tag, although it could be a candidate for further analysis. Hence we apply the supervised classifier to those that contain a descendant term.
2.6. Summary of Our Approaches
Some of our methods in Sections 2.1–2.5 involve multiple classifiers that use different training data subsets. For clarity, here we summarize all the classifiers involved and how they are used in different methods using the corresponding section numbers. We use the following notation to represent different training data subsets for our classifiers.
A: set of all training examples annotated with the main non-leaf term but none of its descendants
D: set of examples not tagged with the main non-leaf term but with at least one of its descendants
AD: examples tagged with both the non-leaf term and at least one of its descendants
O: training examples that are not in sets A, D, or AD.
Based on this notation, in Table 3(a), we identify the five different types of classifiers L1–L5 used in our methods and the corresponding training data subsets used. All classifiers are binary except L2, which is a three way classifier. For L4 and L5, the D set is sampled as elaborated in the second paragraph of Section 2. In Table 3(b), we specify the classifiers used for the baseline approach (Section 1.2) and our proposed FP handling methods in Sections 2.1–2.5.
Table 3:
Classifiers Used in Different Methods
| (a) Classifier-Dataset Map | |||||
|---|---|---|---|---|---|
| Dataset | L1 | L2 | L3 | L4 | L5 |
| A | + | 1 | + | + | |
| D | − | 2 | + | − | − |
| AD | + | + | + | ||
| O | − | 3 | − | ||
| (b) Method-Classifier Map | |||||
|---|---|---|---|---|---|
| Method | L1 | L2 | L3 | L4 | L5 |
| Baseline | ✓ | ||||
| Sec 2.1 | ✓ | ||||
| Sec 2.2 | ✓ | ✓ | |||
| Sec 2.3 | ✓ | ✓ | |||
| Sec 2.4 | ✓ | ✓ | ✓ | ||
| Sec 2.5 | ✓ | ✓ | |||
3. Results and Discussion
To ensure a natural distribution of terms, we chose a set of 1 million already annotated citations with dates of publication between 2005–2014 from around 1900 well known journals used in the BioASQ competition to index biomedical articles. Out of this dataset, 850,000 citations were used for training, 100,000 were used for validation, and the remaining 50,000 were used for testing. The testing dataset instances’ dates of publication are chronologically later than the training and validation dataset instances. Among all non-leaf terms in the MeSH hierarchy, we chose those that had at least 100 instances in the training dataset, which resulted in 4395 non-leaf terms that we considered for our experiments. Before we proceed we outline the performance measures used.
3.1. Performance Measures
Since the task of assigning multiple MeSH terms to a citation is the multi-label classification problem, there are multiple complementary methods [17] for evaluating automatic approaches for this task. Here, we focus on label based measures since we are only concerned about the performance of non-leaf MeSH terms.
For each MeSH heading Tj in the set of terms T being considered, we have label-based precision P(Tj), recall R(Tj), and F1-score F(Tj) defined as
where TPj, FPj, and FNj are true positives, false positives, and false negatives, respectively, of term Tj. Given this, the label-based macro average F1-score is
The label-based micro precision, recall, and F1-score are defined as
While macro measures consider all labels as equally important, micro measures tend to give more importance to more frequent labels. Here we are interested in improving both the micro and macro averages over non-leaf terms.
3.2. Application of Hierarchical Classifiers
We first applied our hierarchical strategies (Section 2) to the set of six terms in the motivating scenario in Section 1.2. All approaches showed some improvement except the two-stage hierarchical approach in Section 2.2, which suffered a major loss in precision. This is not surprising given recent attempts by other researchers [18] also show that top-down hierarchical approaches may not be very suitable with very large terminologies with thousands of labels. Hence all our large scale experiments described in this section are conducted with all the methods except the two-stage approach. We note that all approaches we consider here are thus essentially filters on the positive instances of the baseline. That is, we are not attempting to obtain better recall than baseline but are willing to trade off some recall for precision increases that could yield us better micro and macro F-measures.
Instead of using the default approach of making predictions based on SVM scores, we use a more suitable approach for multi-label classification based on the meta-labeler [19] method where, in addition to a ranked list of labels (based on probability estimates) for each artifact to be classified, a threshold on the number of labels is also predicted using the same set of features used for predicting the labels. This generally helps because some binary classifiers for labels with low frequencies do not always produce high scores for the positive class. We used this approach and built over 27,000 binary classifiers (one for each MeSH term even if it is not a non-leaf term) using the training dataset. We also developed another model that predicts the number of terms for an input citation based on the training dataset. Next, we applied each of these classifiers to each instance in the validation dataset. We ranked the MeSH terms for each instance based on the corresponding SVM classifier scores and chose the threshold for the number of terms to retain based on the thresholding model. Next, for each of our hierarchical strategies, if one of the 4,395 non-leaf terms that we consider in our experiments shows up in the top terms predicted using the baseline approach and the thresholding, we apply the hierarchical strategy as an additional filter. If the output of the hierarchical strategy also ends up in the positive class, we take it as a positive classification for the non-leaf term. If the strategy outputs a negative prediction, we treat the prediction to be negative for the non-leaf term. Thus, we reverse some positive non-leaf term predictions by the base classifier using the hierarchical strategies.
3.3. Results on Validation Dataset
We used the validation dataset to identify non-leaf MeSH terms for which we notice an improvement over the baseline when the hierarchical strategies are used. We then applied the strategies for only such non-leaf terms on the test dataset. This is to ensure that our choice of the terms is not influenced by the test data. Here we present our results on the validation dataset. In Table 4, for each strategy (first column, indicated by the section number), we show how many terms had better F1-score (compared with the baseline), the average increase in macro-F1 and micro-F1 over such terms, and average number of descendants and average depth for such terms. The final row is a hybrid classifier that chooses the best hierarchical strategy over the baseline for each non-leaf term.
Table 4:
Validation Dataset Results
| Method | # terms | macro-F impr | micro-F impr | #desc | #depth |
|---|---|---|---|---|---|
| Sec 2.1 | 131 | 0.019 | 0.011 | 51.59 | 3.39 |
| Sec 2.3 | 341 | 0.018 | 0.011 | 48.96 | 3.55 |
| Sec 2.4 | 342 | 0.018 | 0.018 | 48.15 | 3.55 |
| Sec 2.5 | 317 | 0.012 | 0.007 | 69.32 | 3.46 |
| Hybrid | 536 | 0.017 | 0.015 | 60.00 | 3.48 |
From the table, we see that the FP filtering approach with a descendant classifier and an FN recovery classifier (Sec 2.4) had the largest improvement when considering both macro and micro averages. Although the NER based approach (Sec 2.5) improved results for over 300 terms, the improvement is smaller compared to other methods. The average depth of the full MeSH hierarchy is 4.5 with an average of 17 descendants per non-leaf term. From Table 4, it is evident that the hierarchical strategies seem to do well when the terms are slightly higher in the hierarchy with many descendants. The absolute improvement is around 1.5% in both micro and macro averages for a total of 536 terms, which constitute about 12% of the set of non-leaf terms we considered. We will now apply the hierarchical approaches as discussed in Section 3.2 to the test dataset.
3.4. Results on Test Dataset
The test set results in Table 5 are obtained by considering all those non-leaf terms that had at least 1% improvement in F1 score in the validation dataset and ranking them in the descending order of their frequency in the validation dataset. This is to analyze if we can find a large set of terms for which the performance holds across different datasets. From the table it is clear that both micro and macro averages increase for the top 500 non-leaf terms. We also display relative improvements (= (hybrid − baseline)/baseline) in the table which become prominent at the macro level as we consider fewer frequently occurring terms. But overall were able to identify 500 terms for which we improved micro-F1 score by 1.6% and macro-F1 score by 7.5% relative to the baseline scores. These results indicate the strong potential of exploring hierarchical information in improving the state-of-the-art in automated indexing of biomedical articles.
Table 5:
Test Dataset Results
| # Terms | Micro-F
|
Macro-F
|
||||
|---|---|---|---|---|---|---|
| Baseline | Hybrid | Rel. Impr. | Baseline | Hybrid | Rel. Impr. | |
| Top 100 | 0.6304 | 0.6421 | 1.5% | 0.3835 | 0.4206 | 12.8% |
| Top 250 | 0.6106 | 0.6201 | 1.5% | 0.3651 | 0.3979 | 9.9% |
| Top 500 | 0.5959 | 0.6034 | 1.6% | 0.3274 | 0.3515 | 7.5% |
3.5. Remarks on Class Imbalance
As noted in Section 1.2, there is extreme imbalance between the positive and negative instances of MeSH terms, a situation which is more prominent in non-leaf terms. We conducted additional experiments with different class weighting schemes for all the strategies discussed in Section 2. To handle high negative class bias in a methodical way we also tried a combination of boosting and random under sampling [20]. Although there were marginal improvements for a few terms, we do not think they warrant reporting in this manuscript and so we leave them out. However, this also leads us to believe that this problem is not only important but also very difficult to solve, especially under the assumption that only the abstract and title are available. There is some evidence [21] that points to small improvements if additional sections of the article full text, when available, are considered. The class imbalance in the MeSH training data can also result from the so called “Rule of Three”5 for indexing biomedical articles – “If more than 3 related concepts are discussed in an article but are not a major topic, the more cthe false positives could have been due to more than three specific MeSH headings predicted for a citation with a common immediate parent. So we conducted additional experiments where we replaced such term sets with their immediate ancestor in the final candidate set of predicted terms (an extension to the method in Section 3.2). However, we observed only marginal improvements; incorporating this particular manual indexing guidance into the automation process did not seem to help. The rule-of-three also states that when three or more related concepts are identified as major MeSH terms (terms that are deemed central to the article), their immediate common ancestor is coded as a major topic, while the descendants are retained but not as major terms. Thus this aspect of the rule-of-three is also essentially responsible for situations where both a non-leaf term and several of its descendants are assigned to an article.
For our initial study we used a dataset of one million citations although there are currently about 21 million citations made available through NLM for training purposes. Although we chose the smaller dataset for computational tractability in building over 27,000 baseline models and four sets of 4,395 hierarchical models for non-leaf terms, increasing the dataset size to 22 million citations may not necessarily yield improved performance. This is because of the extreme class imbalance that will persist given the unusually high number of negative examples for each non-leaf term. In such cases, it is not clear whether rare event adjustments to supervised learning approaches [22, 23] will help the situation for MeSH term prediction because of the limitation of considering only the title and abstract. However, there is recent evidence that micro averages (over all MeSH terms) can be improved by using more efficient learning methods and considering all available citations [24]. Our main purpose in this paper is to show that descendant instances play an important role in improving results for many MeSH terms. We believe their role will only increase in relevance, if extreme class imbalances are handled, because of the hierarchical nature of MeSH.
4. Conclusion
Assigning MeSH headings to biomedical articles is an important task at the NLM due to the search flexibility it gives biomedical researchers in satisfying their information needs. Many automated attempts have been developed in the recent past to predict MeSH terms from the title and abstract text of an article. In this paper, we focus on the problem of building binary classifiers for terms corresponding to non-leaf nodes in the MeSH hierarchy. Based on the observation that many false positives for terms with several descendants could be attributed to the descendants, we experimented with approaches that exploit descendant training instances. To our knowledge, this is the first effort to investigate the suitability of hierarchical information toward improving MeSH term prediction. Our results show good promise with a relative improvement of 7.5% in macro-F1 score and 1.6% in micro-F1 score over the baseline non-hierarchical approach for a set of 500 non-leaf terms that occur at least 100 times in our training dataset of one million citations.
Given that there are over 23 million citations and the recent evidence [24] of computational tractability for learning using all citations, we will conduct additional experiments to identify other non-leaf terms whose performance can be improved using hierarchical approaches. We will also conduct additional error analysis to identify factors that make certain non-leaf terms more suitable for hierarchical classification compared to others. Our current effort in this paper, nevertheless, demonstrates that hybrid hierarchical approaches that exploit descendant instances play a key role in assigning MeSH terms to biomedical articles.
Acknowledgments
We are grateful to anonymous reviewers for their careful assessment and constructive criticism of the manuscript, which helped improve it for the final camera-ready version. This publication was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, US National Institutes of Health (NIH), through Grant UL1TR000117. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
For the full architecture of MTI’s processing flow, please see: http://skr.nlm.nih.gov/resource/Medical_Text_Indexer_Processing_Flow.pdf
If some descendants do not have enough examples (which was rare in our experiments), we randomly choose examples from the other descendants until the total instance set size was equal to the size of the main term training instance set
References
- [1].Funk ME, Reid CA. Indexing consistency in MEDLINE. Bulletin of the Medical Library Association. 1983;71(2):176. [PMC free article] [PubMed] [Google Scholar]
- [2].Huang M, Névéol A, Lu Z. Recommending MeSH terms for annotating biomedical articles. Journal of the American Medical Informatics Association. 2011;18(5):660–667. doi: 10.1136/amiajnl-2010-000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Aronson AR, Bodenreider O, Chang HF, Humphrey SM, Mork JG, Nelson SJ, et al. Proceedings of AMIA Symposium. American Medical Informatics Association; 2000. The NLM Indexing Initiative; p. 17. [PMC free article] [PubMed] [Google Scholar]
- [4].Jimeno-Yepes A, Mork JG, Demner-Fushman D, Aronson AR. A One-Size-Fits-All Indexing Method Does Not Exist: Automatic Selection Based on Meta-Learning. Journal of Computing Science and Engineering. 2012;6(2):151–160. [Google Scholar]
- [5].Jimeno-Yepes A, Mork JG, Wilkowski B, Demner-Fushman D, Aronson AR. MEDLINE MeSH indexing: lessons learned from machine learning and future directions; Proceedings of the first international conference on healthcare informatics; 2012. pp. 737–742. [Google Scholar]
- [6].Mork JG, Jimeno-Yepes A, Aronson AR. The NLM Medical Text Indexer System for Indexing Biomedical Literature; Proceedings of the first Workshop on Bio-Medical Semantic Indexing and Question Answering, a Post-Conference Workshop of Conference and Labs of the Evaluation Forum 2013 (CLEF 2013); Valencia, Spain. Sep, 2013. pp. 1–6. [Google Scholar]
- [7].Vasuki V, Cohen T. Reflective random indexing for semi-automatic indexing of the biomedical literature. Journal of Biomedical Informatics. 2010;43(5):694–700. doi: 10.1016/j.jbi.2010.04.001. [DOI] [PubMed] [Google Scholar]
- [8].Yetisgen-Yildiz M, Pratt W. Proceedings of AMIA Symposium. American Medical Informatics Association; 2005. 2005. The Effect of Feature Representation on MEDLINE Document Classification; pp. 849–853. [PMC free article] [PubMed] [Google Scholar]
- [9].Sohn S, Kim W, Comeau DC, Wilbur WJ. Optimal training sets for bayesian prediction of MeSH assignment. Journal of the American Medical Informatics Association. 2008;15(4):546–553. doi: 10.1197/jamia.M2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kavuluru R, He Z. Unsupervised Medical Subject Heading Assignment Using Output Label Co-occurrence Statistics and Semantic Predications In: Natural Language Processing and Information Systems NLDB. Springer; 2013. pp. 176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kavuluru R, Lu Y. Leveraging output term co-occurrence frequencies and latent associations in predicting medical subject headings. Data & Knowledge Engineering. 2014;94(Part B):189–201. doi: 10.1016/j.datak.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tsoumakas G, Laliotis M, Markantonatos N, Vlahavas IP. Large-Scale Semantic Indexing of Biomedical Publications. BioASQ; Conference and Labs of the Evaluation Forum; 2013. [Google Scholar]
- [13].Fan RE, Chang KW, Hsieh CJ, Wang XR, Lin CJ. LIBLINEAR: A Library for Large Linear Classification. Journal of Machine Learning Research. 2008;9:1871–1874. [Google Scholar]
- [14].Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- [15].Silla CN, Jr, Freitas AA. A survey of hierarchical classification across different application domains. Data Mining and Knowledge Discovery. 2011;22(1–2):31–72. [Google Scholar]
- [16].Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. Journal of the American Medical Informatics Association. 2010;17(3):229–236. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tsoumakas G, Katakis I, Vlahavas IP. Mining Multi-label Data. Data Mining and Knowledge Discovery Handbook. 2010:667–685. [Google Scholar]
- [18].Ribadas FJ, de Campos LM, Darriba VM, Romero AE. Proceedings of the CLEF BioASQ Workshop. 2014. CoLe and UTAI participation at the 2014 BioASQ semantic indexing challenge; pp. 1361–1374. [Google Scholar]
- [19].Tang L, Rajan S, Narayanan VK. Large scale multi-label classification via metalabeler; Proceedings of the 18th international conference on World wide web. ACM; 2009. pp. 211–220. [Google Scholar]
- [20].Seiffert C, Khoshgoftaar TM, Van Hulse J, Napolitano A. RUSBoost: A Hybrid Approach to Alleviating Class Imbalance. Systems, Man and Cybernetics, Part A: Systems and Humans, IEEE Transactions on. 2010;40(1):185–197. [Google Scholar]
- [21].Gay CW, Kayaalp M, Aronson AR. Semi-automatic indexing of full text biomedical articles. AMIA Annual Symposium Proceedings. 2005;2005:271. [PMC free article] [PubMed] [Google Scholar]
- [22].Saerens M, Latinne P, Decaestecker C. Adjusting the outputs of a classifier to new a priori probabilities: a simple procedure. Neural computation. 2002;14(1):21–41. doi: 10.1162/089976602753284446. [DOI] [PubMed] [Google Scholar]
- [23].Wallace BC, Dahabreh IJ. Improving class probability estimates for imbalanced data. Knowledge and Information Systems. 2013:1–20. [Google Scholar]
- [24].Wilbur WJ, Kim W. Stochastic Gradient Descent and the Prediction of MeSH for PubMed Records. Proceedings of the AMIA Symposium. 2014:1198–1207. [PMC free article] [PubMed] [Google Scholar]


