Abstract
More than 100 young captive and wild Asian elephants are known to have died from a rapid-onset, acute hemorrhagic disease caused primarily by multiple distinct strains of two closely related chimeric variants of a novel herpesvirus species designated elephant endotheliotropic herpesvirus (EEHV1A and EEHV1B). These and two other species of Probosciviruses (EEHV4 and EEHV5) are evidently ancient and likely nearly ubiquitous asymptomatic infections of adult Asian elephants worldwide that are occasionally shed in trunk wash secretions. Although only a handful of similar cases have been observed in African elephants, they also have proved to harbor their own multiple and distinct species of Probosciviruses—EEHV2, EEHV3, EEHV6, and EEHV7—found in lung and skin nodules or saliva. For reasons that are not yet understood, approximately 20% of Asian elephant calves appear to be susceptible to the disease when primary infections are not controlled by normal innate cellular and humoral immune responses. Sensitive specific polymerase chain reaction (PCR) DNA blood tests have been developed, routine monitoring has been established, the complete large DNA genomes of each of the four Asian EEHV species have now been sequenced, and PCR gene subtyping has provided unambiguous evidence that this is a sporadic rather than epidemic disease that it is not being spread among zoos or other elephant housing facilities. Nevertheless, researchers have not yet been able to propagate EEHV in cell culture, determine whether or not human antiherpesvirus drugs are effective inhibitors, or develop serology assays that can distinguish between antibodies against the multiple different EEHV species.
Keywords: calves, elephant endotheliotropic herpesvirus (EEHV), elephant hemorrhagic disease, Elephas maximus, Loxodonta Africana, lung and skin nodules, Probosciviruses
History of Hemorrhagic Disease Recognition in Zoo and Wild Asian Elephants
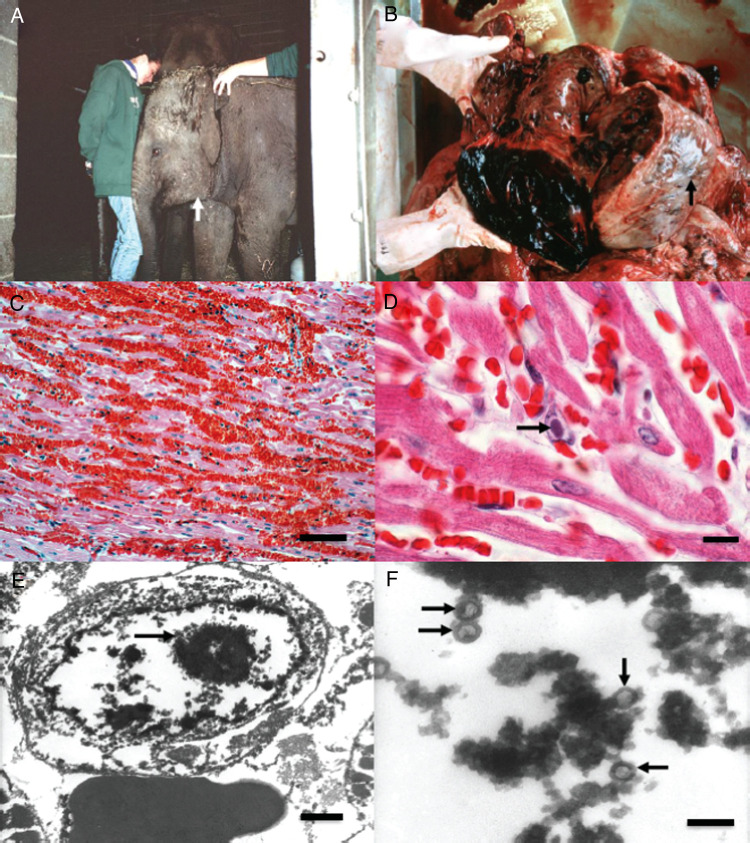
The death of a young Asian elephant (Elephas maximus) from acute hemorrhagic disease of unknown etiology was first reported from a circus in Switzerland in 1988 (Ossent et al. 1990). Nearly 20 apparently similar cases of rapidly developing lethal hemorrhagic disease were then observed in both European and North American zoos during the 1990s, with the signal case considered to be Kumari, a 16-month-old, female Asian calf at the Smithsonian's National Zoo in Washington, DC in 1995. In the latter case, the attending veterinary pathology resident, Laura Richman, recognized herpesvirus-like particles by electron microscopy within nuclear inclusion bodies in vascular endothelial cells from heart and liver necropsy samples that exhibited extensive tissue hemorrhaging (Figure 1, B–F). Following this observation, several young Asian zoo calves with similar early overt disease symptoms of purple tongue, lethargy, or facial and neck edemic swelling (Figure 1A) were apparently saved by systemic treatment with the drug famciclovir (FCV), which is used to treat human herpes simplex virus infections. In 1999, Richman and colleagues (1999) published a report in Science about detecting small diagnostic segments of the DNA of a novel herpesvirus called “elephant endotheliotropic herpesvirus” or EEHV in all necropsy tissue samples and blood of Kumari and 10 other lethal or surviving cases of the disease, including from retrospective archival tissue blocks going back to the early 1980s. All but one of these cases were in young Asian elephants and had the same virus type (called EEHV1), whereas the other, a 1-year-old African elephant calf, had a related but different virus DNA (EEHV2). A more extensive review of the pathology of these cases, including the use of a diagnostic blood test by standard polymerase chain reaction (PCR) showing clearing of EEHV1 DNA from the blood during several weeks of convalescence in two drug-treated survivors, was also published the next year (Richman, Montali, and Hayward 2000). Apparent detection of DNA from the same virus in archival tissue blocks from imported orphan African elephant calves in Florida that suffered an outbreak of skin nodules provided the impetus to initially suggest a model (later invalidated) whereby an endogenous African elephant virus may have been transmitted to susceptible Asian elephant calves, which could explain the surprisingly severe pathology (Richman et al. 1999; Richman, Montali, Cambre, et al. 2000; Richman, Montali, and Hayward 2000).
Figure 1.
Clinical and pathological features of elephant endotheliotropic herpesvirus (EEHV)–positive elephant hemorrhagic disease. (A) Surviving 16-month-old Asian elephant calf showing early clinical symptoms of EEHV1-associated acute hemorrhagic disease including subcutaneous edema of the head, neck (white arrow), and thoracic limbs. This was the first survivor treated with FCV. (B) Gross pathological features at necropsy from a lethal Asian elephant case showing subepicardial ecchymotic hemorrhages of the heart (black arrow) attributed to EEHV1. (C) Low-power photomicrograph of typical extensive extravascular hemorrhage and separation of the myocardial fibers attributed to EEHV1. Hematoxylin and eosin, bar = 150 µm. (D) Higher-power photomicrograph of a diagnostic herpesvirus-like intranuclear inclusion body (black arrow) in a capillary endothelial cell from necropsy heart tissue. Hematoxylin and eosin, bar = 25 µm. (E) Transmission electron micrograph (TEM) of the heart from an African elephant calf that died of EEHV hemorrhagic disease demonstrating an intranuclear inclusion body (black arrow) within a capillary endothelial cell. Bar = 500 nm. (F) Higher magnification TEM of the same intranuclear inclusion body from E, showing greater detail of the unenveloped herpesvirus nucleocapsids (black arrows). Bar = 200 nm. All panels from Richman (2003).
In the 15 years since then, more than 100 potential or proven cases of the disease have been recognized, both in the wild and in elephants under human care, and a number of management and monitoring changes have been implemented that appear to have reduced the incidence in North American zoos, but not elsewhere. Overall, we recognized a total of 29 lethal cases of the disease within North American calves, nearly all of which have been confirmed by a diagnostic test involving PCR-based DNA sequencing, plus some 28 lethal cases in Europe, about half of which have been DNA sequence confirmed. In the United States, these 29 cases represent 58% of all of the deaths of captive-born Asian elephants between the ages of 4 months and 15 years that were born between 1962 and 2007. In comparison, there were 78 other remaining unaffected individuals that were born over that same time period. Before 2010, an additional eight Asian calves developed significant disease symptoms and had the disease proven through positive DNA with moderately high systemic virus loads in the blood, and thus have been categorized as true survivors (Schmitt et al. 2000). Since then, improved sensitivity of detection to include asymptomatic subclinical low-level viremias has clouded the situation with regard to defining cases of disease versus mere “normal” primary infections (Stanton et al. 2010, 2013). All eight of these survivors were treated with one of two drugs, either FCV or ganciclovir (GCV), known to be effective inhibitors of certain human herpesviruses, but many other severely ill elephants that were similarly treated did not recover. Before the latest instance in May 2015, there had been only one lethal case of the disease in North America between 2008 and 2014, but at least 10 calf deaths occurred in European zoos over the same time period. The majority of the proven cases involved Asian elephants between 1 and 8 years of age with a peak between 1 and 4 years and with only one (a 40-year-old) being older than 18 years (Hayward 2012; Richman and Hayward 2011). No more than two proven lethal cases and two survivors of virus-positive viremias are known among African elephants (Loxodonta africana). Elephant calves that died of this disease have included the first ever Asian calves born at the National Zoo, the Oakland Zoo, and the Bronx Zoo, as well as the first live-born Asian calves conceived by artificial insemination methods in both North America and Europe.
A point of considerable importance has been whether or not the same hemorrhagic disease occurs in wild-range countries in Asia. In fact, anecdotal evidence had accumulated of up to eight possible cases in India between 1996 and 2004, as well as one published report of a DNA-positive lethal case from Cambodia (Reid et al. 2006). However, it was not until wildlife veterinarian Arun Zachariah performed field necropsies and collected frozen tissue samples from calves that died suddenly in Kerala, Southern India, that major progress was made. In collaboration with our groups at the National Elephant Herpes Laboratory (NEHL) at the Smithsonian's National Zoo and The Johns Hopkins University, he set up a diagnostic PCR laboratory and confirmed the presence of EEHV1 DNA in 9 of 15 potential cases to firmly establish the existence of both the virus and the disease in India (Zachariah et al. 2013). These nine positive Southern India cases included several observed in wild free-ranging calves in Kerala, Karnataka, and Tamilnadu forest reserves, as well as a privately owned calf, an orphan at the Madras Zoo, and others from orphan training camps (Zachariah et al. 2013). The disease pathology included all of the same hallmarks as reported in Western zoos, including sudden-onset lethargy, cyanotic (purple) tongue, edema, organ and tissue hemorrhaging (Figure 1), and high levels of viral DNA in most major internal organs. Several published cases have also been reported from Thailand (Sripiboon et al. 2013) and Laos (Bouchard et al. 2014), and unpublished lethal cases have also been confirmed by PCR DNA sequencing in Sumatra (two) and Nepal (one). Anecdotal reports have also appeared of between 8 and 20 recent unconfirmed cases in calves from the last working logging camps in Myanmar (Zaw Min Oo, www.asesg.org/PDFfiles/2012/Gajah%2036/36-21-MinOo.pdf) and four more at an orphan camp in Sumatra (from November 2014 to February 2015). Most significantly, 22 more DNA sequence–positive cases from Thailand were described just recently at the Eighth International EEHV Workshop in Houston, Texas (S. Sripiboon, 2015, personal communication, Murdoch University, Perth, Western Australia).
Identification of Multiple Herpesvirus Types in Both Asian and African Elephants
The small EEHV2 viral POL and TER gene DNA segments found in the first African elephant zoo calf known to have died of this disease proved to be nearly 20% different from that found in the 10 Asian calf tissue and blood samples (EEHV1) studied by Richman and colleagues (1999). This result clearly implied that African and Asian elephants may harbor or become infected with different species of this novel herpesvirus type, which were designated as the prototypes of a new Proboscivirus genus. Furthermore, Garner and colleagues (2009) published evidence for two more viruses of this type (named EEHV3 and EEHV4) that were found at high levels in the blood or necropsy tissues from two Asian elephant calves that died of hemorrhagic disease in 2006 and 2002, respectively (with the latter having been found earlier to be negative for EEHV1 DNA). Later, Latimer and colleagues (2011) described evidence for two more different EEHV types in the blood of, first, a mildly ill elderly Asian zoo elephant (EEHV5) and, second, in a moderately ill surviving African zoo calf (EEHV6). Virus particles resembling herpesviruses, as well as diagnostic nuclear inclusion bodies, had originally been reported from lung nodules observed in culled, mostly adult African elephants at Kruger National Park back in the 1970s (McCully et al. 1971). Similar features were also seen in skin nodules from the imported Zimbabwean calves in Florida in the mid-1980s (Jacobson et al. 1986). Examples of both types of nodules are shown in Figures 2 and 3. Curiously, no such nodules have ever been reported or studied from Asian elephants, despite our alerts to wildlife and zoo pathologists over the past 10 years or more.
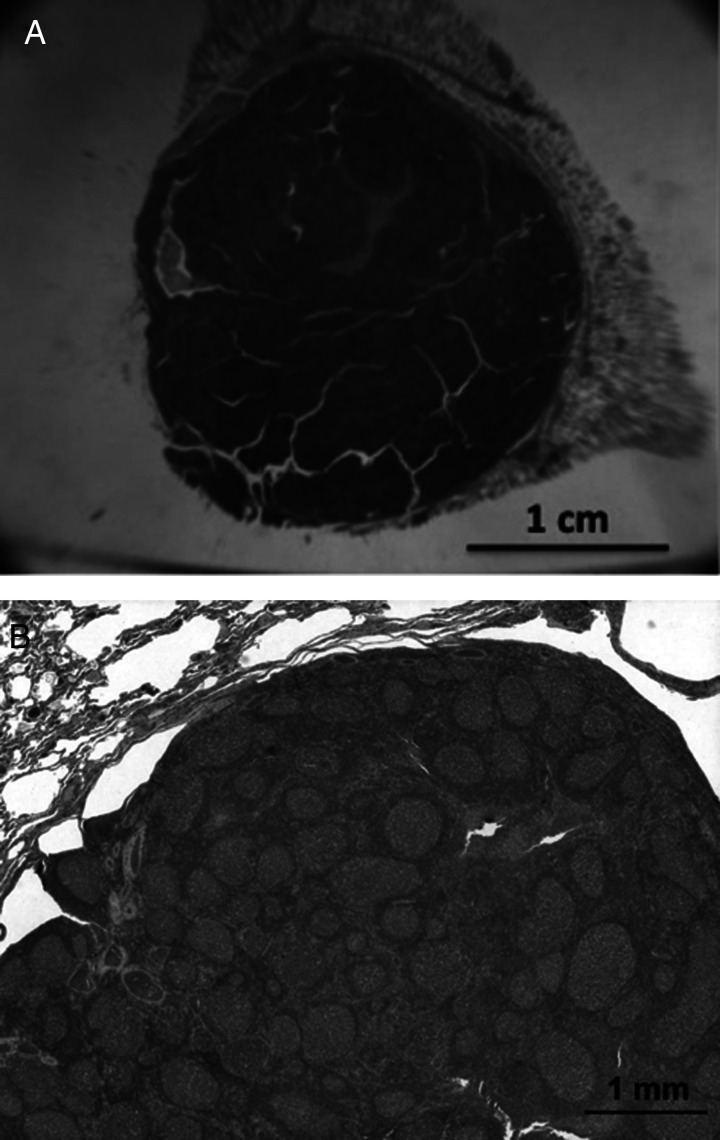
Figure 2.
African elephant lung nodules. Photomicrographs of follicular lymphoid nodules from culled healthy wild adult African elephants from Kruger National Park in South Africa in 1995. Two stored frozen nodules from this study in the National Elephant Herpes Laboratory collection contained very high levels of EEHV3 DNA or EEHV2 plus EEHV3 DNA. According to McCully and colleagues (1971), similar lung nodules were found in 75% of random culled African elephants in Zimbabwe. (A) Low-power photomicrographs of a large follicular lymphoid nodule found at necropsy in the lung of a wild adult Loxodonta africana. (B) Higher-power photomicrograph of a similar lung nodule showing multiple follicles composed of lymphocytes.
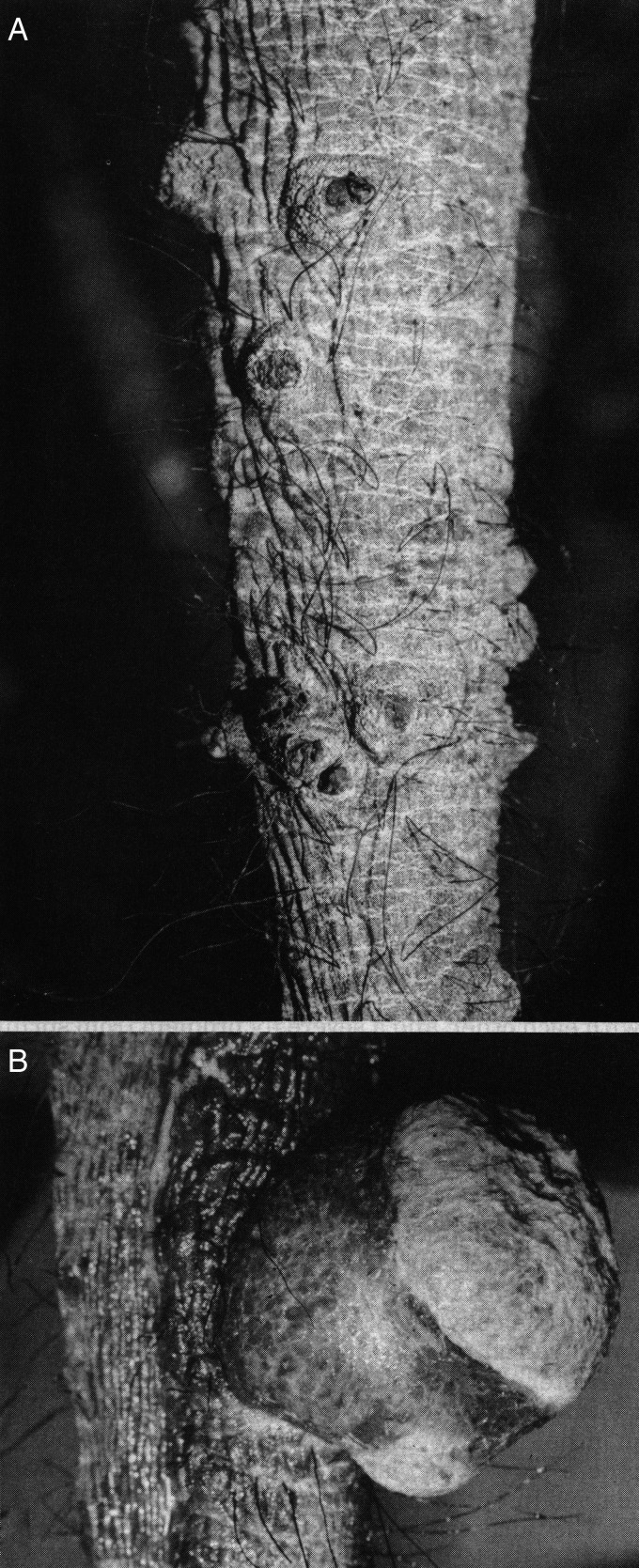
Figure 3.
African elephant skin nodules. Typical herpesvirus skin nodules observed on the trunk and head of 1% of healthy juvenile African elephants in Africa, as well as in the outbreak among a large group of young orphan African calves imported from Zimbabwe to Florida (Jacobson et al. 1986). Similar skin nodules biopsied from Kenyan elephants have been found to contain DNA from multiple viruses, including EEHV2, EEHV3, EEHV7, several types of elephant gammaherpesviruses (EGHVs), and African elephant polyomavirus (V.R. Pearson, personal communication, 2013. Fox Chase Cancer center, Philadelphia; Finding EEHVs in Wild African Elephants by Virginia Pearson https://elephantmanagers.com/…/ElephantManagersArticleKWS__1_.doc. August 2012). (A) Photograph of regressed skin nodules from the trunk of an African elephant. (B) Photograph of a proliferative skin nodule from the trunk of an African elephant. Reproduced with permission from Elliot Jacobson and Journal of the American Veterinary Medical Association.
In an initial effort to learn which of the multiple EEHV types might be present in African elephant tissues, we located two stored frozen lung nodules from healthy wild adults in the NEHL collection that had been culled at Kruger National Park in the mid-1990s. We also dissected out four small (2 mm), white, shiny, spherical lung nodules occurring within a palpable section of fresh unfrozen necropsy lung tissue recovered from a U.S. adult African zoo elephant euthanized in 2012 (that had been among those imported as a young orphan calf to Florida). Surprisingly, both of the large lung nodules from the culled Kruger elephants proved to have very high levels of EEHV3 DNA present, and one also contained even more EEHV2 DNA (J-C Zong, SY Heaggans, SY Long, EM Latimer, SA Nofs, MD Fouraker, VR Pearson, LK Richman and GS Hayward, unpublished data). Even more remarkable, the four small lung nodules all contained DNA from several EEHV types, with a total of four different EEHV species altogether, but distributed in different patterns and levels. These included a single novel strain of EEHV6 at high levels, plus lower levels of one strain of EEHV2, two strains of EEHV3, and another novel virus of this type that we named EEHV7. Subsequently, multiple simultaneous infections by different strains of EEHV2, EEHV3, and EEHV6 have also been detected in random fresh spleen or lung tissue from two other African elephants that died from unrelated causes (one at a zoo in Spain and the other as a result of poaching in Samburu Reserve in Kenya). Notably, we could not detect EEHV1 in any of these African elephant samples, nor could we detect it when we re-evaluated several of the original archival paraffin blocks containing the skin nodule tissue from the old 1980s Florida outbreak. A summary of the identification of the prototype examples of all seven known types (= species) of EEHV, as well as several additional major subtypes, is presented in Table 1 (upper section).
Table 1.
Historical summary of the identification of multiple elephant herpesviruses (listing of the 12 species and 20 subtypes known)
| Species | Case No. | Source | Host | Discovery date (investigator) |
|---|---|---|---|---|
| Proboscivirus Genus (Deltaherpesvirus Sub-Family): | ||||
| EEHV1A | NAP#11a, #19a | Blood/Nec, TW | EM | Richman 1999; Stanton 2009 |
| EEHV1B | NAP#14a, #18a | Blood/Nec, TW | EM | Ehlers 2001; Fickel 2001 |
| EEHV2 | NAP#12a, #42 | Nec, Lung, and SkinNod | LA | Richman 1999; Zong 2010; Pearson 2011 |
| EEHV3A EEHV3A |
NAP#27a NAP#42,SAM6 |
Blood/Nec LungNod, SkinNod |
(EM) LA |
Garner 2007 [not natural host] Zong 2010; Pearson 2011 |
| EEHV3B | SAM5,NAP#63 | SkinNod, Blood, TW | LA | Pearson 2012; Bronson 2013 |
| EEHV4A EEHV4B |
NAP#22a NAP#69 |
Blood/Nec Blood, TW |
EM EM |
Garner 2007 Feury 2014 |
| EEHV5A | NAP#29,EP24a | Blood, TW | EM | Latimer 2008; Atkins 2011, Denk 2012 |
| EEHV5B | NAP#58 | Blood, TW | EM | Atkins 2011 |
| EEHV6 | NAP#35, #42 | Blood, Lung Nodule | LA | Latimer 2009; Zong 2010; Pearson 2011 |
| EEHV7A | NAP#42,SAM2 | Lung, Skin Nodule | LA | Zong 2010; Pearson 2011 |
| EEHV7B | SAM8 | Skin Nodules | LA | Pearson 2012 |
| Gammaherpesvirus Sub-Family: | ||||
| EEHV1A | Swab | EM, LA | Wellehan 2007; Ehlers 2007 | |
| EEHV1B | Saliva | EM | Pearson 2012 | |
| EEHV2 | NAG#1, 3, 4 | Swab, Blood | LA | Wellehan; Latimer; Pearson 2012 |
| EEHV3A | NAG#2, 7 | Swab, Blood | EM, LA | Wellehan 2007 |
| EEHV3B | NAG#5 | Blood | EM | Latimer 2008 |
| EEHV4 | Swab | LA | Wellehan 2007; Pearson 2012 | |
| EEHV5A | NAG#6 | Trunk Papilloma | EM | Latimer 2008; Masters 2011 |
| EEHV5B | NAG#8 | Trunk Papilloma | LA | Long 2011; Pearson 2013 |
A listing of the year of discovery, investigator names, and sources of the prototype examples of all of the major different species and subtypes of elephant herpesviruses (elephant endotheliotropic herpesvirus [EEHVs] and elephant gammaherpesviruses [EGHVs]) known. Bl, blood; EM, Elephas maximus; EP#, European Proboscivirus case/sample number; LA, Loxodonta africana; NAG#, gammaherpesvirus case/sample ID number; NAP#, North American Proboscivirus case/sample number; Nec, necropsy; Nod, nodule; SAM, Samburu, Kenya; TW, trunk washing.
aLethal acute hemorrhagic disease.
Table 2 summarizes some of the most typical features, such as host elephant species and clinical or pathological sample types containing them, for each the designated seven species and five major subtypes of the EEHVs.
Table 2.
Elephant endotheliotropic herpesvirus host species and disease or tissue sources
| Strains | Host type | Lethal acute HD | Mild viremia | TWShed | Saliva | Lung nodules | Skin nodules |
|---|---|---|---|---|---|---|---|
| EEHV 1A | EM | + | + | + | + | ||
| EEHV 1B | EM | + | + | + | + | ||
| EEHV 2 | LA | + | + | + | |||
| EEHV 3Aa | LA | + | + | + | + | ||
| EEHV 3B | LA | + | + | + | + | ||
| EEHV4A | EM | + | |||||
| EEHV4B | EM | + | + | ||||
| EEHV 5A | EM | + | + | + | |||
| EEHV 5B | EM | + | + | ||||
| EEHV 6 | LA | + | + | + | |||
| EEHV 7A | LA | + | + | ||||
| EEHV 7B | LA | + |
Summary of the host species and pathological tissue or clinical sample distribution of twelve types of elephant endotheliotropic herpesviruses (EEHVs) as deduced from polymerase chain reaction DNA analysis of clinical samples.
HD, hemorrhagic disease; TW, trunk wash.
aIndex case was LAHD in an EM calf (Hansa).
To complicate matters even further, both Wellehan and colleagues (2008) and our group (Latimer et al. 2011) have described the presence of PCR-amplifiable POL gene DNA from several types of another subfamily of mammalian herpesviruses, referred to as elephant gammaherpesviruses (EGHVs). Once again, a total of five highly diverged EGHV species have now been identified overall (Table 1, lower section), with at least four of these also having distinctive Asian (A) versus African (B) variants (which differ by between 3.5% and 5% at the nucleotide level). None of these latter EGHVs have yet been associated with any particular disease conditions, but they were found commonly in conjunctival swabs from healthy asymptomatic elephants, as well as occasionally in genital swabs, biopsied genital lesions, or blood. One species (EGHV5A and EGHV5B in Asians and Africans, respectively) has been found exclusively in a form of mucosal skin nodules (Latimer et al. 2011; Masters et al. 2011) (S.Y. Long, unpublished data), and extensive ongoing collaborative studies with Virginia Pearson revealed that the other EGHVs could all be detected frequently in skin nodules biopsied from juvenile wild African elephants in Africa, as well as in saliva swabs collected from a majority of randomly sampled healthy adult Asian and African elephants within U.S. zoos (V.R. Pearson, Finding EEHVs in Wild African Elephants by Virginia Pearson https://elephantmanagers.com/…/ElephantManagersArticleKWS__1_.doc). Finally, Table 3 provides a summary of the elephant host species, and observed tissue location and sources found for the five known species and four A/B subtype pairs of EGHVs.
Table 3.
Elephant gammaherpesvirus host species and tissue sources
| Virus type | Host | Saliva swabs | Skin nodules | Blood |
|---|---|---|---|---|
| EGHV 1A | EM | + | ||
| EGHV 1B | LA | + | + | |
| EGHV 2 | Both | + | + | + |
| EGHV 3A | EM | + | ||
| EGHV 3B | LA | + | ||
| EGHV 4A | EM | + | ||
| EGHV 4B | LA | + | ||
| EGHV 5A | EM | + | ||
| EGHV 5B | LA | + | + |
Summary of host species and source tissue distribution of nine types of elephant gammaherpesviruses (EGHVs) as deduced from POL codehops locus polymerase chain reaction DNA sequence analysis of clinical samples. Some have also been detected in conjunctival and genital swabs.
Novel DNA Genomes of Both the AT-rich and GC-rich Branch Proboscivirus
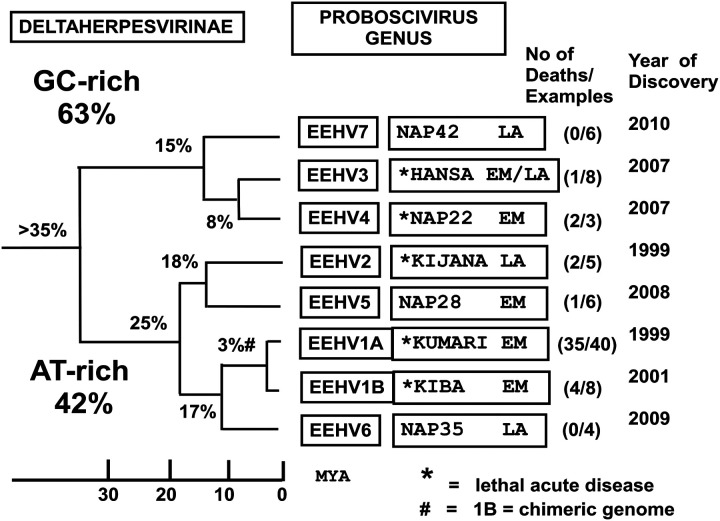
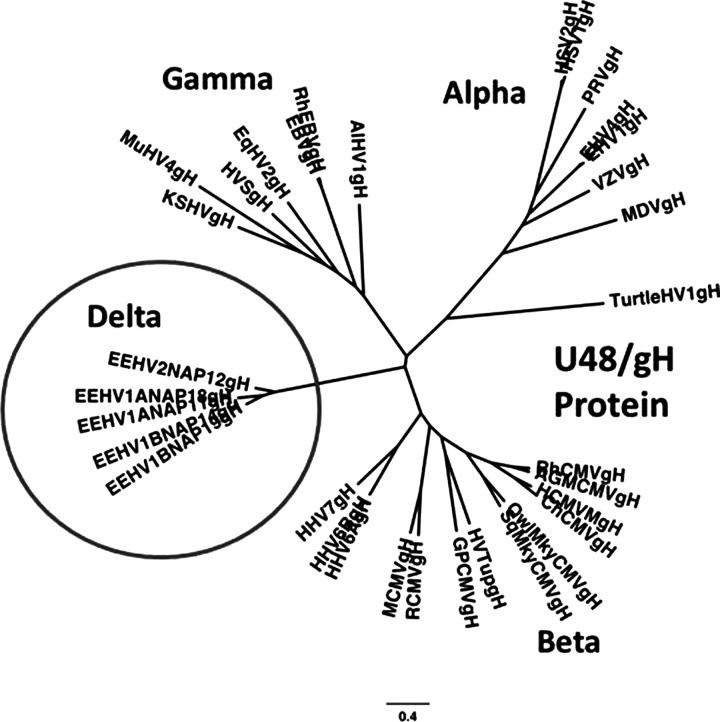
Because the EEHVs have so far proven refractory to all efforts to grow or propagate them in laboratory cell culture conditions (including in white blood cells, primary fibroblasts, and umbilical cord vascular endothelial or placental epithelial cells from both Asian and African elephants), our ability to analyze their genomes, to study their host cell targeting and replication properties, and to understand some of their cellular and molecular biology, as well as to test the efficacy of potential antiviral drugs, has been extremely limited. Nevertheless, the advent and application of very sensitive and specific PCR amplification techniques and DNA sequencing approaches has made it possible to generate a huge amount of selective genetic DNA sequence information about the multiple different EEHV species or strains and their many hypervariable genes (Ehlers et al. 2006; Fickel et al. 2003; Richman 2003; Richman et al. 2014; Zachariah et al. 2013; Zong et al. 2014). A comparative cartoon illustrating the major phylogenetic features of each of the seven EEHV species, including the division into AT-rich and GC-rich branches, the name or case code of the prototype example, the estimated ages of their last common ancestors in millions of years ago (MYA), and the number of lethal cases and total number of thoroughly evaluated samples, as well as the original year of identification, is given in Figure 4. An actual distance-based radial phylogenetic tree comparing the divergence at the protein level of the core U48(glycoprotein-H) of several EEHVs (encircled Delta group) with the orthologous gH proteins of selected examples representing all three other subfamilies (Alpha, Beta, and Gamma) of mammalian herpesviruses, including all nine human herpesvirus species, is shown in Figure 5.
Figure 4.
Genetic relationships among seven distinct species of elephant endotheliotropic herpesvirus (EEHV). Phylogeny-based classification of all major EEHV species within both the AT-rich and GC-rich branches showing average nucleotide divergence levels, approximate estimated ages of last common ancestors (in millions of years ago), name or case code of the prototype example, number of associated lethal cases, total number of examples of each type, and the year of first discovery.
Figure 5.
Elephant endotheliotropic viruses (EEHVs) represent a novel subfamily of mammalian herpesviruses (circled). Radial distance-based phylogenetic tree comparing the amino-acid identity values of the intact U48(glycoprotein-H) proteins of several EEHVs with the orthologous core proteins representative of all major mammalian herpesvirus subfamilies. Because of the lack of a close match to any of the existing defined subfamilies (alpha, beta, and gamma), the EEHV clade (Proboscivirus genus) has been proposed to form a new deltaherpesvirus subfamily (Pellett 2014; Richman et al. 2014; Zong et al. 2014).
More recently, the use of random, next-generation, high-throughput DNA sequencing approaches directly from necropsy tissue has resulted in the assembly and annotation of the complete published 180-kb double-stranded DNA genomes of four examples of the AT-rich branch EEHVs from diseased Asian elephants. These include the strains EEHV1A(Kimba) (Ling et al. 2013), EEHV1A(Raman), EEHV1B (Emelia) (Wilkie et al. 2013), and EEHV5(Vijay) (Wilkie et al. 2014). In addition, we have also now sequenced and annotated the complete 206-kb genome from the GC-rich branch virus EEHV4(Baylor) from an Asian elephant trunk wash DNA sample (PD Ling, SY Long, A Feury, R-S Peng, SY Heaggans, X. Qin, KC Worley, S. Duggan and GS Hayward, unpublished data). Prior to this latter work, only five small PCR loci (totaling just 5-kb in length) had ever been identified by standard, redundant, primer-based PCR sequencing approaches from the GC-rich branch viruses EEHV3, EEHV4, and EEHV7 (Zong et al. 2014).
The most striking features and implications of the genomic analyses of the EEHVs compared with the current three well-defined subfamilies of mammalian herpesviruses, the alphaherpesviruses (HSV1, HSV2, and VZV-like), betaherpesviruses (HCMV, HHV6, and HHV7-like), and gammaherpesviruses (EBV and KSHV-like) are that this group of elephant viruses forms a distinct highly diverged clade, as illustrated in the Figure 5 “family” tree. Very similar results were obtained with all of the core genes of these viruses and imply that they have evolved separately from all other mammalian herpesviruses for approximately 100 million years in the ancestors of modern elephants (Ehlers et al. 2006; Richman et al. 2014; Zong et al. 2014). The EEHV genomes all encode between 115 and 120 total genes, of which only 35 are core genes shared with all other mammalian herpesviruses, 60 are completely novel genes, and the rest include several genes that share homology with orthologues within the alphas and gammas but not the betas (three), or in just the betas (eight), or in both the gammas and betas but not the alphas (seven).
Despite being somewhat closest in overall gene content and organization to the roseoloviruses (i.e., human HHV6 and HHV7) within the Betaherpesvirus subfamily, only one of the 35 shared core proteins (TER) has as much as 50% amino acid identity to its orthologues in any other herpesvirus. The EEHVs also lack many of the genes that are considered characteristic of and unique to betaherpesviruses, and therefore they are not just elephant cytomegaloviruses. Instead they use alphaherpesvirus-like dyad symmetry domains and a UL9-like OBP protein for lytic DNA replication, rather than the large complex lytic origins typical of cytomegaloviruses, and their major immediate-early-like nuclear transcriptional regulators have no direct homology at all to those of any of the other three currently defined subfamilies. They also have a large 40-kb inversion of the central core gene block compared with the cytomegaloviruses, muromegaloviruses, and roseoloviruses. In fact, based on both their overall unique biology and the many novel genomic features, we have proposed that the entire EEHV family should be classified as members of the Proboscivirus genus within a newly named fourth mammalian herpesvirus subfamily, the deltaherpesviruses (Ling et al. 2013; Pellett 2014; Richman et al. 2014; Zong et al. 2014).
There are two separate highly diverged branches of the Proboscivirus (Figure 4). EEHV1A, EEHV1B, EEHV2, EEHV5, and EEHV6 all fall into the AT-rich branch, with 43% overall G plus C content, which is estimated to have separated from the GC-rich branch containing EEHV3, EEHV4, and EEHV7 approximately 35 to 40 million years ago (Zong et al. 2014) or close to the time that old-world primates diverged from new-world primates and the elephantids diverged from mastodons. Among their most conserved core genes, EEHV1 and EEHV6 differ by an average of 17%, EEHV2 and EEHV5 by 18%, and EEHV3 and EEHV4 by approximately 12% at the nucleotide level, but most of their novel genes diverge by twofold to threefold more than that (Zong et al. 2014). EEHV2 and EEHV5 lack one of the large novel glycoproteins (ORF-Q) compared with EEHV1 and EEHV6, have four rather than three inserted vOX2 genes, and also have a totally rearranged version of the 10-kb vIgFam plus vGPCR gene block at one end of the genome. Although EEHV1A and EEHV1B are quite distinctive, they are not separate species, but rather partially chimeric variants with 12 proteins (mapping in three small loci covering a total of 15 kb) that differ by an average of 40% at the amino acid level, but they also have large areas that are almost indistinguishable (Richman et al. 2014; Wilkie et al. 2013). EEHV5A and EEHV5B also display rather similar chimeric features. However, EEHV1B lacks a likely inhibitory vCXCL1 chemokine gene found in all other AT-rich branch viruses (Richman et al. 2014; Zong et al. 2014).
The additional novel genes not shared with other herpesviruses fall primarily into a large related set of between 24 and 28 members of the 7xTM domain protein family, of which between 8 and 10 have significant homology with classical host G-protein coupled receptors (GPCRs), including a subset that especially resembles the C-5-A orphan receptors, such as the retinoic acid induced protein 3 (vRAIP3). Other new EEHV-encoded proteins include up to five membrane-bound immunoglobulin (IgFam) family members (but in EEHV1 and EEHV5 only, not in EEHV4) and several large, novel, group-specific envelope glycoproteins or tegument proteins. There are also two more vGPCRs that are chemokine receptors and a small number of isolated evidently relatively recently “captured” or “pirated” cellular genes, including an acetyl glucosamine transferase (vGCNT1), a fucosyl transferase (vFUT9), and three or four variants of vOX2s in both EEHV1 and EEHV5.
With regard to the GC-rich branch compared with AT-rich branch viruses, at 206 kb, EEHV4 is 28 kb larger than the others and displays a novel pattern of GC-rich ORF coding regions interspersed with very unusual large noncoding regions full of 5- to 12-bp–long A or T tracts. Therefore, although the EEHV4 coding genes themselves nearly all average approximately 62% to 65% GC content (with the third wobble position of the codons often above 90% GC content), the overall genome has 55% GC content. The core genes of EEHV4 differ by, on average, 40% to 50% at the amino-acid identity level from those of the AT-rich branch, with about half the novel subfamily limited genes being still related but highly diverged by an average of 70% or more, whereas the other half of their novel genes are either absent, rearranged, or duplicated. Compared with the other viruses, EEHV4 lacks a total of 25 genes, including the captured fucosyl transferase gene, the ORF-P and ORF-Q glycoproteins, and three of the four vOX2 genes, but harbors an extra novel UDP-acetyl glucosamine transferase (vOGT), a novel ORF-R glycoprotein, a C-type lectin gene (vECTL), at least six more copies of the 7X TM family, and up to a dozen additional unidentified genes. The single large ORF-L immediate-early-like nuclear transcriptional transactivator, the MTA multifunctional post-transcriptional regulator, and the genus-specific ORF-C, ORF-K, and ORF-M proteins are all, on average, some 50% larger than their counterparts in EEHV1 or EEHV5, with each possessing only 15% to 30% overall residual amino-acid identity. EEHV4 also lacks the entire complex 10-gene block of membrane-bound vIgFam plus vGPCR family genes at the right-hand end that we have characterized as the most hypervariable segment among EEHV1 strains.
Epidemiological Implications of Detecting Numerous Distinct Strains of EEHV1 in Viremic Disease Cases
Until recently, some very vocal animal rights activists used the pretext of EEHV disease as one of their strongest points against breeding elephants in captivity in zoos and circuses, arguing that the transport of animals (or sperm) among breeding facilities and the existence of contaminated facilities, together with the stress of captivity, caused the spread of this unnatural infection. There was even a published paper (Ryan and Thompson 2001) that purportedly traced the transmission of a single causative entity for nearly all known cases of EEHV between and among elephant housing facilities in North America. However, the problem with this scenario that they proposed is that there has not actually been any movement of EEHV strains among different breeding facilities. The disease is sporadic, not epidemic—that is, it involves numerous different strains of multiple species of EEHVs, and each facility has its own strains (Zong et al. 2008, 2009). There is not a single documented example of the same strain being present at more than one facility in either Europe or North America. Indeed, there have been several examples of multiple infections with identical viruses in different animals at the same facility. These have involved either lethal cases or survivors and usually occur at nearly the same time. But otherwise each facility has its own strains, and where examined in detail, most (and probably all) facilities have proven to have multiple strains and species of EEHV already present naturally within their herds. How do we know this? Simply by carrying out DNA sequencing analysis on PCR products from multiple regions of these viruses from every case of disease or of virus infection detected for which clinical tissues or blood samples can be collected, as well as by random screening of trunk washes or saliva swabs from closely monitored herds.
Right from the beginning, we designed precise analytical PCR DNA tests that would distinguish between EEHV1 and EEHV2, then also for EEHV1A versus EEHV1B, and later also for EEHV3, EEHV4, EEHV5, EEHV6, and EEHV7 (Latimer et al. 2011). With these reagents we learned that “active” infections in U.S. elephants involved up to seven different species or types of EEHVs, with at least five of these leading to lethal cases. In addition, the DNA sequence data (even for common well-conserved core genes such as POL, U71/gM, and HEL) among just the EEHV1 samples revealed (as expected) low levels of polymorphisms typical of the known range of sequence variations found among different strains of human and animal herpesvirus species (Stanton et al. 2010; Zong et al. 2008). For the POL, TER, and HEL loci, all EEHV1A strains displayed approximately 3% common nucleotide variations from all EEHV1B strains, although for some other genes, including those involved in the chimerism as well as those referred to as hypervariable, the differences are very much greater (Zachariah et al. 2013). However, even among EEHV1A strains, there are low levels of sporadic but stable strain-defining polymorphisms found across all of the core genes (0.1% to 0.5%) (Zong et al. 2008; Zong et al. 2009). This latter value is quite similar to the level of DNA polymorphism found in the same gene among different individual humans (unless very closely related or identical twins). Sometimes multiple different EEHV strains just happen to be identical in some of their 500- to 1000-bp loci examined, but once 10 or so of even these most conserved loci have been sequenced (as we have done for all known North American cases after 1995), all independent strains show clear characteristic polymorphisms, whereas, on the other hand, closely epidemiologically connected strains show no divergence at all (often even over a total of up to 10,000 bp sequenced). The results revealed that the EEHV1A strains present in every single case occurring at a different facility, as well as even several from the same facility, and even among multiple progeny with the same parents, were distinct.
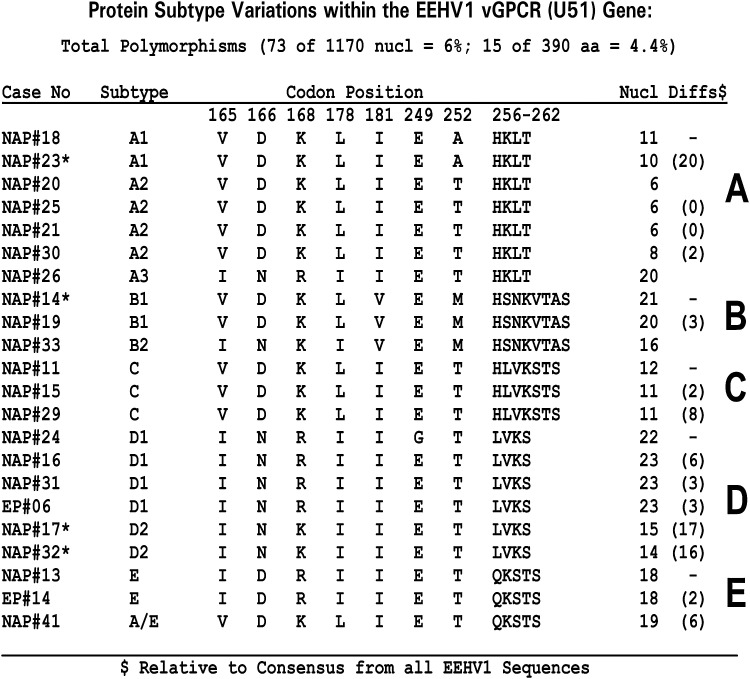
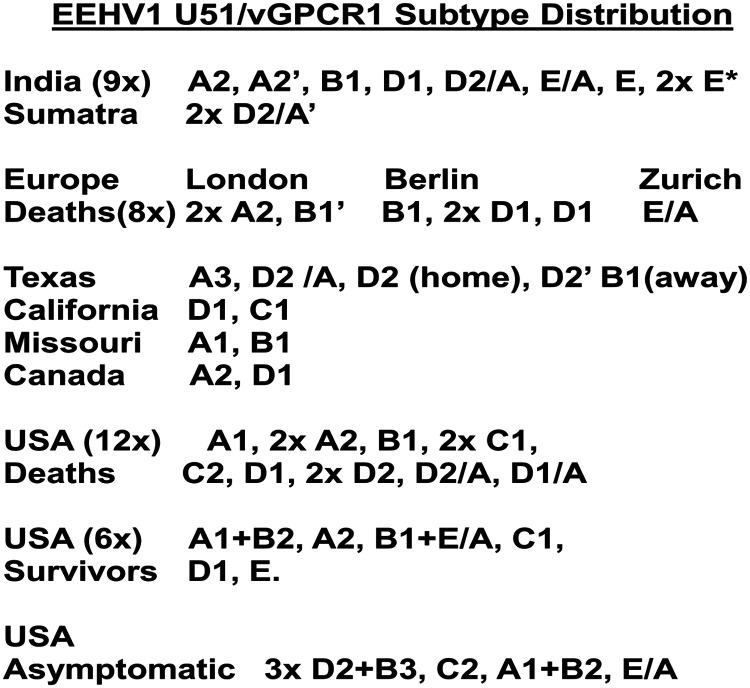
Later this type of analysis was extended to all known cases in North America, as well as 10 cases from Europe and nine cases from India over three or more additional hypervariable gene loci (Zachariah et al. 2013) (J-C Zong, EM Latimer, SY Heaggans, SY Long, LK Richman, GS Hayward, unpublished data). The results of these studies revealed that individual EEHV1 strains all fall into between five and seven clustered subtypes at the vGPCR1 and TK-gH loci that form quite distinct and individually well-conserved clades. Accordingly, we can define clusters of strains as having A, B, C, D, or E subtype vGPCR1s (Figure 6), often with additional variant structure such as B1, B2, B3, or C/D chimerism (Figure 6) and A, B, C, D, E, F, or G subtype TK-gH genes. For both loci, the B subtypes occur only within those strains defined elsewhere in their genomes as being EEHV1B chimeras, but otherwise the other subtypes show no linkage between the two loci. A summary of the different vGPCR1 subtypes found in lethal EEHV1 cases worldwide, as well as those in survivors or asymptomatic shedders, is detailed in Figure 7. Except for a small number of pairs of identical strains at the same facility (see below), all EEHV1 strains examined have been distinguishable, implying distinct epidemiologic origins. Furthermore, nearly all subtypes were represented in Asia as well as in Europe and North America and also found among both survivors and lethal cases.
Figure 6.
Wide range of genetic variation among elephant endotheliotropic herpesvirus (EEHV) strains. Observed amino-acid polymorphisms among polymerase chain reaction–analyzed DNA sequences for the U51(vGPCR1) protein from 22 examples of EEHV1-associated hemorrhagic disease from North American (NAP#) or European (EP#) Proboscivirus cases. The particular characteristic motif between positions 252 to 260 defines the major subtype designation (A, B, C, D, or E). * = Four cases born at the same U.S. facility.
Figure 7.
Lack of spread of specific EEHV1 strains between elephant housing facilities and broad distribution of EEHV1 strains worldwide. Summary of the U51(vGPCR1) subtypes identified up to the year 2012 among lethal and surviving or asymptomatic elephant endotheliotropic herpesvirus (EEHV) hemorrhagic disease cases worldwide. Home and away refer, respectively, to calves born at a single facility that died at the same facility or after transfer to another facility.
When we did find samples from two or more diseased elephants that were identical EEHV1 strains at all of the core gene PCR loci, they have always also proven to be identical at all of the hypervariable loci examined as well. Therefore, we consider it reasonable to assume that these samples are indeed truly identical throughout the rest of their genomes. In all six occasions where identical pairs of strains were identified among infected calves with either viremia or hemorrhagic disease (one pair each at zoos in England and Germany and at orphan camps in Sumatra and Kerala, as well as two pairs at the same U.S. zoo), the cases occurred within just a few days or weeks of each other at the same facility. Furthermore, the pair at the German zoo were also identical to a third lethal case that occurred there 11 years earlier.
Sequential Transient Shedding of Multiple EEHVs Detected in Trunk Wash Screening Assays
After the development of faster, more sensitive, and quantitative real-time assays for detection of EEHVs (Stanton et al. 2012), we also applied the gene subtyping analyses to many examples of identified positive trunk wash shedding, as described in Stanton and colleagues (2013). These types of studies revealed several other examples of identical viruses involving both a surviving calf with moderate viremic disease symptoms and one or more asymptomatic adults shedding high levels of the same virus in trunk washes several years later (but these always occurred at the same facility). Close monitoring at a zoo in Texas also revealed exactly the same strain of EEHV1A in both blood and trunk washes from two young surviving Asian calf herdmates with minimal disease that matched exactly to the virus strain found in necropsy tissues from a calf that died there three years earlier (Stanton et al. 2010; Stanton et al. 2013). Most intriguingly, not only were a pair of surviving young calves at another facility found to harbor identical EEHV1B strains at the same time, but they both also underwent and survived viremic episodes with identical EEHV1A strains about a year later. This type of phenomenon of multiple sequential infections with EEHV1A then EEHV1B (or vice versa) has been described in a total of five closely monitored Asian elephants (at three different facilities) and includes the calf with the adult herdmate shedding an identical EEHV1A strain (Stanton et al. 2013). We note that no examples of a single elephant displaying shedding of more than one strain of either EEHV1A or EEHV1B have yet been detected.
Later, in 2012 during similar close monitoring of the same zoo herd in Texas, all herdmates were observed to undergo sequential transient infection episodes with EEHV5 involving just mild viremia, followed by a later peak of high-level trunk wash shedding (Atkins et al. 2013). Curiously, this event proved to involve two distinct subtypes of EEHV5A and EEHV5B that we subsequently characterized as another example of partial chimerism (Zong et al. 2014). Finally, in summer 2014, three Texas zoo calves also underwent similar viremic episodes, followed again by transient trunk wash shedding with the same identical strain of EEHV4B (Fuery et al. 2015). In fact, this Texas zoo herd, which has been monitored very closely in recent years and currently consists of just eight Asian elephants, has been shown to have had a total of three lethal cases (between 2000 and 2008) of EEHV1A disease caused by different strains of EEHV1A, as well as subsequent nonlethal, mildly viremic infection episodes by EEHV1A, EEHV1B, EEHV4B, EEHV5A, and EEHV5B (each involving multiple adults and juveniles). Furthermore, over the past five years, several currently healthy herdmates there (including juveniles) have been observed to undergo successive active infections by EEHV1A, EEHV1B, EEHV5A (or EEHV5B), and EEHV4B (but never at the same time).
African Skin Nodules, Saliva Shedding, and Natural Hosts
Clearly the findings by Zachariah and colleagues (2013) that the nine Southern India cases of hemorrhagic disease also involved EEHV1, together with the detection of multiple other EEHV types but not EEHV1 within lung and skin nodules of African elephants (J-C Zong, SY Heaggans, SY Long, EM Latimer, SA Nofs, M Fouraker, VR Pearson, LK Richman, GS Hayward, unpublished data) (V.R. Pearson, unpublished data), have major negative implications for the originally proposed concept that EEHV1 may have crossed species from African elephants to Asian elephants in zoos. Furthermore, our extensive genetic analysis showing that the nine strains in India involved both one case of EEHV1B and seven distinct strains of EEHV1A also just about completely eliminates any possible scenarios involving introductions of EEHV1 from Africa up to a thousand years ago via Hannibal's, or Alexander's, or the Assyrian army's war elephants (in which case the strains found now would likely be very few and nearly identical). In fact, just two orphan calves affected at the same camp in Kerala at the same time were identical, with the other six strains showing just as much variation in their hypervariable vGPCR1 and TK-gH gene loci as detected across the whole range of EEHV1 strains studied in both North American and European cases (Figure 6). Importantly, the same wide range of vGPCR subtypes was also found in the very recent large Thailand study (S. Sripiboon, personal communication, 2015, Chang Mai, Thailand). This result implies that probably all of the strains causing EEHV disease in zoos were derived from a large variety of strains that were originally carried from Asia to Western countries in a likely unapparent latent state within previous generations of wild-caught Asian elephants.
Therefore, if EEHV1s are, in fact, all natural ancient infections of Asian elephants, which (if any) EEHV species are found within African elephant skin nodules, such as those originally observed in the Florida calf outbreak in the 1980s, and were the original claims for EEHV1 there incorrect? Three questions needed to be answered: (1) Could EEHV1 be found in those same archival skin nodules specimens when they were reexamined? (2) If not, which other EEHVs might be present within the Florida nodules? and (3) Which viruses can be found in new samples of skin nodules obtained from wild African elephants within Africa? The answer to the first question was negative; upon re-examination we could not detect EEHV1 in DNA extracted from paraffin block sections from several of the same archival skin nodules used in the original PCR studies. However, we did not at that time search for other EEHVs, although Virginia Pearson is now doing so, having located more such samples. Paraffin block–derived DNA samples are notorious for being difficult to work with, especially for sequence analyses, including being very susceptible to contamination (which we presume occurred in the original studies), although interestingly the only viral DNA found so far in the new studies is that from two distinct strains of African elephant polyomavirus).
However, Virginia Pearson, working together with Save the Elephants and the Kenya Wildlife Service and other groups in Botswana, has now collected and tested more than a dozen skin nodule biopsies (and saliva swabs) that were field preserved by a variety of different methods from immobilized juvenile African elephants exhibiting the typical kind of “herpes” skin nodules (V.R. Pearson, personal communication, 2012; Finding EEHVs in Wild African Elephants by Virginia Pearson https://elephantmanagers.com/…/ElephantManagersArticleKWS__1_.doc). Her initial PCR analyses were carried out in a completely EEHV–naive laboratory at Princeton University, and most of her results were later independently confirmed in our Johns Hopkins laboratory. Similar to our results above with lung nodules, the majority of her biopsied skin nodules proved to contain multiple examples each of unique and novel strains of EEHV2, EEHV3, EEHV6, and EEHV7. These are mostly of low abundance and difficult to amplify and sequence until the second or even third round of conventional PCR. In addition, even although only five PCR loci were available for study for the GC-rich branch genomes, the results have revealed an even greater complexity of two distinct subtypes of EEHV3 and two of EEHV7 (sometimes with both EEHV3A and EEHV3B or both EEHV7A and EEHV7B or even all four together within the same nodule). Remarkably, many of these same biopsied skin nodules also contain one or more EGHV DNAs as well as the elephant polyomavirus DNA. Her extensive studies have also revealed the presence of often high levels of one or more EGHVs and sometimes also EEHV2, EEHV3, EEHV6, or EEHV7 in many randomly collected saliva swabs from the majority of tested healthy adult African elephants in the wild in Africa and under human care in U.S. zoos. As an important control, when the saliva studies with EGHV1 primers were carried out on both Asian and African zoo elephants, EGHV1A was found in eight Asian elephant salivas, whereas only EGHV1B was detected in saliva from numerous wild and captive African elephants. New African elephant–specific examples of EGHV3B and EGHV5B have also been identified, whereas both Asian and African elephants appear to carry and shed the same single subtype of EGHV2.
From the combined results of all of these genetic analysis studies, it is now inescapable to conclude unambiguously that EEHV2, EEHV3A, EEHV3B, EEHV6, EEHV7A, and EEHV7B are all natural, predominantly innocuous infections of African elephants. Whereas in contrast—although without lung or skin nodules the numbers of examples are much smaller—EEHV1A, EEHV1B, EEHV4, EEHV5A, and EEHV5B are most likely also all natural infections of Asian elephants worldwide. Furthermore, EEHV infections in African elephants can also be concluded to be largely ubiquitous, with many animals even having multiple EEHV species (and EGHVs) detectable within the same nodules or saliva samples. On the other hand, although we have described several examples of multiple sequential infections detectable at different times in blood or trunk washes of individual Asian zoo elephants, few, if any, examples of multiple simultaneous EEHV infections have yet been observed in any Asian elephants. Finally also, as yet, only a very small total number of EEHV4 and EEHV5 infections have been identified, with the majority of those being nonlethal even when involving viremias. It will be important to determine whether the latter distinctions are valid comparisons (or not) between the situations pertaining to Asian versus African elephants, or whether they are just the outcomes of the different sample sources and methods used, or perhaps in the case of EEHV4 and EEHV5, just reflect the lower frequency of testing that has been carried out.
Finally, in this regard, the case of a 7-year-old Asian elephant (NAP27) at a zoo in the United States that died in 2006 from a very heavy load acute EEHV3A infection (Garner et al. 2009; Latimer et al. 2011) is currently the only known example of what must (by implication) have been a cross-species infection from an unknown African elephant source. However, the single adult African elephant that was housed with this calf tested negative for EEHV in the blood both before and after the calf died.
Only one episode of EEHV viremia and shedding in an African elephant has been thoroughly evaluated by the techniques we applied to Asian elephants. In this case, a young male L. africana was observed to have typical EEHV symptoms, and after testing revealed the presence of moderately high blood levels of an EEHV3B strain, he was treated and recovered. Subsequently, a large series of DNA studies from blood, trunk washes, and saliva collected throughout his illness have been evaluated in as yet unpublished collaborative studies between the Johns Hopkins (G Hayward), NEHL (E Latimer), Baylor (P Ling) and Fox Chase groups (V Pearson) with Ellen Bronson and Mike McClure at the Maryland Zoo. In essence, the results have shown that after the clinical symptoms and viremia subsided, exactly the same strain of EEHV3B was shed for six months in both trunk wash and saliva samples before complete clearing. One of this elephant's female adult herdmates also shed lower levels of the same strain of EEHV3B in saliva for a short time during this period. Interestingly, all four herdmates were also found to periodically shed one or more of four different types of EGHVs in saliva samples collected before, during, and after this elephant's illness (E. Bronson et al., unpublished data). Overall, we conclude that African elephants can and do undergo similar, although probably usually far less severe, patterns of primary infections with at least EEHV3 as detected in Asian elephants with EEHV1, EEHV4, and EEHV5.
Clinical Samples, Monitoring, Serology Assays, and Drug Treatment
A major component of the very successful and informative EEHV genetic analyses described herein has been the diligence of the dedicated veterinary staff, elephant keepers, and pathologists who have collected and shipped appropriate fresh and frozen clinical or necropsy tissue samples for submission to the NEHL for DNA testing, virus and strain gene subtyping, and long-term archiving. We would still know next to nothing about these viruses without this essential resource. Most involved facilities in the United States have in place appropriate local IACUC-approved protocols for routine or emergency blood, serum, and trunk wash or saliva collections and blood draw training for their calves. The NEHL can usually give an emergency DNA blood test result with a turnaround time of 12 hours after receipt and has programs in place for both initial and continuous routine DNA test screening by a variety of techniques. In addition, both Paul Ling's group at Baylor College of Medicine and Virginia Pearson at Princeton and Fox Chase have organized and carried out large routine screens for monitoring by real-time or conventional PCR procedures at several participating facilities. Together with the Johns Hopkins research group, they have advised and assisted in such screening, collecting, or testing of samples from potential cases or from nodule biopsies in both Asian and African range countries and continue to do so.
Routine monitoring of Asian and African calves is recommended by the EEHV Advisory Group and the American Zoological Association Taxological Group (AZA-TAG) and is described on the eehvinfo.org website. Detailed and frequently updated regimens for recommended high-quality treatment of suspected cases are available, including antiviral drug administration, and also, when necessary, for detailed pathological examination. Thorough necropsy sample collections have been organized by the EEHV Advisory Group, AZA-TAG, and the International Elephant Foundation, with detailed written protocols and recommendations being made widely available on several Internet sites. Although there is no question that close monitoring and good immediate medical care have helped to save a number of acutely infected Asian calves in recent years, the issue of whether the administration of human antiherpesviral drugs such as FCV, GCV, or even aciclovir (ACV) has helped or not is unresolved. Although they were initially tried and seemed to be effective before it was even known whether the EEHVs encoded a viral TK enzyme or not, the fact is that the targeting of these antiviral drugs is highly selective toward the relatively few virus species that are known to be susceptible. Although all of the sequenced EEHV types do encode both a viral TK enzyme (the usual target for FCV and ACV) and the conserved protein kinase enzyme (CPK or UL97 equivalent, the target for GCV), both have only 25% amino-acid identity to their orthologues in HSV and HCMV (where those drugs work best). Direct testing for the efficacy of these drugs is not possible without, as yet (despite considerable efforts), a successful laboratory cell culture model for the growth and propagation of EEHV. A few indirect surrogate in vitro assays have been carried out with the cloned expressed EEHV enzymes (C-J Chiou, C ap Rhys, GS Hayward, unpublished data), but the results have not been very encouraging, with the CPK perhaps having some weak activity targeting GCV (M. Ackerman and P. Ling, personal communication), but with the TK especially being almost totally inactive in GCV cell-killing assays at least. Although additional in vitro scientific approaches and funding resources are needed to obtain definitive answers, it seems most likely the 9 or 10 young Asian calves that were treated with drugs and survived proven symptomatic EEHV1-positive acute viremic infections represent relatively mild cases that either were diagnosed very early and able to recover because of rapid good medical care or would have survived without the drug treatment anyway. Certainly, the evidence from DNA testing shows that none of the survivors had anywhere near the extremely high levels of viral DNA in blood and serum as did most of the lethal cases (by an order of magnitude or more). Nevertheless, one can hardly question the common practice of keeping sufficient quantities of these very expensive drugs on hand just in case at many facilities, and we have even seen them (especially ACV) within camp pharmacies in India and Nepal. Sensibly, some enterprising facilities have created cooperatives for the sharing of drugs where and when needed and of exchanging them for nonoutdated samples with local hospitals when the shelf life has nearly expired. The problem is that the disease can strike so fast and no one wants to lose a calf or be accused of not doing everything possible to save them.
Routine prospective archival collection of Asian and African elephant sera have also been organized and carried out by the International Elephant Foundation together with NEHL for more than 10 years now. However, compared with the precision and high sensitivity of DNA sequence testing, the results for EEHV antibody testing have been disappointing and relatively unhelpful. At first, a simple enzyme-linked immunosorbent assay–based serology assay test using a synthetic epitope fragment of the gB protein antigen from EEHV1A was developed (Richman 2003) and used for testing a large set of banked adult Asian elephant sera. Although this did identify a small number of individual elephants with consistently high antibody titers, it became apparent when screening selected African elephant sera as well that it must be giving cross-reacting results with other similar viruses carried by African elephants and presumably by extension in Asians as well. In addition to the lack of specificity, this single epitope assay proved to be quite sensitive to heat denaturation in serum samples that had suffered freeze–thaw episodes (a common occurrence in range countries). Another similar serology assay using the large intact multiepitope gB protein of EEHV1, which may be more robust with regard to the quality and history of the sera but unfortunately likely provides even greater opportunities for cross-reactions with the many other EEHV types that were later identified, was also developed in Europe. We have a project in progress to attempt to develop more robust, more sensitive, and more specific antibody tests, especially using a fluorescent microarray chip approach with multiple sets of protein antigens cloned and expressed in yeast from several different EEHV types, but interruptions in both the funding and scientific personnel involved have kept progress rather slow, and the task has become more and more complex as new EEHV types have continually been discovered. Considering our current anticipation that almost all adult Asian and African elephants are already latently and asymptomatically infected with multiple EEHV types, such assays are most likely to be primarily useful (if indeed they can discriminate between the different EEHV types) in monitoring susceptible young calves for prior infections, as well as perhaps in the future for testing of any candidate vaccines that might become available, although the task of generating and properly testing either live or attenuated vaccines in this situation faces almost impossible barriers.
Pathogenesis, Proposed Disease Models, Order and Timing of Primary Infections, and Fragmentation of Wild Habitats
The original proposal in 1999 of EEHV hemorrhagic disease in Asian elephant calves as a cross-species infection acquired recently from African elephants seemed to make sense of the unexpected high severity of this disease, based on the precedent of fulminant herpes B virus infections in humans (and vice versa for the closely related human HSV in rhesus macaques). However, now that it is very clear that EEHV1 is a natural endogenous (and probably nearly ubiquitous) infection of Asian elephants and one that must have been present in this host species for at least as long as Asian elephants have been a distinct species from African elephants, the disease itself becomes a major problem to understand.
Generally, lethal disease within the natural host is a very rare situation among the very slowly evolving mammalian herpesviruses, in which both host and virus have had eons of time to adapt and form a benign interaction. Furthermore, no other herpesviruses are known to produce such a rapidly progressing disease of this type. Overall, hemorrhagic disease does not seem to be a generic feature of EEHVs themselves: after all, benign infections by multiple EEHV species occur possibly even more frequently and ubiquitously in African elephants than in Asian elephants. Yet, whereas up to 20% of all Asian elephant calves appear to be susceptible to EEHV disease, in contrast, severe hemorrhagic disease is extremely rare in African elephants (which presumably undergo all of the same proposed stresses in both the wild and in captivity as Asian elephants might do so). Admittedly, six of the seven known species of the EEHVs have been observed to produce viremia, including at least one lethal case of hemorrhagic disease each, but more than 90% of all cases have involved just a very large variety of distinct strains of EEHV1A. Furthermore, the EEHV1 cases seem to have involved just about all known subtypes, without there being any particular preference for certain subtypes in disease compared with mild infections. Certainly, it is also clear that the exact same EEHV1A strain can be lethal in one calf but not in other herdmates at the same facility.
These points tend to argue against any notion of a particularly highly pathogenic mutant or variant being responsible, as is the case for equine herpesvirus disease, for example. One could perhaps argue that because of considerably more mixed breeding from parents of diverse Asian origins in zoos, the current generation of captive-born Asian elephants are far more genetically heterogenous than even their most recent ancestors would have been. This, in turn, could conceivably affect their susceptibility to EEHV1 disease, which presumably must not have been anywhere near as great in the past or the E. maximus species as a whole could hardly have survived and flourished for four million years. However, this idea is clearly somewhat mitigated by the high rates of disease now also being detected in Asian herds, as well in the captive Western diaspora. Assessments of whether EEHV disease is newly emerging in Asia itself are hard to judge. On the one hand, there was apparently no indication of this sort of hemorrhagic disease among the ancient written records of early Indian elephant culture, but on the other hand there were many recorded die-offs of Asian elephants back in the 1800s, which were sometimes attributed to anthrax, for example. It has also been pointed out that even the recent cases now recognized as EEHV disease within Myanmar and Thailand were dismissed originally (without necropsies being performed) as just natural events or septicemia or even poisoning.
Another alternative scenario could be that the disease is actually caused instead by some other unidentified agent, with the observed associated high EEHV viral loads being a reactivation event in response to immunosuppression. However, one can make several major arguments against this possibility. First, the EEHV1 viral DNA loads in the blood in the severest disease cases can reach as high as 10 to 75 million viral genome equivalents per milliliter (which is 1,000-fold to 10,000-fold higher than in even immunosuppressed humans with either EBV or HCMV viremia that is considered severe enough to be treated with antiviral drugs). Second, until the last stages of acute disease, the EEHV DNA remains associated with mononuclear white blood cells rather than spilling out as free virions into the serum. Third, in the relatively few cases studied, there has been a clear correlation between levels of virus in the blood and the severity of symptoms, with all the known survivors never reaching the same extremely high levels of viral DNA in the blood or having any free viral DNA in the serum as found in the lethal cases. In fact, viral DNA loads above approximately 200,000 per milliliter in the blood are fully predictive of associated symptoms and vice versa (whereas, intriguingly and conversely, no clinical signs have ever been observed in elephants shedding even the highest measured trunk wash viral DNA loads). Fourth, if the EEHVs were just passengers, one would not expect every observed disease case to also have high levels of EEHV viremia, nor for 85% to 90% of the reactivated EEHVs to be just the one species. Also some would surely have multiple reactivated EEHV species at the same time, whereas not a single example of viremia involving substantial amounts of a second EEHV species as well has been documented. Fifth, considering the six occasions where two calves fell ill at nearly the same time with exactly the same virus strain, how could that be the case if the EEHVs were just passengers rather than the actual causative agent (previously infected calves would have reactivated different strains)?
This leads to the final most telling point, which is that our current evidence and theory strongly suggest that all cases of severe viremic disease involve primary infections. Indeed, no examples exist of a proven reactivating virus that is the same strain as detected in an earlier viremic episode from the same animal, even in the most closely monitored herds. In fact, only two potential surmised reactivated infections that apparently initiated a subsequent series of downstream primary infections in herdmates have been documented. In both instances, this interpretation is based on evidence that the trunk wash shedding observed in the initially infected reactivating animal was not associated with an immediately prior viremia, whereas there was both a transient viremia and a subsequent trunk wash shedding event in all of the other later primary infections within the other herdmates (Atkins et al. 2013; Stanton et al. 2013).
What then is the mechanism of pathogenesis here? One likely telling point is the nearly complete absence of severe or lethal disease in calves under one year of age. Obviously, this suggests that maternal antibodies could be providing immune protection, not against infections necessarily but certainly against disease. Furthermore, if primary EEHV1 or infections by other EEHV types occur during the period when maternal antibodies are available and then the calf develops its own antibodies (or cellular immunity) against these mild EEHV infections, it is likely that the antibodies against at least the most closely related EEHVs could be protective against disease (but not infections) caused by future exposures to either EEHV1 or the other EEHVs. In this regard, the evidence given above clearly tells us that prior—even symptomatic—infections by EEHV1B do not appear to protect against infection by the other subtype (or vice versa), whereas infections by other strains of the same subtype probably are protected against in this case.
Conclusions
These observations all lead to a fairly obvious proposed working model whereby the timing and order of multiple primary EEHV infections are the keys to disease protection or not, and if one or more of these are significantly delayed, especially until after maternal antibodies are no longer available, then susceptibility to disease may become manifested in accordance with the peak of disease susceptibility that has been observed (between one and four years of age). It seems reasonable to interpret that there are relatively fewer and reduced opportunities to transmit those key early primary infections in smaller, less interactive herds in captivity versus in conditions that normally should pertain (or that used to pertain) within the wild with higher chances to be exposed to numerous viruses and that similar captivity-like conditions would also be present among small groups of orphans in Asia. It is also likely that these same types of altered conditions, involving loss of habitat and mobility and smaller herds, perhaps with reduced host genetic diversity, must likely also be coming into play among the dwindling remaining wild herds and are likely to be further exacerbated in the future.
Acknowledgments
We thank our colleagues Jiang-Chao Zong and Sarah Y. Heaggans for assistance in carrying out extensive PCR sequencing and phylogenetic analyses, as well as our collaborators in Paul D. Ling's group at Baylor College of Medicine. We also thank Virginia R. Pearson at Princeton University and Fox Chase Cancer Center and Arun Zachariah of the Wayanad Forest and Wildlife Service and the Veterinary and Animal Sciences University in Kerala for open exchange of materials or protocols and sharing research activities and results. Studies at Johns Hopkins University were supported at various times by research grant R01 AI24576 to G.S.H. from NIAID, National Institutes of Health, DHEW, the International Elephant Foundation, and a subcontract to Lauren Howard at the Houston Zoo under an IMLS National Leadership Collections Stewardship Program. S.Y.L. is the recipient of a Morris Animal Foundation Postdoctoral Fellowship D14ZO-411. Studies at the NEHL were supported by research grants to Erin Latimer, Laura K. Richman, and Timothy Walsh from the Smithsonian Institution, the National Zoo, the International Elephant Foundation, the Morris Animal Foundation, and the Ringling Bros & Barnum and Bailey Center for Elephant Conservation. We are especially grateful to all of the other wildlife and zoo veterinarians and elephant managers worldwide who provided clinical or pathological samples for diagnosis and research purposes.
References
- Atkins L, Zong J-C, Tan J, Mejia A, Heaggans SY, Nofs SA, Stanton JJ, Flanagan JP, Howard L, Latimer E, Stevens MR, Hoffman DS, Hayward GS, Ling PD. 2013. EEHV-5, a newly recognized elephant herpesvirus associated with clinical and subclinical infections in captive Asian elephants (Elephas maximus). J Zoo Wildl Med 44:136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard B, Xaymountry B, Thongtip N, Lertwatcharasarakul P, Wajjwalku W. 2014. First reported case of elephant endotheliotropic herpes virus infection in Laos. J Zoo Wildl Med 45:704–707. [DOI] [PubMed] [Google Scholar]
- Ehlers B, Dural G, Marschall M, Schregel V, Goltz M, Hentschke J. 2006. Endotheliotropic elephant herpesvirus, the first betaherpesvirus with a thymidine kinase gene. J Gen Virol 87:2781–2789. [DOI] [PubMed] [Google Scholar]
- Fickel J, Lieckfeldt D, Richman LK, Streich WJ, Hildebrandt TB, Pitra C. 2003. Comparison of glycoprotein B (gB) variants of the elephant endotheliotropic herpesvirus (EEHV) isolated from Asian elephants (Elephas maximus). Vet Microb 91:11–21. [DOI] [PubMed] [Google Scholar]
- Fuery A, Tion J, Ling PD, Long SY, Heaggans SY, Hayward GS. 2015. EEHV infection in surviving USA Asian elephant calves. J Zoo Wildl Med In press. [Google Scholar]
- Garner MM, Helmick K, Ochsenreiter J, Richman LK, Latimer E, Wise AG, Maes RK, Kiupel M, Nordhausen RW, Zong J-C, Hayward GS. 2009. Clinico-pathologic features of fatal disease attributed to new variants of endotheliotropic herpesviruses in two Asian elephants (Elephas maximus). Vet Pathol 46:97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward GS. 2012. Conservation: clarifying the risk from herpesvirus to captive Asian elephants. Vet Rec 170:202–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson ER, Sundberg JP, Gaskin JM, Kollias GV, O'Banion MK. 1986. Cutaneous papillomas associated with a herpesvirus-like infection in a herd of captive African elephants. J Am Vet Med Assoc 189:1075–1078. [PubMed] [Google Scholar]
- Latimer E, Zong J-C, Heaggans SY, Richman LK, Hayward GS. 2011. Detection and evaluation of novel herpesviruses in routine and pathological samples from Asian and African elephants: Identification of two new probosciviruses (EEHV5 and EEHV6) and two new gammaherpesviruses (EGHV3B and EGHV5). Vet Microbiol 147:28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling PD, Reid JG, Qin X, Muzny DM, Gibbs R, Petrosino J, Peng R, Zong J-C, Heaggans SY, Hayward GS. 2013. Complete genome sequence of elephant endotheliotropic herpesvirus 1A. Genome Announc 1:e0010613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters NJ, Stidworthy MF, Everest DJ, Dastjerdi A, Baulmer S. 2011. Detection of EGHV-5 in a self-limiting papilloma-like lesion in the trunk of an Asian elephant (Elephas maximus). Vet Rec 169:209. [DOI] [PubMed] [Google Scholar]
- McCully RM, Basson PA, Pienaar JG, Erasmus BJ, Young E. 1971. Herpes nodules in the lung of the African elephant (Loxodonta africana (Blumebach, 1792)). Onderstepoort J Vet Res 38:225–235. [PubMed] [Google Scholar]
- Ossent P, Guscetti F, Metzler AE, Lang EM, Rübel A, Hauser B. 1990. Acute and fatal herpesvirus infection in a young Asian elephant (Elephas maximus). Vet Pathol 27:131–133. [DOI] [PubMed] [Google Scholar]
- Pellett PE. 2014. Trunkloads of viruses. J Virol 88:13520–13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid CE, Hildebrandt TB, Marx N, Hunt M, Thy N, Reynes JM, Schaftenaar W, Fickel J. 2006. Endotheliotropic elephant herpes virus (EEHV) infection. The first PCR-confirmed fatal case in Asia. Vet Q 28:61–64. [DOI] [PubMed] [Google Scholar]
- Richman LK. 2003. Pathological and molecular aspects of fatal endotheliotropic herpesviruses of elephants. PhD dissertation, The Johns Hopkins University. [Google Scholar]
- Richman LK, Hayward GS. 2011. Elephant herpesviruses. In: Miller RE, Fowler M, eds. Fowler's Zoo and Wild Animal Medicine Current Therapy. St. Louis, MO: Elsevier/Saunders Co. p 496–502. [Google Scholar]
- Richman LK, Montali RJ, Cambre RC, Schmitt D, Hardy D, Hildbrandt T, Bengis RG, Hamzeh FM, Shahkolahi A, Hayward GS. 2000. Clinical and pathological findings of a newly recognized disease of elephants caused by endotheliotropic herpesviruses. J Wildl Dis 36:1–12. [DOI] [PubMed] [Google Scholar]
- Richman LK, Montali RJ, Garber RL, Kennedy MA, Lehnhardt J, Hildebrandt T, Schmitt D, Hardy D, Alcendor DJ, Hayward GS. 1999. Novel endotheliotropic herpesviruses fatal for Asian and African elephants. Science 283:1171–1176. [DOI] [PubMed] [Google Scholar]
- Richman LK, Montali RJ, Hayward GS. 2000. Review of a newly recognized disease of elephants caused by endotheliotrophic herpesviruses. Zoo Biol 19:383–392. [DOI] [PubMed] [Google Scholar]
- Richman LK, Zong J-C, Latimer E, Lock J, Fleischer RC, Heaggans SY, Hayward GS. 2014. Elephant endotheliotropic herpesviruses EEHV1A, EEHV1B, and EEHV2 from cases of hemorrhagic disease are highly diverged from other mammalian herpesviruses and may form a new subfamily. J Virol 88:13523–13546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan SJ, Thompson SD. 2001. Disease risk and inter-institutional transfer of specimens in cooperative breeding programs: Herpes and the elephant species survival plans. Zoo Biol 20:89–101. [DOI] [PubMed] [Google Scholar]
- Schmitt DL, Hardy DA, Montali RJ, Richman LK, Lindsay WA, Isaza R, West G. 2000. Use of famciclovir for the treatment of endotheliotrophic herpesvirus infections in Asian elephants (Elephas maximus). J Zoo Wildl Med 31:518–522. [DOI] [PubMed] [Google Scholar]
- Sripiboon S, Tankaew P, Lungka G, Thitaram C. 2013. The occurrence of elephant endotheliotropic herpesvirus in captive Asian elephants (Elephas maximus): first case of EEHV4 in Asia. J Zoo Wildl Med 44:100–104. [DOI] [PubMed] [Google Scholar]
- Stanton JJ, Nofs SA, Peng R, Hayward GS, Ling PD. 2012. Development and validation of quantitative real-time polymerase chain reaction assays to detect elephant endotheliotropic herpesviruses-2, 3, 4, 5, and 6′. J Virol Meths 186:73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton JJ, Zong J-C, Eng C, Howard L, Flanagan J, Stevens S, Schmitt D, Wiedner E, Graham D, Junge R, Weber MA, Fischer M, Mejia A, Tan J, Latimer E, Herron A, Hayward GS, Ling PD. 2013. Kinetics of viral loads and genotypic analysis of elephant endotheliotropic herpesvirus-1 infection in captive Asian elephants (Elephas maximus). J Zoo Wildl Med 44:42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton JJ, Zong J-C, Latimer E, Tan J, Herron A, Hayward GS, Ling PD. 2010. Detection of pathogenic elephant endotheliotropic herpesvirus in routine trunk washes from healthy adult Asian elephants (Elephas maximus) by use of a real-time quantitative polymerase chain reaction assay. Am J Vet Res 71:925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellehan JF, Johnson AJ, Childress AL, Harr KE, Isaza R. 2008. Six novel gammaherpesviruses of afrotheria provide insight into the early divergence of the gammaherpesvirinae. Vet Microbiol 127:249–257. [DOI] [PubMed] [Google Scholar]
- Wilkie GS, Davison AJ, Kerr K, Stidworthy MF, Redrobe S, Steinbach F, Dastjerdi A, Denk D. 2014. First fatality associated with elephant endotheliotropic herpesvirus 5 in an Asian elephant: Pathological findings and complete viral genome sequence. Sci Rep 4:6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie GS, Davison AJ, Watson M, Kerr K, Sanderson S, Bouts T, Steinbach F, Dastjerdi A. 2013. Complete genome sequences of elephant endotheliotropic herpesviruses 1A and 1B determined directly from fatal cases. J Virol 87:6700–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariah A, Zong J-C, Long SY, Latimer EM, Heaggans SY, Richman LK, Hayward GS. 2013. Fatal herpesvirus (EEHV) hemorrhagic disease in wild and orphan Asian elephants in southern India. J Wildl Dis 49:383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong J-C, Latimer EM, Heaggans SY, Richman LK, Hayward GS. 2008. Pathogenesis and Molecular Epidemiology of Fatal Elephant Endotheliotropic Disease Associated with the Expanding Proboscivirus Genus of the Betaherpesvirinae. Proceedings of the Internat Elephant Conservation and Research Symposium, Florida. [Google Scholar]
- Zong J-C, Latimer E, Heaggans SY, Richman LK, Hayward GS. 2009. Viral gene subtyping of eighteen North American cases of EEHV hemorrhagic disease. Proceedings of the Internat Elephant Conservation and Research Symposium, Bangkok, Thailand. [Google Scholar]
- Zong J-C, Latimer EM, Long SY, Richman LK, Heaggans SY, Hayward GS. 2014. Comparative genome analysis of four elephant endotheliotropic herpesviruses, EEHV3, EEHV4, EEHV5, and EEHV6, from cases of hemorrhagic disease or viremia. J Virol 88:13547–13569. [DOI] [PMC free article] [PubMed] [Google Scholar]