Abstract
Personal health records (PHRs) provide patient-centric healthcare by making health records accessible to patients. In China, it is very difficult for individuals to access electronic health records. Instead, individuals can easily obtain the printed copies of their own medical records, such as prescriptions and lab test reports, from hospitals. In this paper, we propose a practical approach to extract structured data from printed medical records photographed by mobile phones. An optical character recognition (OCR) pipeline is performed to recognize text in a document photo, which addresses the problems of low image quality and content complexity by image pre-processing and multiple OCR engine synthesis. A series of annotation algorithms that support flexible layouts are then used to identify the document type, entities of interest, and entity correlations, from which a structured PHR document is built. The proposed approach was applied to real world medical records to demonstrate the effectiveness and applicability.
Introduction
Personal health record (PHR) is an electronic application through which individuals can access, manage and share their health information1, which can help individuals take a more active role in their own health, and provide decision support to assist patients in managing chronic diseases2. Data collection is a base functionality of a PHR system, and it is time-consuming and error-prone for individuals to manually enter their PHR data3. Hence, a more practical manner for collecting PHR data is to give individuals access to their own electronic health records (EHRs), which has been or is being implemented in some developed countries. For example, in the United States, many individuals can access and download their own EHR data using Blue Button4. And in England, all patients will have online access to the EHR data that their general practitioners hold by 20155.
In China, even though PHR has been given much attention by health authorities and researchers6, at the present stage it is still difficult for Chinese individuals to access their EHR data, especially in underdeveloped areas. So far, China has not released a privacy standard like HIPAA7 (which mandates that patients can access their health records), and many Chinese individuals are regarded as lack of security and privacy concerns. Therefore, China health institutions tend to be conservative in giving patients access to their EHR data. Moreover, there are dozens of EHR vendors in China, and the EHR systems from different vendors widely vary due to the lack of implementation guidelines. So it is also difficult to develop unified interfaces for individuals to access their data in EHRs. Instead, individuals can easily obtain from hospitals the printed copies of their medical records, such as prescriptions, lab test reports, etc. This situation is not unique to China, but is widespread in many regions, especially in the developing countries.
Smartphones are very popular nowadays. China now has more than 500 million smartphone users, most of whom are accustomed to use phone cameras to take pictures of paper-based documents, including printed medical records, for the purpose of storage. Figure 1 shows some printed Chinese medical records, which were photographed using mobile phones. Because the data in photographed medical records are not structured, they cannot be directly used to build PHRs. A potential solution is to identify structured data in them using optical character recognition (OCR) techniques.
Figure 1.
Sample prescriptions and lab test reports in China
However, it is a challenging problem to build structured PHR from the photographs of printed medical records. First of all, the images are shot by ordinary mobile phones rather than high-quality scanners. Compared with the scanned images, they tend to have uneven light and shade, lower clarity, more noise, as well as paper skewing. Secondly, different types of medical records are involved to create PHRs. Even for a specific type, as shown in Figure 1, the layouts of printed medical records are not standardized in China, which may obviously vary in different time and different hospitals. Furthermore, various categories of entities, such as diagnoses, medications, test items names and values, are supposed to be extracted from medical records, where different types of characters can be observed, including Chinese characters, English characters, digits and punctuation. Existing OCR systems in clinical settings8,9,10 focus on the identification of structured clinical or privacy information from scanned documents, and do not consider photographed images. Moreover, these systems were designed for standardized documents, where the entities of interest are written in English and Arabic numerals. None of these systems can support unstandardized documents that may have very flexible layouts or have complex character sets as Chinese medical records do.
In this paper, we address these issues by proposing an approach to extract structured data from the photographs of printed medical records. Before OCR, several image processing algorithms, including denoising, binarization and deskewing, are performed to reduce the influence of low image quality of photographs shot by mobile phones. Multiple OCR engines are then applied to recognize text, and the results of the engines are synthesized to achieve a higher performance in recognizing characters of a complex character set. Furthermore, a series of flexible annotation algorithms that support diverse document layouts are designed to identify document types, entities of interest and entity correlations, using machine learning, pattern matching, and slot-based techniques. Based on these algorithms, we developed a system pipeline to automatically build structured PHR documents from printed medical records. The proposed approach has been applied to build PHRs from real world prescriptions and lab test reports, with high precision and sensitivity.
Methods
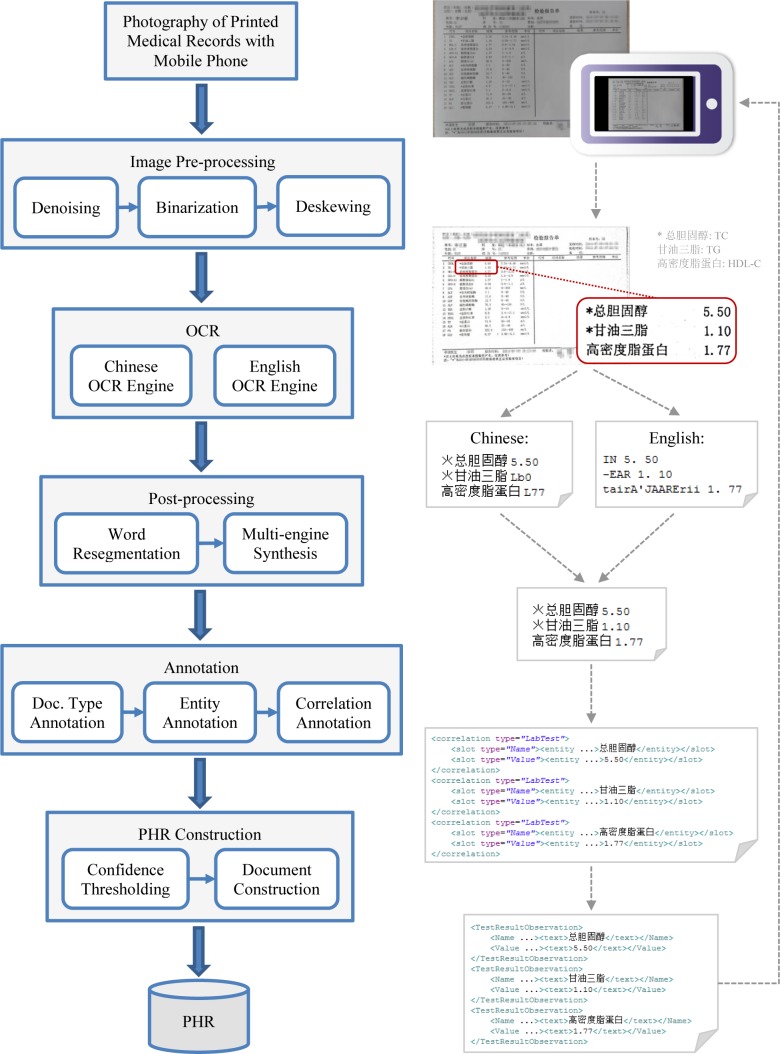
The goal of the proposed approach is to accurately extract entities of interest and entity correlations from photographs of printed medical records as many as possible. Therefore, both precision and recall (sensitivity) of the approach should be taken into account when designing the system and algorithms. The system pipeline and sample data flow are shown in Figure 2. A user first shoots a printed medical record using an ordinary mobile phone camera, and sends it to the system. Then the system performs image pre-processing on the photo. The processed image is sent to multiple OCR engines to build documents with recognized text, which are then resegmented and synthesized in the post-processing step. After that, the document type, entities of interest and entity correlations of the document are automatically annotated. The credible entities are adopted to create a structured PHR document, which is finally sent back to the user for further editing. In this study, two types of documents, prescriptions and lab test reports, were involved. And four categories of entities were focused on, including diagnosis names, medication names, test item names and test item values, which are critical to collect for building PHRs.
Figure 2.
The system pipeline and sample data flow
Corpus
The corpus of this study consists of 100 printed medical record documents (including 54 prescriptions and 46 lab test reports, see Figure 1) from 19 different hospitals in China, which were voluntarily provided by individuals. These documents were respectively photographed by several volunteer users using their own mobile phones, including Apple iPhone 4S, iPhone5C, iPhone 5S, LG Nexus 5, Samsung SCH-I779, Xiaomi HM Note, etc. All records were mainly written in Chinese, with many numerical values and a small amount of English words. The corpus includes 1476 entities to recognize, including 84 diagnosis names, 111 medication names, 650 test item names and 631 test item values. For evaluation purpose, these entities were manually annotated by three human readers.
Image Pre-processing
Document photographs that are shot by mobile phone cameras tend to have lower quality than scanned images due to uneven light and shade, photo noise and skewing. For obtaining reliable OCR results, image pre-processing of the photographs needs to be first performed to enhance image quality.
To reduce the negative impacts of light and shade on OCR, image binarization, which converts an image into a black-and-white image, was performed. Since a photo shot by phone usually has different lighting conditions in different areas, the binarization algorithms that use a global value as threshold cannot generate a reasonable binary image. Thus, we applied the adaptive thresholding algorithm11 that calculates the local threshold for a small region of the image. For a pixel, its local threshold is the weighted sum of neighborhood grayscale values of the pixel, where the weights are computed in a Gaussian window. The pixel whose grayscale value is greater than the local threshold is assigned white, otherwise it is assigned black. Figure 3(c) shows a binarized image derived from the photo in Figure 3(a).
Figure 3.
Sample results of the image pre-processing algorithms
As shown in Figure 3(c), the binarized result of a photo shot by phone can have much salt-and-pepper noise, which may lead to additional OCR errors in some cases. Notice that most of the salt-and-pepper noise in the binarized image is derived from the Gaussian noise in the original photos. So we used the non-local means denosing algorithm12, which replaces a pixel with the average color of the most resembling pixels in a search window, to remove the Gaussian noise in an original image before it is binarized (see Figure 3(b)). And as shown in Figure 3(d), a majority of the salt-and-pepper noise in the binarized image were removed accordingly. In this study, we utilized the implementation of the binarization algorithm and the denoising algorithm provided in the OpenCV 2.4.9 library13.
Since it is difficult for users to exactly align their phone screens with paper-based documents during photographing, documents in photos often become skewed, which also reduces OCR accuracy. Therefore, we performed a deskewing algorithm that rotates an image to make text run as horizontally across the document as possible (Figure 3(e)). In this study, we utilized the deskewing algorithm in the image process toolkit provided by IBM Datacap Taskmaster Capture 8.114. We also tried some image cleanup algorithms such as line removal and dot shading removal offered by Datacap. However, in our experiments, these cleanup algorithms did not significantly improve the OCR performance because they also removed useful strokes in text. So these algorithms were not integrated in our current system pipeline.
Optical Character Recognition
For building PHRs, multiple types of characters were supposed to be recognized in medical records, such as Chinese characters, English characters, digits and punctuation. In general, a single OCR engine cannot achieve a satisfactory accuracy in recognizing characters of this complex character set. For example, the accuracy of a Chinese OCR engine in recognizing English characters and digits is usually relatively low, while an English OCR engine normally identifies Chinese characters as garbled text. To address this problem, we applied multiple OCR engines and synthesized the results of the engines in the following post-processing phase. In this study, the OCR was performed using IBM Datacap Taskmaster Capture 8.114, which embeds the Nuance OmniPage OCR engines15 and supports OCR in multiple languages. We applied the embedded Chinese OCR engine and the English OCR engine, and Figure 4(b) and 4(c) show the results of these two engines respectively for the processed image shown in Figure 4(a). The OCR errors of the examples are highlighted, including the character recognition errors (red circle) and the word segmentation errors (red blank). Note that here we show the results of the OCR engines in plain text for simplicity. Actually, the output of each OCR engine was formatted as a XML document with a line-word-character structure, where each line, word or character has its minimum bounding box B and a confidence value v ∊ [0, 1].
Figure 4.
Sample results of the OCR engines and the post-processing algorithms
Post-processing
As mentioned above, the results of the multiple OCR engines were synthesized in the post-processing phase. Before doing that, the word segmentation errors from the OCR engines should be corrected, because both the synthesis and annotation approaches work on word-level, and incorrectly segmented words can cause extraction errors.
OCR engines normally segment characters into words based on inter-character intervals, and rarely consider the syntax information such as punctuation. For example, non-numeric characters separated by a full point “.” should be segmented, while two integers separated by a point “.” (e.g., “5.50”) usually represent a decimal number that should not be split. As these criteria are not captured by the OCR engines, we performed two rules sequentially to resegment the words more accurately: 1) Punctuation segmentation, which splits words using a set of pre-defined punctuation, including English punctuation such as “.” and “(”, as well as Chinese punctuation such as “∘” and “(”; 2) Decimal recombination, which concatenates two integers (e.g., “5”, “50”) that appear in a same line and have none but a point “.” or “.” between them. Figure 4(d) and 4(e) show the resegmented results of Figure 4(b) and 4(c) respectively, where the decimal numbers were correctly recombined (except for a misrecognized number “O” in Figure 4(e)). Note that in Figure 4(d), an incorrect resegmentation occurred, where “脂蛋白(a)” is a single term that should not be spitted. However, the rules correctly segmented words in most cases and improved the overall performance.
After the resegmentation, we synthesized the results of the Chinese OCR engine and the English OCR engine to achieve an optimal recognition result. We first initiated a synthesis document by simply copying the result recognized by the Chinese engine. For each word wcn with a bounding box Bcn in the document, we found the word wen recognized by the English engine whose bounding box Ben has the largest overlap with Bcn. Then we replaced wcn with wen in the document if wcn and wen fulfill the following three criteria: 1) wcn has at least one digit character; 2) the confidence value of wen is not less than that of wcn; and 3) the string length of wen is not shorter than that of wcn. Using this rule, we adopted the correct numerical values recognized by the English engine while avoiding its garbled Chinese text. Figure 4(f) shows a synthesized result of 4(d) and 4(e), where the wrong numbers of the two engines were corrected.
Annotation
From the recognized text, we then annotated relevant medical data. Multiple types of medical records were involved, and in general different categories of data entities can be identified in different types of documents. For example, medication names can only be detected in prescriptions and lab test items can only be identified in lab test reports. Therefore, a document classifier was first built to determine the document type. Besides, since the layouts of Chinese medical records are not standardized and can be very flexible, the location-based methods8,9 cannot be used to annotate entities of interest and build entity correlations. Thus, we applied flexible entity annotation algorithms such as fuzzy term matching and regular expression matching to locate and identify the entities of interest, and used a slot-based correlation annotation algorithm to locate the related entities (e.g., pairs of test item names and test item values).
1) Document type annotation: keyword-based approach and machine learning approach
Document type annotation is a general problem in the document classification/categorization area. In this study, we focused on two document types, prescription and lab test report. And in each type of documents, there indeed exist some typical keywords which are able to differentiate each other, therefore a keyword-based document classification approach was used. Having explored hundreds of prescriptions and lab test reports, we collected typical keywords for them. For example, the keywords for prescription include {处方(prescription), 药房(pharmacy), 药费(medical fee), 发药(drug dispensing), …}; and those for lab test report include {检验(test), 标本(specimen), …}. Each keyword set was then written as one regex (regular expression) pattern to facilitate the matching process. Once a medical records is matched against a regex pattern, the document type is determined accordingly.
However, since medical records are multifarious and OCR is error-prone, for some documents, neither of the keywords can be matched. In these cases, we also applied the Naïve Bayes16 model, which is a machine learning model commonly used for document classification, to annotate the documents. The Naïve Bayes approach constructs a classifier by calculating the probability distribution of selected features from training dataset, and then assigns document labels to problem instances. In this study, the training dataset came from an EHR system in China, which contains 2319 prescriptions and 2803 lab test reports. We used bag-of-words as features, and took “tf-idf”16 (term frequency and inverse document frequency) instead of “tf” (term frequency) to minimize the noise of common words.
2) Entity annotation: dictionary-based approach and regex-based approach
Named entity recognition (NER) is a subtask in information retrieval, which seeks to locate and classify elements in text into pre-defined categories, such as medical terms (e.g., diagnosis names, medication names and test item names) and numerical values, using linguistic-based approaches and statistical models17. Differing from general-purposed NER approaches, there is little context information in medical records; instead, hint information like entity inner structures and closed sets of medical terminologies can be used to develop specific annotation algorithms.
For medical term entities, a dictionary-based approach was used to detect the matches of dictionary terms in text, which required a dictionary with high coverage and good quality. By collecting the standard medical terminologies like ICD-10 and the local medical terminology systems in China, we developed a dictionary-based engine to annotate medical terms. Considering the non-standard terminology usage and inevitable OCR errors, a fuzzy matching algorithm was used by calculating the similarity between a recognized word and a dictionary term:
| (1) |
where w1, w2 ∈ [0, 1], and w1 + w2 = 1. Unigram similarity is the ratio of intersected single words to the total number of single words, and bigram similarity is the ratio of intersected two consecutive words to the total number of two consecutive words. If the similarity is above a pre-defined threshold, the term is taken as an entity candidate. Multiple entity candidates for one word are allowed, which can be finally determined during correlation annotation.
Besides, a regex-based approach was used to annotate the entities with obvious structural patterns, such as numerical value, date and identifier. For example, a numerical value can be identified using the regex pattern “[0–9]+(\.[0–9]+)?”.
3) Correlation annotation: slot-based approach
Correlation annotation aims at locating the relationship among entities (e.g., the correspondence between test item names and test item values). Correlation annotation is usually solved using syntax parser or semantic parser18 within a given context. Since little syntax information is available in printed medical records, we adopted the semantic approach and leveraged layout and entity type information, where the correlations were represented in pre-defined slot-and-filler formats. A pivot entity (e.g., test item name) must be first identified from the entity annotation results (i.e., entity candidates), then the correlated entity (e.g., test item value) of the pivot can be located according to the entity type. Since there are usually multiple correlations of a same type, to avoid incorrect correlation annotation, two entity-span constraints were applied: 1) the correlated entities must occur in a same line or in two consecutive lines; 2) the correlated entities cannot be interrupted by another pivot entity with the same type.
PHR Construction
Before the annotated entities were adopted to construct PHR documents, we checked whether each entity is credible and rejected the entities with low confidence. Both the confidence value v that represents the OCR engine’s certainty and the similarity value s were considered. For a medical term entity, s is the similarity between the word recognized by the Chinese OCR engine and the annotated dictionary term. And for a numerical value, s is the similarity between the words recognized by the two OCR engines. If the product v × s is not less than a pre-defined confidence threshold θ, then this entity is accepted. Otherwise, it is rejected and not be adopted in building PHR documents.
Finally, we built structured PHR documents from the accepted entities. For each document type such as prescription and lab test report, a template PHR document was defined, as well as mappings from correlated entities to elements in the template document. The template documents are based on the interface XML format used in our EHR system19, which can be easily transformed into the standard CDA (HL7 Clinical Document Architecture) format20. Given the annotation results of a document, a template PHR document is first determined according to the annotated document type. Then the accepted entities are mapped to the elements in the PHR document. As shown in the example in Figure 2, three pairs of correlated entities (“Name” and “Value” in the “Lab Test” correlations) were transformed to three elements (“TestResultObservation”) in a PHR document of lab test report.
Results
To evaluate the performance of our methods, we validated the PHR documents automatically built by our approach against the gold standard created by human readers using the corpus described above which has 100 testing documents. The exact match evaluation was performed, where an automatically extracted entity is correct if its text exactly matches that of the gold standard entity (i.e., “5.5” and “5.50” are not regarded as equal). For each experiment configuration, precision (positive predictive value), recall (sensitivity) and F-measure (harmonic mean of precision and recall) were computed.
In our study, we first configured our system pipeline as described in the above section, and performed the evaluation for the confidence threshold value θ = 0.0 (i.e., all the recognized and annotated entities are adopted to generate PHR documents). Table 1 demonstrates the results for every entity category and for all entities. Here the category of “Term” means the union set of the categories of diagnosis name, medication name and test item name. As shown in Table 1, without constraining the confidence value, our system could identify over 80% (recall) entities that were annotated by human readers, and over 90% (precision) of our extracted entities were precise. The performance for the term entities, with a recall of 0.88, outweighed that for the value entities (i.e., test item values), with a recall of about 0.75.
Table 1.
Evaluation on the system using the confidence threshold = 0.0 (P: precision; R: recall; F: F-measure)
| Category | #entity | P | R | F | Category | #entity | P | R | F |
|---|---|---|---|---|---|---|---|---|---|
| Diagnosis name | 84 | 0.973 | 0.869 | 0.918 | Test item value | 631 | 0.867 | 0.746 | 0.802 |
| Medication name | 111 | 0.916 | 0.784 | 0.845 | Term | 845 | 0.945 | 0.880 | 0.912 |
| Test item name | 650 | 0.946 | 0.898 | 0.922 | All | 1476 | 0.914 | 0.823 | 0.866 |
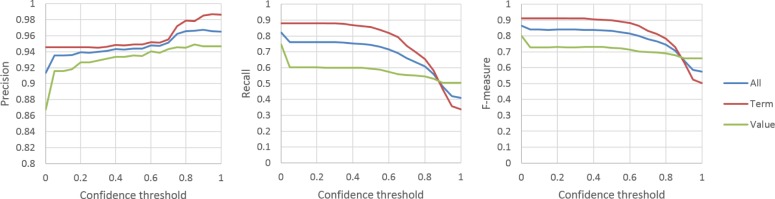
To demonstrate the influence of the confidence threshold, we kept the same configuration as above but gradually changed the threshold θ from 0.0 to 1.0. As shown in Figure 5, with the increasing of the threshold, the precisions of the system slowly rose and could achieve over 0.98 for the term entities and 0.94 for the value entities, whereas the recalls and the F-measures dramatically descended accordingly.
Figure 5.
Precision, recall and F-measure of the system with respect to the confidence threshold
Besides, we also performed the evaluation on different pre-processing and post-processing algorithms for our system. Table 2 shows the results of the PHR document evaluation on image pre-processing approaches, where the configurations of the other modules except pre-processing were set in the same manner as the experiment shown in Table 1. Compared with the results of directly using original images, the image processing algorithms we used did not significantly improve the precision of the system. However, all the three algorithms promoted the recall to some extent, and the combination of these algorithms achieved the best performance.
Table 2.
Evaluation results for pre-processing
| Pre-processing algorithm | P | R | F |
|---|---|---|---|
| None | 0.892 | 0.736 | 0.807 |
| Binarization | 0.911 | 0.783 | 0.842 |
| Denoising + Binarization | 0.910 | 0.812 | 0.858 |
| Denois.+ Binar.+ Deskew. | 0.914 | 0.823 | 0.866 |
Table 3 shows the results of the PHR document evaluation on different configurations of post-processing approaches. Both the word resegmentation algorithm and the multi-engine synthesis algorithm obviously improved the precision and recall of the system, and their combination obtained the best results.
Table 3.
Evaluation results for post-processing
| Post-processing algorithm | P | R | F |
|---|---|---|---|
| None | 0.760 | 0.629 | 0.689 |
| Resegmentation | 0.874 | 0.781 | 0.825 |
| Multi-engine synthesis | 0.818 | 0.671 | 0.737 |
| Reseg. + Synthesis | 0.914 | 0.823 | 0.866 |
To evaluate the performance of the document type annotation algorithms, we directly compared the types classified by the algorithms with the type tags given by the human readers for the 100 testing documents, and computed the classification accuracy. As shown in Table 4, both the keyword-based and Naïve Bayes algorithms led to a few classification errors, and the combination of these algorithms achieved an absolutely correct result on our testing set.
Table 4.
Evaluation results for doc. type annotation
| Doc. type annotation algorithm | Accuracy |
|---|---|
| Keyword-based | 0.97 |
| Machine learning (Naïve Bayes) | 0.96 |
| Keyword-based + Naïve Bayes | 1.0 |
To evaluate the performance of the term matching approaches, we performed the PHR document evaluation on the exact matching algorithm as well as the fuzzy matching algorithms using unigram similarity and using the combination of unigram and bigram as Equation (1) (w1 = 0.4, w2 = 0.6). As shown in Table 5, though the precision of the exact matching algorithm was the highest, its recall was relatively low. The recalls of the two fuzzy matching metrics had no significant difference, but the precision of the combined metric was obviously higher than that of unigram.
Table 5.
Evaluation results for term entity annotation
| Term matching algorithm | P | R | F |
|---|---|---|---|
| Exact matching | 0.988 | 0.659 | 0.791 |
| Unigram | 0.619 | 0.871 | 0.724 |
| Unigram + Bigram | 0.945 | 0.880 | 0.912 |
To evaluate the applicability of the proposed system, we also performed experiment on same medical record documents photographed by different users different mobile phones. 20 printed documents were randomly selected from the corpus and 5 volunteer users respectively shot 0.907 the 20 documents using their own mobile phones. Figure 6 shows examples of a same document photographed by different users, where the clarity and illumination distribution of the images are obviously different. The quality of the photos shot by user #5 is on the low side in comparison to others. We performed our pipeline on these 20 documents for the 5 users respectively, using the same configuration as the experiment shown in Table 1, and the evaluation results are shown in Table 6. For the images from each user, our system achieved a precision of over 0.85. And the recalls for the images from all the users except #5 were also close (from 0.75 – 0.79).
Figure 6.
Examples of a document photographed by different users using different mobile phones
Table 6.
Results for same documents shot by different users
| User | Mobile phone | P | R | F |
|---|---|---|---|---|
| #1 | Apple iPhone 5S | 0.891 | 0.779 | 0.831 |
| #2 | Apple iPhone 4S | 0.886 | 0.767 | 0.822 |
| #3 | Samsung I779 | 0.905 | 0.791 | 0.844 |
| #4 | LG Nexus 5 | 0.896 | 0.747 | 0.815 |
| #5 | Huawei Honor 5 | 0.854 | 0.648 | 0.737 |
Finally, we also analyzed the extraction errors of our system based on the results of the experiment shown in Table 1. We detected total 284 errors including 169 missing entities that were annotated by the human readers but not identified by the system, 23 superfluous entities that were not manually annotated but extracted by the system, and 92 incorrect entities of which the results recognized by the system were different with the gold standard. The reasons of the errors were also classified by the human readers, which are shown in Table 7. More than a half of the errors were caused by the OCR engines, and the annotation algorithms also contributed to a large proportion of errors.
Table 7.
Statistics of the extraction errors
| Error type | #errors | Percentage |
|---|---|---|
| Image defect | 22 | 7.7 % |
| Pre-processing errors | 9 | 3.2 % |
| OCR errors | 149 | 52.5 % |
| Post-processing errors | 3 | 1.1 % |
| Annotation errors | 101 | 35.6 % |
Discussion
Admittedly, the image quality of photographed medical records significantly affects the performance of OCR and annotation. There are two primary image problems that may cause extraction errors: 1) printing and typesetting defects in original printed documents; 2) problems caused by camera or careless photographing. As shown in the experimental results, a majority of the image problems could be solved by the image pre-processing approach and the fuzzy term matching algorithm. The overall system had good tolerance to uneven light and shade, Gaussian noise, and skewing in images, and was adaptable to photos shot by different users using different phones. However, as shown in Table 7, there were still some extraction errors caused by image defects that were not eliminated. Figure 7(a) gives some examples of printing defects where some strokes were not completely printed, and Figure 7(b) shows a typesetting problem where two entities (test item name and value) are concatenated with each other. Besides, more errors tended to occur when the system handled images with low clarity (e.g., some images shot by user #5).
Figure 7.
Examples of image defects and pre-processing errors
As shown in Table 2, the image pre-processing approach significantly improved the sensitivity of our system, because the uneven illumination distribution, Gaussian noise and skewing problems could be relieved. However, as shown in Figure 7(c), these image processing algorithms also made a few additional errors, where some extremely thin strokes disappeared in the binarized images. Even so, we still applied these algorithms in the current system because the overall performance could be promoted. And a detail-preserving image processing approach can be used to solve this problem in the future.
The limitation of the OCR engines caused more than a half of the extraction errors. Although we did not attempt to modify the algorithms of the OCR engines, we observed that reasonable post-processing algorithms, such as word resegmentation, could greatly improve the overall performance (see Table 3). More importantly, we observed that with well-defined synthesis criteria, the results of multiple OCR engines could be combined to correct some errors from each OCR engine alone. In this study, we only synthesized the results of one Chinese OCR engine and one English OCR engine. It can be predicted that if more OCR engines are integrated in a reasonable manner, OCR errors can be further reduced. As shown in Table 7, the post-processing algorithms produced very few additional errors (one of which has been shown in Figure 4(f)), and the benefits significantly outweigh the drawbacks.
Another key factor that contributed to the extraction errors is the annotation algorithms. We observed two main types of annotation errors. The first type was the superfluous term entities caused by the fuzzy term matching algorithm. Compared with the exact matching algorithm, the fuzzy similarity metric greatly improved the recall of term identification, but indeed incorrectly matched some entities that were not regarded by human readers as terms of interest, and therefore reduced the precision as shown in Table 5. Since the performance goal of the system was not only precision but also recall, we finally used the fuzzy matching algorithm in the system. Another type of errors was the item values that were exactly recognized by the OCR engines and correctly annotated as numerical value entities, but were not correctly linked to their corresponding item names by the correlation annotation algorithm. That is primarily because the location information of the entities is not adequately used by the slot-base algorithm. For the value entities that were correctly identified, the current slot-based algorithm could achieve an approximately 86% recall of correlation annotation, which can probably be further improved by combining statistical learning models such as conditional random field. Besides, though we achieved a 100% accuracy of document classification on our testing set by combining the keyword-based and Naïve Bayes approaches, more testing data in the document type level is still needed to further prove the effectiveness of the classification algorithm.
Although the confidence threshold of the system can be configured, we did not finally use it in the current system configuration (i.e., θ = 0.0). That is because the performance goal was to achieve both high precision and high recall, and we observed that with the increasing of the threshold, the recall descended more dramatically than the precision increased (see Figure 5). However, if a case requires a higher precision, we can accordingly raise the threshold (and can also use the exact term matching algorithm instead of the fuzzy matching algorithm as discussed above).
Conclusion
Data collection, which is the basis of building PHRs, is very difficult in a developing country like China due to the policy and technical obstruction to giving patients access to EHR data. Considering the easy availability of printed medical records and the high popularity of smartphones, we proposed a practical approach to build structured PHRs from printed medical records photographed by mobile phones. By combining a series of image processing, OCR, pattern matching and machine learning techniques in a reasonable system pipeline, we addressed the problems of low image quality, layout diversity and content complexity of photographed medical records. The proposed approach was applied to build PHRs from real world prescriptions and lab test reports, and the results showed that our approach can automatically extract structured medical data with relatively high precision and recall, and has wide applicability.
As discussed above, the limitations of the current approach caused some errors that could be eliminated by improving the architecture and algorithms in the future, including detail-preserving image processing, synthesis of more OCR engines, as well as layout-aware and statistic-based annotation. This paper focused on the extraction of medical information, such as diagnosis, medication and test item, from medical records. Actually, the privacy information specified by HIPAA7, such as patient information and dates, can also be located using our methodology for entity and correlation annotation, which can be then de-identified for data transfer and distribution purpose. Besides, though the current system was designed for printed Chinese medical records, our methodology can also be adapted to those of other languages or hand-written medical records by extending the OCR and annotation modules.
References
- 1.Connecting for Health. The personal health working group final report. Markle Foundation. 2003 Jul 1; [Google Scholar]
- 2.Tang PC, Ash JS, Bates DW, Overhage JM, Sands DZ. Personal health records: definition, benefits, and strategies for overcoming barriers to adoption. J Am Med Inform Assoc. 2006;13(2):121–6. doi: 10.1197/jamia.M2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer N, Fevrier-Thomas U, Lokker C, McKibbon KA, Straus SE. Personal health records: a scoping review. J Am Med Inform Assoc. 2011;18(4):515–22. doi: 10.1136/amiajnl-2011-000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.HealthIT Blue button. [cited 2015 Match 8] http://www.healthit.gov/patients-families/blue-button.
- 5.Kmietowicz Z. Patients will have digital access to GP records by 2015, says NHS England. BMJ. 2014 Nov 12;349:g6805. doi: 10.1136/bmj.g6805. [DOI] [PubMed] [Google Scholar]
- 6.Xie L, Yu C, Liu L, Yao Z. XML-based Personal Health Record system; Proceedings of the 3rd International Conference on Biomedical Engineering and Informatics; 2010. pp. 2536–40. [Google Scholar]
- 7.United States Department of Health and Human Service The health insurance portability and accountability act (HIPAA) privacy rule. [cited 2015 Match 8] http://privacyruleandresearch.nih.gov/
- 8.Biondich PG, Overhage JM, Dexter PR, Downs SM, et al. A modern optical character recognition system in a real world clinical setting: some accuracy and feasibility observations. Proc AMIA Symp. 2002:56–60. [PMC free article] [PubMed] [Google Scholar]
- 9.Rasmussen LV, Peissig PL, McCarty CA, Starren J. Development of an optical character recognition pipeline for handwritten form fields from an electronic health record. J Am Med Inform Assoc. 2012;19:90–5. doi: 10.1136/amiajnl-2011-000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenz S, Heurix J, Neubauer T. Recognition and privacy preservation of paper-based health records. Stud Health Technol Inform. 2012;180:751–5. [PubMed] [Google Scholar]
- 11.Sezgin M, Sankur B. Survey over image thresholding techniques and quantitative performance evaluation. Journal of Electronic Imaging. 2004;13(1):146–65. [Google Scholar]
- 12.Buades A, Coll B, Morel JM. Non-local means denoising. Image Processing On Line. 2011;1 http://dx.doi.org/10.5201/ipol.2011.bcm_nlm. [Google Scholar]
- 13.Itseez Team Open source computer vision library (OpenCV) [cited 2015 Match 8] http://opencv.org/about.html.
- 14.IBM IBM datacap taskmaster capture version 8.1.0. [cited 2015 Match 9]. http://www.ibm.com/developerworks/data/library/techarticle/dm-ind-datacap-taskmaster/
- 15.Nuance OmniPage capture SDK. [cited 2015 Match 8] http://www.nuance.com/for-business/byproduct/omnipage/csdk/index.htm.
- 16.Rennie J, Shih L, Teevan J, Karger D. Tacking the poor assumptions of Naïve Bayes classifiers; Proceedings of the Twentieth International Conference on Machine Learning (ICML); pp. 616–23. [Google Scholar]
- 17.Nadeau D, Sekine S. A survey of named entity recognition and classification. Lingvisticae Investigationes. 2007;30(1):3–26. [Google Scholar]
- 18.Grune D, Jacobs CJH. Parsing techniques: a practical guide. Springer Science & Business Media; 2007. [Google Scholar]
- 19.Liu S, Zhou B, Xie G, Mei J, Liu H, Liu C, Qi L. Beyond regional health information exchange in China: a practical and industrial-strength approach. AMIA Annu Symp Proc. 2011:824–33. [PMC free article] [PubMed] [Google Scholar]
- 20.Dolin RH, Alschuler L, Boyer S, Beebe C, Behlen FM, Biron PV, Shabo Shvo A. HL7 clinical document architecture, release 2.0. J Am Med Inform Assoc. 2006;13(1):30–9. doi: 10.1197/jamia.M1888. [DOI] [PMC free article] [PubMed] [Google Scholar]