Abstract
Carbapenem-resistant Enterobacteriaceae (CRE) and carbapenem-resistant Pseudomonas aeruginosa (CRPAE) are globally a major medical issue, especially in intensive care units. The digestive tract is the main reservoir for these isolates; therefore, rectal swab surveillance is highly recommended. The purpose of this study was to detect the prevalence of gastrointestinal tract colonization of CRE and CRPAE in patients admitted to intensive care units in Saudi Arabia. This project also aimed to characterize carbapenem-hydrolyzing enzyme production in these isolates. From February to May 2015, 200 rectal swab specimens were screened by CHROMagar KPC. Organism identification and susceptibility testing were performed using the Vitek 2 system. One CRE and 13 CRPAE strains were identified, for a prevalence of 0.5% (1/200) and 6.5% (13/200) respectively. Strains showed high genetic diversity using enterobacterial repetitive intergenic consensus sequence-based PCR. NDM type and VIM type were detected by PCR in four and one CRPAE isolates respectively. ampC overexpression was detected in eight CRPAE isolates using Mueller-Hinton agar containing 1000 μg/mL cloxacillin. CTX-M-15 type was detected in 1 CRE by PCR. The prevalence of CRE strain colonization was lower than that of CRPAE isolates. The detection of NDM and VIM in the colonizing CRPAE strains is a major infection control concern. To our knowledge, this is the first study in Saudi Arabia and the gulf region focusing on digestive tract colonization of CRE and CRPAE organisms and characterizing the mechanisms of carbapenem resistance.
Keywords: Carbapenem resistance, colonization, Enterobacteriaceae, infection control, Pseudomonas aeruginosa
Introduction
Enterobacteriaceae and Pseudomonas aeruginosa are among the most common organisms causing nosocomial infections worldwide, especially in intensive care units (ICUs) [1], [2], [3], [4], [5]. The treatment of these organisms became more difficult because of their production of extended-spectrum β-lactamases (ESBLs), such as blaCTX-M, plasmid-mediated AmpC (pAmpC) or overexpression of chromosomal ampC gene [6], [7], [8], [9], [10]. Carbapenems became the last source for treatment of such organisms, which in turn has led to the emergence of carbapenem resistance under antibiotic selective pressure [8], [11], [12]. The global spread of carbapenem-resistant, Gram-negative rods is significantly increasing [1], [2], [13]. Carbapenem resistance in Gram-negative rods is mainly due to two mechanisms: first the production of carbapenem-hydrolyzing enzymes (i.e. serine-carbapenemases and metallo-β-lactamases), and second the combination of membrane impermeability with production of ESBLs, pAmpC or ampC overexpression [2], [8], [11], [13]. Carbapenem-hydrolyzing enzymes is the most alarming mechanism because genes encoding carbapenem resistance (i.e. blaNDM, blaOXA-48, blaIMP, blaVIM, blaKPC) are carried out on mobile DNA elements [8], [14], [15]. Therefore, they are transmissible from one organism to another, thus creating a major infection control concern worldwide [8], [14], [15]. Generally carbapenem resistance is associated with resistance to other antibiotics such as aminoglycosides and fluoroquinolones in these organisms [11], [15].
Carbapenem-resistant Enterobacteriaceae (CRE) and carbapenem-resistant Pseudomonas aeruginosa (CRPAE) cause ICU-acquired infections and are associated with high morbidity and mortality rates [16], [17]. Gastrointestinal flora is the main source of these organisms and can play a key role in the spread of antibiotic resistance and ICU infections caused by CRE and CRPAE [18], [19], [20]. In order to control their spread, good surveillance and infection control programmes must be established to identify the digestive colonization of these carbapenem-resistant organisms. There are scanty data in the literature characterizing the gastrointestinal colonization of these organisms [11], [21]. In a previous study, our group found that the prevalence of digestive tract colonization of carbapenem-resistant Acinetobacter baumannii was 6.2% [11]. To our knowledge, there are no studies focusing on the prevalence of gastrointestinal tract colonization of CRE or CRPAE in Saudi Arabia or in the other Gulf Cooperation Council (GCC) countries.
The aim of the current study was to evaluate the prevalence of intestinal carriage of CRE and CRPAE in patients admitted to ICUs in Saudi Arabia. We also sought to characterize the carbapenem-hydrolyzing genes produced by these organisms.
Materials and Methods
Study population and setting
This project was conducted in two hospitals in the Eastern Province of Saudi Arabia: a 450-bed general hospital, King Fahad University Hospital, in Khobar and a 650-bed tertiary-care hospital, King Fahad Specialist Hospital, in Dammam. From February 2015 to May 2015, nonrepetitive rectal swab specimens were collected from ICU patients at admission in both hospitals. Specimens were screened for CRE and CRPAE using CHROMagar KPC as recommended by the manufacturer (CHROMagar, Paris, France).
Organism identification and antimicrobial susceptibility testing
All colonies with colours specified by the manufacturer (pink, red, cream, metallic blue, translucent) growing on CHROMagar KPC were subcultured on blood agar plates and MacConkey agar plates for further assessment. Organism identification and antimicrobial susceptibility testing for these strains were carried out using the Vitek 2 automatic system (bioMérieux, Marcy l’Étoile, France) according the manufacturer's recommendation. The Vitek 2 system was considered the reference standard method in this study. Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 strains were used as controls for antimicrobial susceptibility testing. Minimum inhibitory concentrations (MIC) and breakpoints were determined according to the guidelines of Clinical and Laboratory Standards Institute (CLSI). Antibiotics tested in this study included imipenem (IMP), meropenem (MEP), gentamicin (GM), amikacin (AK), ciprofloxacin (CIP), cefepime (FEP), cefotaxime (CTX), ceftazidime (CAZ) and colistin (CO). In addition, ertapenem (ERT) was tested for Enterobacteriaceae.
All strains confirmed to be carbapenem resistant using the Vitek 2 system were further tested phenotypically and by PCR. For phenotypic detection of carbapenemases, Etest strips for detection of metallo-β-lactamases (Etest MBL) were performed as previously described [22], [23]. Phenotypic testing for ESBL production in CRE isolates was performed using double disk synergy testing as previously described [16]. In addition, the overexpression of the chromosomally encoded ampC gene in the CRPAE organisms was tested by restoration of susceptibility for ceftazidime using Mueller-Hinton agar plates containing 1000 μg/mL of cloxacillin [8].
Molecular characterization of β-lactamase-resistant genes
PCR methodology was used for molecular characterization of β-lactamase genes. All carbapenem-resistant strains were tested for the presence of carbapenem-resistant genes (OXA-48, NDM, IMP, VIM, KPC, SIM, SPM and GIM). CRE strains were also tested for ESBL production including TEM, SHV and CTX-M β-lactamases. In addition, CMY, DHA, FOX, EBC, MOX and ACC β-lactamases were tested to detect the production of pAmpC in CRE. Primers and PCR parameters were used as previously described [21], [24], [25], [26], [27], [28]. Positive controls were used for all genes tested in this study. Molecular-grade water was also used as a negative control to detect contamination. Amplified amplicons were then sequenced using an ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA) as previously described [25], [26], [28].
Epidemiologic analysis of carbapenem-resistant strains
The relatedness of carbapenem-resistant strains was studied using enterobacterial repetitive intergenic consensus sequence-based PCR (ERIC-PCR) methodology using a protocol and primers described by Rivera et al. [29]. Briefly, genomic DNA template was extracted using a standard heat boiling lysis method. A pure single colony was inoculated in 10 mL of Luria-Bertani broth with 0.5% NaCl (w/v) and incubated in incubator shaker at 37°C with rotary speed at 250 rpm for 18 hours. The overnight-grown bacterial culture was pelleted, resuspended in 500 μL of sterilized distilled water, boiled in a water bath at 100°C for 15 minutes, and centrifuged. A volume of 2 μL of the supernatant was used for DNA amplification. The amplification conditions were performed as follows: an initial denaturation cycle at 94°C for 5 minutes, 35 cycles at 95°C for 1 minutes, 52°C for 1 minute and 72°C for 5 minutes, then a final extension at 72°C for 10 minutes. The amplified products were analysed on a 1.5% agarose gel stained with ethidium bromide. All gels were analysed using the gel Compar II 6.6 software package (Applied Maths, Sint-Martens-Latem, Belgium). Dendrograms for ERIC-PCR gels were generated using the Dice similarity coefficient and the unweighted-pair group method using arithmetic (unweighted pair group method with arithmetic mean, UPGMA) averages, with 1% optimization and 1% position tolerance [30].
Statistical analysis
Categorical variable comparison was determined by two-tailed Fisher's exact test; p <0.05 was considered statistically significant.
Results
A total of 200 nonduplicated rectal swab specimens were screened at ICU admission during the 4-month period of the study. The male/female ratio was 2.1, and the median age was 43.8 years (range, 1–84 years). A total of 31 strains were isolated using CHROMagar KPC as a screening method. Of these 31 strains, only 14 strains (one Klebsiella pneumoniae and 13 P. aeruginosa) were carbapenem resistant using the reference standard method, the Vitek 2 system (Table 1). Seventeen strains screened by CHROMagar KPC out of the 31 strains were carbapenem susceptible by Vitek 2 (Table 1). Therefore, the overall prevalence of CRE and CRPAE strains colonizing the patients admitted to the ICU units was 7% (14/200). The prevalence of CRE and CRPAE was 0.5% (1/200) and 6.5% (13/200) respectively. The rate of carbapenem resistance in these isolates was 45.1% (14/31). The highest susceptibility rate was for colistin (100%, 31/31), followed by aminoglycosides (amikacin 23/31, 74.2%, and gentamicin 22/31, 71%) (Table 2). The susceptibility rates were 32.3% (10/31), 29% (9/31), 25.8% (8/31) and 13% (4/31) for cefepime, ceftazidime, ciprofloxacin and cefotaxime respectively. Using Fisher's exact test, carbapenem resistance was statistically significantly associated with more resistance to amikacin, gentamicin, cefepime and ceftazidime with p values of less than 0.05 (Table 2). However, it was not the case for ciprofloxacin, cefotaxime and colistin, where p values were more than 0.05 (Table 2).
Table 1.
Epidemiology and antimicrobial susceptibility testing
| Sample no. | Organism ID | Age (years) | Sex | IMP | MEP | ERT | FEP | CTX | CAZ | GM | AK | CIP | CO | Carbapenemase |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbapenem-resistant Enterobacteriaceae and Pseudomonas aeruginosa | ||||||||||||||
| 1 | Klebsiella pneumoniae | 8 | M | R | R | R | R | R | R | S | S | S | S | CTX-M-15 type |
| 2 | P. aeruginosa | 30 | F | R | R | NA | R | R | R | R | R | R | S | VIM type |
| 3 | P. aeruginosa | 35 | M | R | I | R | R | R | R | R | R | S | ampC overexpression | |
| 4 | P. aeruginosa | 61 | M | R | R | S | R | S | S | S | S | S | ||
| 5 | P. aeruginosa | 19 | M | R | R | R | R | R | S | S | S | S | ||
| 6 | P. aeruginosa | 2 | F | R | R | R | R | R | S | S | R | S | ||
| 7 | P. aeruginosa | 3 | F | R | R | R | R | R | S | S | S | S | ||
| 8 | P. aeruginosa | 40 | M | R | R | R | R | R | R | R | R | S | NDM type | |
| 9 | P. aeruginosa | 2 | M | R | R | R | R | R | S | S | S | S | ampC overexpression | |
| 10 | P. aeruginosa | 61 | M | R | R | R | R | R | R | R | R | S | NDM type | |
| 11 | P. aeruginosa | 74 | M | R | R | R | R | R | R | R | R | S | NDM type | |
| 12 | P. aeruginosa | 66 | M | R | R | R | R | R | R | R | R | S | ampC overexpression | |
| 13 | P. aeruginosa | 44 | M | R | R | R | R | R | S | S | R | S | ||
| 14 | P. aeruginosa | 76 | M | R | R | R | R | R | R | R | R | S | NDM type | |
| Carbapenem-susceptible Enterobacteriaceae and P. aeruginosa | ||||||||||||||
| 15 | Escherichia coli | 20 | M | S | S | S | S | S | S | S | S | S | S | NA |
| 16 | E. coli | 56 | F | S | S | S | R | R | R | S | S | R | S | |
| 17 | E. coli | 47 | F | S | S | S | R | R | R | R | S | R | S | |
| 18 | K. pneumoniae | 1 | M | S | S | S | R | R | R | S | S | S | S | |
| 19 | K. pneumoniae | 6 | M | S | S | S | R | R | I | S | S | S | S | |
| 20 | K. pneumoniae | 84 | F | S | S | S | R | R | R | S | S | S | S | |
| 21 | K. pneumoniae | 77 | M | S | S | S | R | R | R | S | S | S | S | |
| 22 | Providencia stuartii | 74 | M | S | S | S | S | R | R | R | S | S | S | |
| 23 | P. aeruginosa | 52 | M | S | S | NA | I | R | S | S | R | I | S | NA |
| 24 | P. aeruginosa | 76 | F | S | S | I | R | R | S | S | S | S | ||
| 25 | P. aeruginosa | 58 | F | S | S | S | R | S | S | S | S | S | ||
| 26 | P. aeruginosa | 30 | M | S | S | S | R | S | S | S | S | S | ||
| 27 | P. aeruginosa | 72 | F | S | S | S | R | S | S | S | S | S | ||
| 28 | P. aeruginosa | 47 | M | S | S | S | S | S | S | S | S | S | ||
| 29 | P. aeruginosa | 84 | F | S | S | S | S | S | S | S | S | S | ||
| 30 | P. aeruginosa | 31 | M | S | S | S | S | S | S | S | S | S | ||
| 31 | P. aeruginosa | 66 | M | S | S | S | R | S | S | S | S | S | ||
AK, amikacin; CAZ, ceftazidime; CIP, ciprofloxacin; CO, colistin; CTX, cefotaxime; ERT, ertapenem; FEP, cefepime; GM, gentamicin; IMP, imipenem; MEP, meropenem; NA, not applicable; R, resistant; S, susceptible.
Table 2.
Association of carbapenem resistance and non-β-lactam antibiotics
| Antibiotic | Carbapenem-resistant Enterobacteriaceae and Pseudomonas aeruginosa (n = 14) |
Carbapenem-susceptible Enterobacteriaceae and P. aeruginosa (n = 17) |
Total susceptibility rate |
p | ||
|---|---|---|---|---|---|---|
| S (%) | R (%) | S (%) | R (%) | S (%) | ||
| Gentamicin | 7 | 7 | 15 | 2 | 22/31 (71) | 0.04 |
| Amikacin | 7 | 7 | 16 | 1 | 23/31 (74.2) | 0.01 |
| Ciprofloxacin | 5 | 9 | 3 | 14 | 8/31 (25.8) | 0.4 |
| Cefepime | 1 | 13 | 9 | 8 | 10/31 (32.3) | 0.008 |
| Cefotaxime | 0 | 14 | 4 | 13 | 4/31 (13) | 0.1 |
| Ceftazidime | 1 | 13 | 8 | 9 | 9/31 (29) | 0.02 |
| Colistin | 14 | 0 | 17 | 0 | 31/31 (100) | 1.0 |
R, resistant; S, susceptible.
Etest MBL was positive for 5 CRPAE strains. The phenotypic confirmatory test for ESBLs was positive for the carbapenem-resistant Klebsiella pneumoniae (CRKP) strain. Susceptibility to ceftazidime was restored for eight of 13 CRPAE strains using Mueller-Hinton agar plates containing cloxacillin.
PCR tests for CRPAE strains revealed a 497 bp amplicon correlating with NDM in four strains and a 382 bp amplicon using VIM primers in one strain. NDM and VIM types were confirmed by sequencing. For the CRKP isolate, a 415 bp PCR amplicon correlating with CTX-M-15 type was detected and revealed by sequencing. PCRs using pAmpC primers were negative for CRKP. No PCR amplification was detected for TEM, SHV, OXA-48, IMP, KPC, SIM, SPM and GIM.
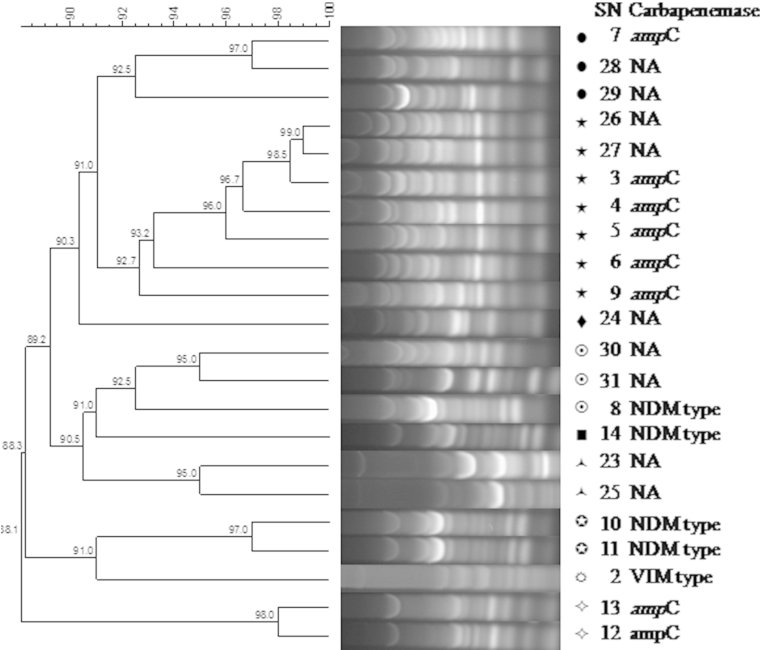
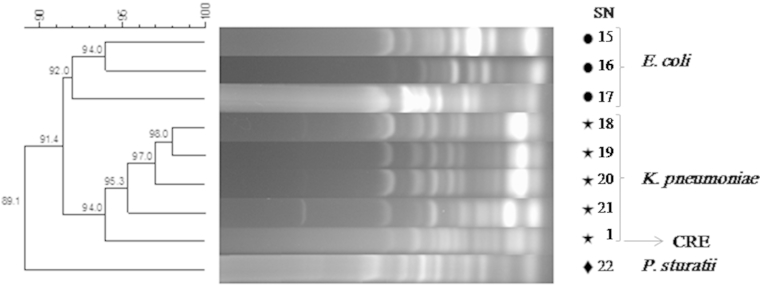
Using ERIC-PCR methodology, the 22 strains of P. aeruginosa were grouped into six clusters and three single isolates at 90% similarity level (Fig. 1). Enterobacteriaceae strains were grouped into two clusters and one single isolate at 91% similarity level (Fig. 2). The dendrogram of Enterobacteriaceae strains exhibited discriminatory capability by producing a species-specific cluster that discriminated K. pneumoniae from E. coli and Providencia stuartii. However, three strains of P. aeruginosa and one strain of Enterobacteriaceae demonstrated no genetic correlation (Fig. 1, Fig. 2). No outbreaks caused by carbapenem-resistant Gram-negative rods were reported in either hospital during the study period. In addition, routine environmental sampling did not detect any carbapenem-resistant organisms.
Fig. 1.
ERIC-PCR methodology for Pseudomonas aeruginosa strains. ampC, ampC overexpression; ERIC-PCR, enterobacterial repetitive intergenic consensus sequence-based PCR; NA, not applicable; SN, strain number.
Fig. 2.
ERIC-PCR methodology for Enterobacteriaceae strains. ERIC-PCR, enterobacterial repetitive intergenic consensus sequence-based PCR; SN, strain number. Klebsiella pneumoniae SN 1 is carbapenem-resistant Enterobacteriaceae.
Discussion
Digestive tract colonization by carbapenem-resistant Gram-negative bacilli is a critical step before the development of nosocomial infections such as bloodstream infections, urinary tract infections and ventilator-associated infections [1], [16], [17], [21]. Therefore, early and accurate detection of colonized patients will help reduce the spread of such organisms. In addition, rectal swab specimens are the most appropriate specimens because the reservoir of these organisms is the gastrointestinal tract [1], [5], [7], [18].
In this study, the prevalence of colonization of CRE in the ICU patients at admission was 0.5%. This is low compared to other countries in the Middle East such as Morocco (13%), India (9.9%) and Pakistan (18.3%). However, it is comparable to studies in Korea (0.3%) [12], [19], [31], [32]. In addition, the prevalence of colonization of CRPAE is 6.5%, which is lower than studies performed in Spain (12%) and higher than those in France (3.4%) [8], [13], [33]. CRPAE is more prevalent than CRE. This may be due to the ability of P. aeruginosa to rapidly develop antimicrobial resistance through acquisition of resistant genes or through modifications of antibiotic targets by mutations [13], [33]. The prevalence study of these organisms is significant from infection-control standpoint; it will also help in developing protocols of antibiotic use, especially for gastrointestinal tract surgeries.
In our study, the production of carbapenem-hydrolyzing enzymes was detected in five isolates of P. aeruginosa strains. Our data correlate with other studies in the GCC states [34], [35], [36], [37]. The production of carbapenemases in Enterobacteriaceae and P. aeruginosa is an emerging problem in the GCC region. OXA-48 and NDM-1 are the most common carbapenemases produced in Enterobacteriaceae, and to a lesser extent KPC, VIM-4 and OXA-181 [34]. MBLs are the most common mechanism causing carbapenem resistance in P. aeruginosa in Saudi Arabia and the GCC states, with VIM being the most prevalent MBL [34], [35]. All these studies reported carbapenemase genes harboured by Enterobacteriaceae and P. aeruginosa strains causing different types of infections. However, our study characterizes the mechanisms of carbapenem resistance in colonizing strains.
Carbapenem resistance was associated with CTX-M-15 production in a CRKP strain and ampC overexpression in eight isolates of CRPAE. It is well known that ESBLs or ampC can lead to carbapenem resistance when coupled with porin mutations [8], [9], [10]. Therefore, it is possible that ESBL and ampC genes in these strains might be associated with porin mutations, resulting in carbapenem resistance. This is supported by the fact that ceftazidime susceptibility was restored in the presence of cloxacillin in eight CRPAE isolates. To our knowledge, ours is the first study in the GCC states to report the involvement of ampC overexpression in CRPAE isolates. The molecular epidemiology using ERIC-PCR demonstrated high genetic diversity among these isolates. These data, taken together, suggest that each strain has a unique pathway of emergence of carbapenem resistance. CRPAE strains producing VIM and NDM are of particular concern because these genes are generally harboured on mobile DNA elements that can be transmitted from patient to patient in a hospital setting.
It is possible that ampC overexpression in these CRPAE isolates was due to overuse of β-lactam agents such as ceftazidime, which can cause derepression of ampC. Buying antibiotics over the counter is a common practice throughout the Middle East, which makes it a big source of the emergence of antimicrobial resistance. It is also possible to develop carbapenem resistance under selective pressure of carbapenem use because it is also common to initiate carbapenem therapy as an empirical treatment in this region. Therefore, carbapenem-susceptible P. aeruginosa strains colonizing ICU patients are a major concern. These strains are able to acquire resistance mechanisms either by horizontal transmission from patient to patient or by selection of mutants. Several reports have confirmed the development of carbapenem resistance in susceptible P. aeruginosa strains in the hospital setting [1], [5], [7], [8]. A study characterizing the determining risk factors for the acquisition of carbapenem resistance in Gram-negative bacilli in ICU patients and other wards of the hospital is in progress.
There are a few limitations to our study. Despite the statistical significance of our findings, the study was conducted on a relatively small number of specimens. In addition, we screened only rectal swabs; other anatomic sites can be colonized by carbapenem-resistant Gram-negative rods, and these sites should not be ignored. Therefore, we are in the process of conducting a larger epidemiologic study collecting specimens from different anatomic sites from patients in the ICU and other hospital wards.
To our knowledge, this is the first study characterizing the digestive tract colonization of CRE and CRPAE isolates in patients admitted to the ICU in Saudi Arabia and the gulf region. The data presented here clearly demonstrate the prevalence of these organisms in our patients. It also emphasizes the need for strong infection-control programs to detect the digestive tract colonization of carbapenem-resistant Gram-negative bacilli; the need to reduce the overuse of carbapenems; and the need to establish good antimicrobial stewardship programs.
Acknowledgements
The authors thank Z. AlQahtani, King Fahad Specialist Hospital, Dammam, for technical support.
Conflict of Interest
None declared.
References
- 1.Marchaim D., Perez F., Lee J. ‘Swimming in resistance’: co-colonization with carbapenem-resistant Enterobacteriaceae and Acinetobacter baumannii or Pseudomonas aeruginosa. Am J Infect Control. 2012;40:830–835. doi: 10.1016/j.ajic.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nordmann P., Gniadkowski M., Giske C.G. Identification and screening of carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect. 2012;18:432–438. doi: 10.1111/j.1469-0691.2012.03815.x. [DOI] [PubMed] [Google Scholar]
- 3.Nordmann P., Poirel L. Strategies for identification of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother. 2013;68:487–489. doi: 10.1093/jac/dks426. [DOI] [PubMed] [Google Scholar]
- 4.Paramythiotou E., Lucet J.C., Timsit J.F. Acquisition of multidrug-resistant Pseudomonas aeruginosa in patients in intensive care units: role of antibiotics with antipseudomonal activity. Clin Infect Dis. 2004;38:670–677. doi: 10.1086/381550. [DOI] [PubMed] [Google Scholar]
- 5.Carlet J. The gut is the epicentre of antibiotic resistance. Antimicrob Resist Infect Control. 2012;1:39. doi: 10.1186/2047-2994-1-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Philippe E., Weiss M., Shultz J.M., Yeomans F., Ehrenkranz N.J. Emergence of highly antibiotic-resistant Pseudomonas aeruginosa in relation to duration of empirical antipseudomonal antibiotic treatment. Clin Perform Qual Health Care. 1999;7:83–87. [PubMed] [Google Scholar]
- 7.Johnson J.K., Smith G., Lee M.S. The role of patient-to-patient transmission in the acquisition of imipenem-resistant Pseudomonas aeruginosa colonization in the intensive care unit. J Infect Dis. 2009;200:900–905. doi: 10.1086/605408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armand-Lefevre L., Angebault C., Barbier F. Emergence of imipenem-resistant Gram-negative bacilli in intestinal flora of intensive care patients. Antimicrob Agents Chemother. 2013;57:1488–1495. doi: 10.1128/AAC.01823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacoby G.A. AmpC beta-lactamases. Clin Microbiol Rev. 2009;22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradford P.A., Urban C., Mariano N., Projan S.J., Rahal J.J., Bush K. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC beta-lactamase, and the foss of an outer membrane protein. Antimicrob Agents Chemother. 1997;41:563–569. doi: 10.1128/aac.41.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aljindan R., Bukharie H., Alomar A., Abdalhamid B. Prevalence of digestive tract colonization of carbapenem-resistant Acinetobacter baumannii in hospitals in Saudi Arabia. J Med Microbiol. 2015;64(pt 4):400–406. doi: 10.1099/jmm.0.000033. [DOI] [PubMed] [Google Scholar]
- 12.Rai S., Das D., Niranjan D.K., Singh N.P., Kaur I.R. Carriage prevalence of carbapenem-resistant Enterobacteriaceae in stool samples: a surveillance study. Australas Med J. 2014;7:64–67. doi: 10.4066/AMJ.2014.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pena C., Guzman A., Suarez C. Effects of carbapenem exposure on the risk for digestive tract carriage of intensive care unit-endemic carbapenem-resistant Pseudomonas aeruginosa strains in critically ill patients. Antimicrob Agents Chemother. 2007;51:1967–1971. doi: 10.1128/AAC.01483-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vrioni G., Daniil I., Voulgari E. Comparative evaluation of a prototype chromogenic medium (ChromID CARBA) for detecting carbapenemase-producing Enterobacteriaceae in surveillance rectal swabs. J Clin Microbiol. 2012;50:1841–1846. doi: 10.1128/JCM.06848-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez-Zorrilla S., Camoez M., Tubau F. Antibiotic pressure is a major risk factor for rectal colonization by multidrug-resistant Pseudomonas aeruginosa in critically ill patients. Antimicrob Agents Chemother. 2014;58:5863–5870. doi: 10.1128/AAC.03419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villar H.E., Baserni M.N., Jugo M.B. Faecal carriage of ESBL-producing Enterobacteriaceae and carbapenem-resistant Gram-negative bacilli in community settings. J Infect Dev Ctries. 2013;7:630–634. doi: 10.3855/jidc.2900. [DOI] [PubMed] [Google Scholar]
- 17.Snyder G.M., O’Fallon E., D'Agata E.M. Co-colonization with multiple different species of multidrug-resistant Gram-negative bacteria. Am J Infect Control. 2011;39:506–510. doi: 10.1016/j.ajic.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris A.D., Johnson J.K., Thom K.A. Risk factors for development of intestinal colonization with imipenem-resistant Pseudomonas aeruginosa in the intensive care unit setting. Infect Control Hosp Epidemiol. 2011;32:719–722. doi: 10.1086/660763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J., Lee J.Y., Kim S.I. Rates of fecal transmission of extended-spectrum beta-lactamase-producing and carbapenem-resistant Enterobacteriaceae among patients in intensive care units in Korea. Ann Lab Med. 2014;34:20–25. doi: 10.3343/alm.2014.34.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DiNubile M.J., Chow J.W., Satishchandran V. Acquisition of resistant bowel flora during a double-blind randomized clinical trial of ertapenem versus piperacillin–tazobactam therapy for intraabdominal infections. Antimicrob Agents Chemother. 2005;49:3217–3221. doi: 10.1128/AAC.49.8.3217-3221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azim A., Dwivedi M., Rao P.B. Epidemiology of bacterial colonization at intensive care unit admission with emphasis on extended-spectrum beta-lactamase- and metallo-beta-lactamase-producing Gram-negative bacteria—an Indian experience. J Med Microbiol. 2010;59(pt 8):955–960. doi: 10.1099/jmm.0.018085-0. [DOI] [PubMed] [Google Scholar]
- 22.Walsh T.R., Bolmstrom A., Qwarnstrom A., Gales A. Evaluation of a new Etest for detecting metallo-beta-lactamases in routine clinical testing. J Clin Microbiol. 2002;40:2755–2759. doi: 10.1128/JCM.40.8.2755-2759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pournaras S., Poulou A., Tsakris A. Inhibitor-based methods for the detection of KPC carbapenemase-producing Enterobacteriaceae in clinical practice by using boronic acid compounds. J Antimicrob Chemother. 2010;65:1319–1321. doi: 10.1093/jac/dkq124. [DOI] [PubMed] [Google Scholar]
- 24.Perez-Perez F.J., Hanson N.D. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodford N., Fagan E.J., Ellington M.J. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum (beta)-lactamases. J Antimicrob Chemother. 2006;57:154–155. doi: 10.1093/jac/dki412. [DOI] [PubMed] [Google Scholar]
- 26.Monteiro J., Widen R.H., Pignatari A.C., Kubasek C., Silbert S. Rapid detection of carbapenemase genes by multiplex real-time PCR. J Antimicrob Chemother. 2012;67:906–909. doi: 10.1093/jac/dkr563. [DOI] [PubMed] [Google Scholar]
- 27.Grobner S., Linke D., Schutz W. Emergence of carbapenem-non-susceptible extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates at the university hospital of Tubingen, Germany. J Med Microbiol. 2009;58(pt 7):912–922. doi: 10.1099/jmm.0.005850-0. [DOI] [PubMed] [Google Scholar]
- 28.Cicek A.C., Saral A., Iraz M. OXA- and GES-type beta-lactamases predominate in extensively drug-resistant Acinetobacter baumannii isolates from a Turkish university hospital. Clin Microbiol Infect. 2014;20:410–415. doi: 10.1111/1469-0691.12338. [DOI] [PubMed] [Google Scholar]
- 29.Rivera I.G., Chowdhury M.A., Huq A., Jacobs D., Martins M.T., Colwell R.R. Enterobacterial repetitive intergenic consensus sequences and the PCR to generate fingerprints of genomic DNAs from Vibrio cholerae O1, O139, and non-O1 strains. Appl Environ Microbiol. 1995;61:2898–2904. doi: 10.1128/aem.61.8.2898-2904.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones J.H., Lennard-Jones J.E., Morson B.C. Numerical taxonomy and discriminant analysis applied to non-specific colitis. Q J Med. 1973;42:715–732. [PubMed] [Google Scholar]
- 31.Girlich D., Bouihat N., Poirel L., Benouda A., Nordmann P. High rate of faecal carriage of extended-spectrum beta-lactamase and OXA-48 carbapenemase-producing Enterobacteriaceae at a university hospital in Morocco. Clin Microbiol Infect. 2014;20:350–354. doi: 10.1111/1469-0691.12325. [DOI] [PubMed] [Google Scholar]
- 32.Livermore D.M. Current epidemiology and growing resistance of Gram-negative pathogens. Korean J Intern Med. 2012;27:128–142. doi: 10.3904/kjim.2012.27.2.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lepelletier D., Cady A., Caroff N. Imipenem-resistant Pseudomonas aeruginosa gastrointestinal carriage among hospitalized patients: risk factors and resistance mechanisms. Diagn Microbiol Infect Dis. 2010;66:1–6. doi: 10.1016/j.diagmicrobio.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 34.Zowawi H.M., Balkhy H.H., Walsh T.R., Paterson D.L. Beta-lactamase production in key Gram-negative pathogen isolates from the Arabian Peninsula. Clin Microbiol Rev. 2013;26:361–380. doi: 10.1128/CMR.00096-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al-Agamy M.H., Shibl A.M., Tawfik A.F., Radwan H.H. High prevalence of metallo-beta-lactamase-producing Pseudomonas aeruginosa from Saudi Arabia. J Chemother. 2009;21:461–462. doi: 10.1179/joc.2009.21.4.461. [DOI] [PubMed] [Google Scholar]
- 36.Sonnevend A., Ghazawi A., Yahfoufi N. VIM-4 carbapenemase-producing Enterobacter cloacae in the United Arab Emirates. Clin Microbiol Infect. 2012;18:E494–E496. doi: 10.1111/1469-0691.12051. [DOI] [PubMed] [Google Scholar]
- 37.Shibl A., Al-Agamy M., Memish Z., Senok A., Khader S.A., Assiri A. The emergence of OXA-48- and NDM-1-positive Klebsiella pneumoniae in Riyadh, Saudi Arabia. Int J Infect Dis. 2013;17:e1130–e1133. doi: 10.1016/j.ijid.2013.06.016. [DOI] [PubMed] [Google Scholar]