Highlight
Functional analysis of cucumber CsHAN1 showed that it regulates meristem development through WUSCHEL and SHOOT MERISTEMLESS pathways, and mediates leaf development through a complicated gene regulatory network in cucumber.
Key words: CsHAN, CsSTM, CsWUS, cucumber, leaf development, shoot apical meristem.
Abstract
The shoot apical meristem (SAM) is essential for continuous organogenesis in higher plants, while the leaf is the primary source organ and the leaf shape directly affects the efficiency of photosynthesis. HANABA TARANU (HAN) encodes a GATA3-type transcription factor that functions in floral organ development, SAM organization, and embryo development in Arabidopsis, but is involved in suppressing bract outgrowth and promoting branching in grass species. Here the function of the HAN homologue CsHAN1 was characterized in cucumber, an important vegetable with great agricultural and economic value. CsHAN1 is predominantly expressed at the junction of the SAM and the stem, and can partially rescue the han-2 floral organ phenotype in Arabidopsis. Overexpression and RNAi of CsHAN1 transgenic cucumber resulted in retarded growth early after embryogenesis and produced highly lobed leaves. Further, it was found that CsHAN1 may regulate SAM development through regulating the WUSCHEL (WUS) and SHOOT MERISTEMLESS (STM) pathways, and mediate leaf development through a complicated gene regulatory network in cucumber.
Introduction
The shoot apical meristem (SAM) is crucial for continuous organogenesis in higher plants. All the aerial organs including leaves, flowers, and stems are initiated from the SAM. The SAM is generally established during embryogenesis with a dome-shaped morphology, and can be divided into three functional zones: (i) the central zone with self-maintaining stem cells at the centre of the SAM; (ii) the peripheral zone where the lateral organ primordia are initiated from the shoulder of the SAM; and (iii) the rib zone in which stem tissue is specified beneath the central zone of the SAM (Steeves and Sussex, 1989; Fletcher, 2002; Tucker and Laux, 2007). Two independent pathways have been identified to be required for meristem establishment and maintenance in Arabidopsis, one is the WUSCHEL (WUS)–CLAVATA (CLV) pathway (Brand et al., 2000; Schoof et al., 2000). WUS, a homeodomain transcription factor, is expressed in the centre of the SAM, called the organizing centre, and functions to promote meristematic cell fate (Mayer et al., 1998). Mutation in WUS leads to a premature SAM with no ability to self-maintain the stem cells (Laux et al., 1996). CLV3, a signalling peptide, directly binds to the plasma membrane-localized receptor-like kinases CLV1 or CLV2/CRN complex and transmits a signal that restricts WUS expression, while WUS promotes the expression of CLV3 in the stem cells as a feedback loop (Fiers et al., 2005; Ito et al., 2006; Kondo et al., 2006; Ogawa et al., 2008; Bleckmann et al., 2010; Yadav et al., 2011). SHOOT MERISTEMLESS (STM) is the other pathway that is essential for meristem maintenance. STM, a KNOTTED1-LIKE HOMEOBOX (KNOX) gene, is expressed throughout the SAM but is excluded from the organ primordia that function to maintain the undifferentiated cells in the SAM (Endrizzi et al., 1996; Long et al., 1996; Lenhard et al., 2002). KNAT1/BREVIPEDICELLUS (BP), another member of the KNOX family, plays a role in meristem maintenance partially redundant with STM (Byrne et al., 2002; Douglas et al., 2002).
Leaf is the primary source organ, and the leaf shape directly affects the efficiency of photosynthesis (Tsukaya, 2006; Nicotra et al., 2008). The leaf primordium is initiated from the peripheral zone of the SAM, in which STM is down-regulated (Long et al., 1996) and ASYMMETRIC LEAVES 1 and 2 (AS1/2) are up-regulated (Ori et al., 2000; Guo et al., 2008). Leaves of as1 and as2 mutants are downward curling with asymmetric lobes and short petioles (Byrne et al., 2000; Iwakawa et al., 2002; Iwakawa et al., 2007). AS1 and AS2 form a protein complex that directly represses BP and KNAT2 transcription (Guo et al., 2008). Consistently, ectopic expression of KNOX genes results in lobed leaves in simple leaf species, and super-compoundness in compound leaf species such as tomato (Lincoln et al., 1994; Chuck et al., 1996; Janssen et al., 1998; Hake et al., 2004; Belles-Boix et al., 2006). Further functional studies assessed a key role for KNOX genes in leaf shape determination (Hay and Tsiantis, 2010; Di Giacomo et al., 2013; Bar and Ori, 2015). Several additional regulators have been found to mediate leaf shape development. For example, mutation in SERRATE (SE), a zinc finger protein involving in a miRNA gene silencing pathway, results in serrated leaves in Arabidopsis (Prigge and Wagner, 2001). Cap Binding Protein 20 (CBP20) encodes the 20kDa subunit of the nuclear mRNA cap-binding complex (nCBC), and a cpb20 mutant shows a serrated leaf margin (Papp et al., 2004). Mutation of ARGONAUTE1 (AGO1), a key player in transgene-induced post-transcriptional gene silencing, also leads to serrated leaves (Bohmert et al., 1998; Morel et al., 2002). AGO10/PINHEAD (PNH), another AGO protein gene, represses the accumulation of miR165/166, thereby affecting the establishment of leaf polarity (Liu et al., 2009). JAGGED (JAG), a C2H2-type zinc finger transcription factor gene, is expressed in the initiating lateral organ primordia and is essential for proper leaf shape. jag mutants show narrow and serrated leaves, and the gain-of-function mutant jag-5D has bract-like organs subtending most flowers (Dinneny et al., 2004; Ohno et al., 2004). Boundary genes CUP-SHAPED COTYLEDON (CUC1, 2, and 3) were initially identified by defective SAM development and organ fusion (Aida et al., 1999; Aida and Tasaka, 2006). Recently, CUC genes have been shown to play an important role in leaf margin development in both simple and compound leaf species and to act downstream of KNOX transcription factor genes (Nikovics et al., 2006; Blein et al., 2008; Berger et al., 2009; Bilsborough et al., 2011; Hasson et al., 2011; Spinelli et al., 2011). In Arabidopsis, genetic interactions among these different regulators lead to increased dissection of the Arabidopsis leaf margins (Blein et al., 2013).
HANABA TARANU (HAN) is a boundary gene that regulates SAM organization and flower organ development in Arabidopsis (Zhao et al., 2004). HAN encodes a GATA3 transcription factor that is expressed in the boundaries between the meristem and developing organ primordia, the boundaries between different floral whorls, as well as the junctional domain between the SAM and the stem (Zhao et al., 2004). Mutation of HAN leads to fused sepals, reduced floral organs, and a flatter SAM (Zhao et al., 2004). Previous studies showed that HAN and three GATA3 family genes, HANL2 (HAN-LIKE 2), GNC (GATA, NITRATE-INDUCIBLE, CARBON-METABOLISM-INVOLVED), and GNL (GNC-LIKE), form a negative feedback loop to regulate flower development (Zhang et al., 2013). The functions of HAN homologues are divergent in different species. For example, HAN homologues such as TASSEL SHEATH1 (TSH1) in maize (Zea mays), NECK LEAF1 (NL1) in rice (Oryza sativa), and THIRD OUTER GLUME (TRD) in barley (Hordeum vulgare) are involved in repressing bract outgrowth and promoting branching (Wang et al., 2009; Whipple et al., 2010).
In this study, the function of HAN was explored in cucumber (Cucumis sativus L.), a globally cultivated vegetable that is of important economic and nutritional value (Huang et al., 2009). Unlike the model plant Arabidopsis and most crops, cucumber is a typical unisexual plant with indeterminate growth, continuously producing leaves and male or female flowers simultaneously (Malepszy and Niemirowicz-Szczytt, 1991; Kater et al., 2001; Hao et al., 2003; Bai et al., 2004). Two HAN homologous genes were identified in cucumber, and the function of CsHAN1 was characterized in detail. CsHAN1 is predominantly expressed at the junction of the SAM and the stem, and can partially rescue the han-2 floral organ phenotype in Arabidopsis. Overexpression or down-regulation of CsHAN1 in the transgenic cucumber plants led to retarded growth and lobed leaves. Further, it was found that CsHAN1 may regulate SAM development through bridging the WUS and STM pathways, and mediate leaf margin development through a complicated gene regulatory network in cucumber.
Materials and methods
Plant materials and growth conditions
Cucumber (Cucumis stativus L.) inbred line R1407, which is a northern China type cucumber with dark green fruits similar to the sequenced line 9930, was used in this study. The cucumber seedlings were grown in a growth chamber under 16h/8h and 25 °C/18 °C day/night until the two true-leaf stage, and the cucumber plants were then transferred to a greenhouse in the experimental field of China Agricultural University in Beijing. Pest control and water management were carried out according to standard protocols. The Arabidopsis thaliana Landsberg erecta (Ler) and Columbia (Col) ecotypes, and the mutant alleles han-2(Ler) were described previously (Zhao et al., 2004; Zhang et al., 2013) and obtained from the Meyerowitz lab stock collection. The mutant allele han-2(Col) was obtained by crossing han-2(Ler) to Col followed by six generations of selfing. The Arabidopsis plants were grown in a growth chamber under 16h light/8h dark at 22 °C.
Gene cloning and phylogenetic analysis
Total RNA was extracted from the cucumber floral buds using a Quick RNA isolation Kit (Waryoung, China), and cDNA was synthesized using a Promega reverse transcriptase kit (Promega, USA). The coding sequences (CDS) of CsHAN1 and CsHAN2 were obtained using gene-specific primers (Supplementary Table S1 available at JXB online). The gene structure analysis was performed using online software GSDS 2.0 (http://gsds.cbi.pku.edu.cn/). The amino acid sequences of related HAN proteins in other species were obtained by BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST/). Protein alignment of CsHAN and related HANs was performed using ClustalW in the MEGA5 software package, and the boxes were drawn using the BoxShade web site (http://www.ch.embnet.org/software/BOX_form.html). The phylogenetic analysis based on amino acid sequences was performed using the Neighbor–Joining (NJ) method with 1000 bootstrap replicates through MEGA5 software (Saitou and Nei, 1987).
Quantitative real-time PCR
Total RNA was extracted from different cucumber tissues or Arabidopsis inflorescences using a Quick RNA isolation Kit (Waryoung, China), and cDNA was synthesized using a Promega reverse transcriptase kit (Promega, USA). An ABI PRISM 7500 Real-Time PCR System (Applied Biosystems, USA) was used for quantitative real-time reverse transcription–PCR (qRT–PCR) experiments. Three biological and three technical replicates (3×3) were performed for each gene. The cucumber Ubiquitin extension protein (UBI-ep) gene and the Arabidopsis ACTIN2 gene were used as internal references to normalize the expression data. The standard deviation was calculated between three biological replicates, using the average of the three technical replicates for each biological sample. The gene-specific primers are listed in Supplementary Table S1 at JXB online.
In situ hybridization
Cucumber shoot apexes of 6-, 12-, and 15-day-old seedlings, male and female buds, and young fruits from 0.8cm to 2.8cm were fixed in 3.7% formalin–acetic acid–alcohol (FAA), and in situ hybridization was performed as described previously (Zhang et al., 2013). Sense and antisense probes were synthesized by PCR amplification using SP6 and T7 RNA polymerase, respectively. Probes of CsWUS, CsSTM, and CsBP were designed according to the specific gene fragments. The primers for probe generation are listed in Supplementary Table S1 at JXB online.
Ectopic expression of CsHAN1 in Arabidopsis
To make the CsHAN1 overexpression construct, the full-length CsHAN1 CDS were amplified and cloned into the binary vector pBI121 through XbaI and SmaI sites. The recombinant plasmids were introduced into Agrobacterium by electroporation and then transformed into wild-type (WT) and han-2 mutant plants through the floral dip method (Clough and Bent, 1998). The transgenic plants were screened on Murashige and Skoog (MS) medium with 40mg l–1 kanamycin. The primers for vector construction are listed in Supplementary Table S1 at JXB online.
Cucumber transformation
The same CsHAN1 overexpression construct was used for cucumber transformation. To generate CsHAN1-RNAi transgenic plants, the 258bp sense and antisense fragments from the 3′ end of CsHAN1 were amplified using gene-specific primers containing SpeI(5′ end)/SacI(3′ end) and BamHI(5′ end)/KpnI(3′ end) sites, respectively. The two fragments were inserted into the RNAi-1 vector, and the empty RNAi-1 vector was used as a transformation control. The resultant CsHAN1-RNAi construct and empty RNAi-1 vector were then delivered into Agrobacterium by electroporation and transformed into the cucumber inbred line R1407 line using the cotyledon transformation method as previously described (Wang et al., 2014). The primers containing the restriction enzyme cutting sites are listed in Supplementary Table S1 at JXB online.
Paraffin sections
Young cucumber seeds at 16 d after fertilization were fixed, embedded, sectioned, and dewaxed as described (Jiang et al., 2014). Sections of 8 μm thickness were mounted in neutral resins, and images were taken under a light microscope (D72, Olympus, Japan).
Accession numbers
Sequence data in this paper can be found in the Cucumber Genome DataBase, TAIR, or GenBank under the following accession numbers: CsHAN1 (Csa016191), CsHAN2 (Csa012029), CsPNH1 (Csa015921), CsPNH2 (Csa004392), CsAGO1 (Csa000946), CsJAG (Csa008074), CsAS2 (Csa012250), CsBP (Csa009344), CsKNAT2 (Csa013896), CsKNAT6 (Csa011388), CsWUS (Csa000479), CsSTM (Csa000554), AtHAN (AT3G50870), SE (AT2G27100), AGO1 (AT1G48410), AS2 (AT1G65620), KNAT2 (AT1G70510), CPB20 (AT5G44200), BP (AT4G08150), CUC3 (AT1G76420), PNH (AT5G43810), JAG (AT1G68480), GNC (AT5G56860), GNL (AT4G26150), HvTRD (GU722206), OsNL1 (DQ784546), and ZmTSH1 (AC199892.4_FG031).
Results
Isolation of the cucumber CsHAN genes
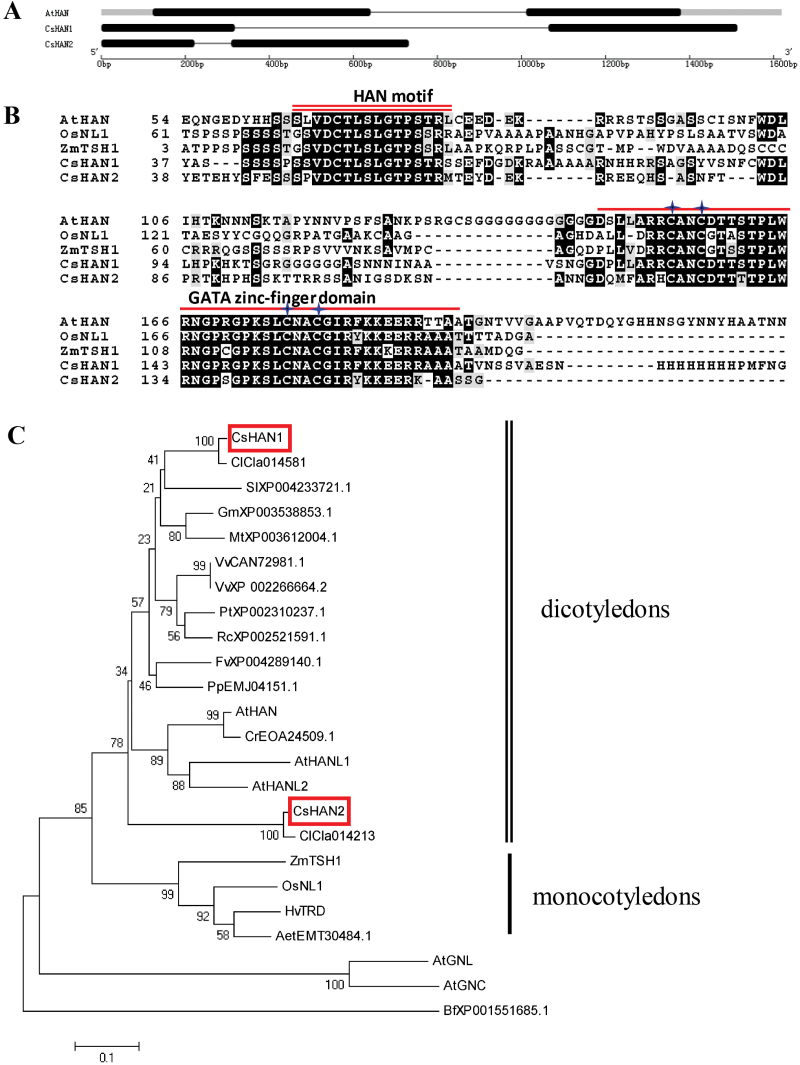
To identify the HAN homologues from cucumber, a BLAST search was performed in the Cucumber Genome DataBase (Huang et al., 2009) based on the amino acid sequence information of Arabidopsis HAN. Two candidate genes, Csa016191 and Csa012029, showed the highest similarity. A further BLAST search was performed in TAIR (http://www.arabidopsis.org/) using the two candidate gene, and both of them got the best hit to Arabidopsis HAN (AtHAN). Thus, Csa016191 was named CsHAN1 and Csa012029 was named CsHAN2, respectively, and their CDS as well as their genomic sequence from the flower buds of cucumber line R1407 were cloned. Gene structure analysis showed that CsHAN1 and CsHAN2, encoding 255 and 214 amino acids, respectively, contain two exons and one intron, consistent with the gene structure of AtHAN and HAN homologues (Zhao et al., 2004; Wang et al., 2009; Whipple et al., 2010) (Fig. 1A). Previous studies showed that HAN encodes a GATA3-like transcription factor with a single zinc finger domain and a HAN motif (Whipple et al., 2010). Protein alignment of HAN homologues from Arabidopsis (AtHAN), rice (OsNL1), maize (ZmTSH1), and cucumber (CsHAN1/2) was performed using ClustalW in the MEGA5 software. Despite CsHAN1 and CsHAN2 showing only 39.55% and 34.46% identity with AtHAN, respectively, the GATA zinc finger domain and the HAN motif are highly conserved (Fig. 1B).
Fig. 1.
Gene structure and phylogenetic analyses of CsHAN. (A) Structural analysis of HAN genes in cucumber and Arabidopsis. Grey boxes represent the 3′- or 5′-untranslated regions, black boxes indicate the exon, and black lines represent the introns. Cs, Cucumis sativus; At, Arabidopsis thaliana. (B) Protein alignment of HANs from Arabidopsis, rice (Oryza sativa), maize (Zea mays), and cucumber. The single and double underlines indicate the conserved GATA zinc finger domain and HAN motif, respectively. The asterisks indicate the conserved cysteine residues present in type IV zinc finger domains (C-X2-C-X17-20-C-X2-C). (C) Phylogenetic analysis of CsHAN genes (boxed) and HAN-like genes. MEGA 5.0 software was used to construct the Neighbor–Joining tree. Homologues of CsHAN genes from 11 dicotyledon species (double underlines) and four monocotyledon species (single underline) were used for the analyses and formed distinct clades (dicotyledon group and monocotyledon group). Vv, Vitis vinifera; Cr, Capsella rubella; Pt, Populus trichocarpa; Rc, Ricinus communis; Fv, Fragaria vesca subsp. vesca; Gm, Glycine max; Pp, Prunus persica; Mt, Medicago truncatula; Sl, Solanum lycopersicum; Bf, Botryotinia fuckeliana; Aet, Aegilops tauschii; Cl, Citrullus lanatus; Hv, Hordeum vulgare, Zm, Zea mays; Os, Oryza sativa; At, Arabidopsis thaliana. (This figure is available in colour at JXB online.)
Phylogenetic analysis of the deduced HAN proteins from various species was performed using the NJ method (Saitou and Nei, 1987). The phylogenetic tree showed that HAN homologues in the eudicot species form a distinct clade from those in the monocotyledon species such as rice, maize, and barley (Fig. 1C). In watermelon (Citrullus lanatus), another Cucurbitaceae species, there are also two HAN homologues (Cla014581and Cla014213) as well, and they formed two different clades with CsHAN1 and CsHAN2, respectively (Fig. 1C), implying that HAN homologues in Cucurbitaceae may have a distinct function from that in the model Arabidopsis plant. Given that CsHAN1 is more closely related to AtHAN than CsHAN2 (Fig. 1C), CsHAN1 was chosen and analysed in this study.
Expression pattern of CsHAN1 in cucumber
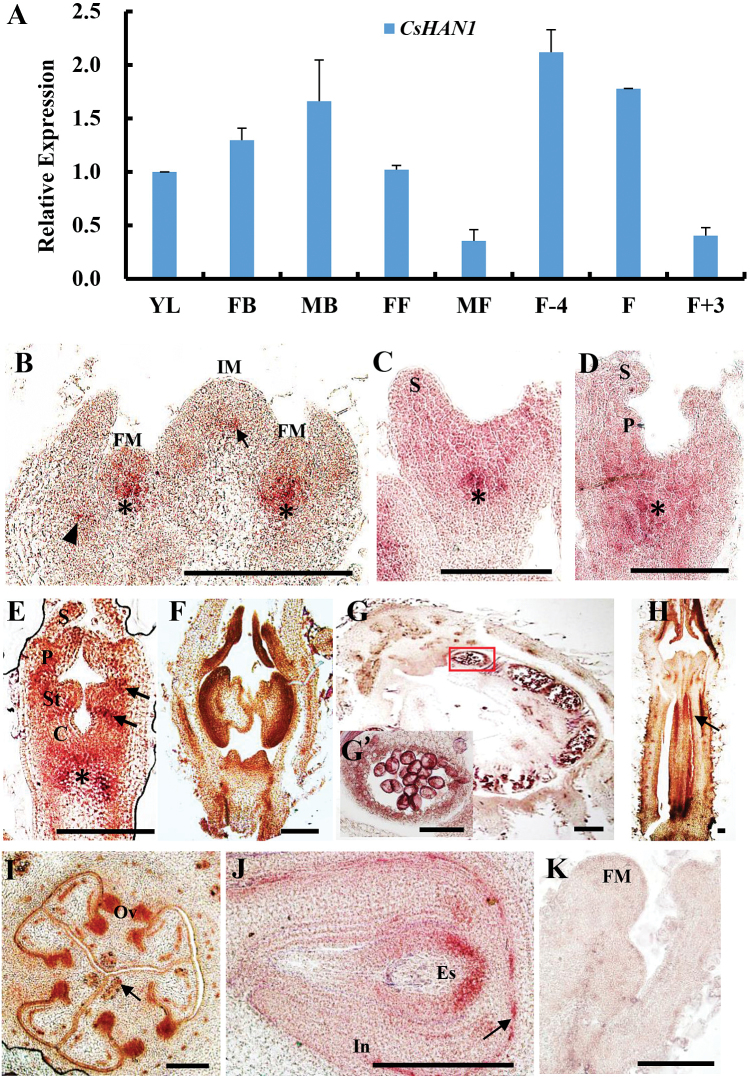
The expression of CsHAN1 was examined in different organs of cucumber through qRT–PCR (Fig. 2A). Total RNA was extracted from young leaves, female flower buds, male flower buds, female opening flowers, male opening flowers, and fruits at three different developmental stages. The data showed that CsHAN1 has the highest level in the young fruits 4 d before anthesis, and exhibited the lowest level in the male opening flowers (Fig. 2A). The expression of CsHAN1 in floral buds is higher than that in the opening flowers (Fig. 2A), suggesting that CsHAN1 is more abundant in young tissues.
Fig. 2.
Expression analysis of CsHAN1 in cucumber. (A) Quantitative RT–PCR (qRT–PCR) analysis of CsHAN1 in different tissues of cucumber. YL, young leaves; FB, female buds; MB, male buds; FF, female flowers; MF, male flowers; F-4, young fruits 4 d before anthesis; F, fruit at anthesis; F+3, fruits 3 d after anthesis. The Ubiquitin extension protein (UBI-ep) gene was used as an internal reference to normalize the expression data. (B–K) In situ hybridization with the CsHAN1 antisense probe (B–J) and sense probe (K). (B) In the ucumber shoot apex, CsHAN1 is expressed in the junction region of the inflorescence meristem (IM) and stem (arrow), the junction regions of the floral meristem (FM) and stem (asterisk), and the axil of leaf primordia (arrowhead). (C–E) Floral buds at stage 2 (C), 3 (D), and 4 (E). Asterisks show the expression domain of CsHAN1 at the junction of the meristem and stem, and arrows indicate the expression of CsHAN1 at the boundary between the petal and stamen, and the boundary between the stamen and initiating carpel primordia. (F–G’) Male flowers at stage 9 (F) and stage 11 (G); (G’) is a high magnification view of the anther in (G). The signal of CsHAN1 was detected in the developing anther, tapetum cell layer, and the uninuclear pollen. (H) Female flower in stage 8. CsHAN1 is expressed in the ovary (arrow). (I, J) Cross-sections of the female ovary in stage 9 (I) and stage 10 (J) showing the expression domain of CsHAN1 in the ovules and the base of the embryo sac. (K) No signal was found on hybridization with the sense CsHAN1 probe. S, sepal; P, petal; St, stamen; C, carpel; Ov, ovule; In, integument; Es, embryo sac. Bar=100 μm. (This figure is available in colour at JXB online.)
To investigate the spatial and temporal expression pattern of CsHAN, in situ hybridization was performed. The signal of CsHAN1 is detected at the junction of the inflorescence meristem (IM) and the stem (arrow in Fig. 2B), the junction regions of the floral meristem (FM) and the stem (asterisks in Fig. 2C–E), and in the axil of leaf primordia (arrowhead in Fig. 2B). In addition, transcripts of CsHAN1 are mainly concentrated in the boundary between petal primordia and stamen primordia, and the boundary between stamen primordia and carpal primordia in the stage 4 flower bud (arrows in Fig. 2E). In the male flower, CsHAN1 is primarily expressed in the developing anthers at stage 9 (Fig. 2F), and then in the tapetum cell layer and uninuclear pollen at stage 11 (Fig. 2G). In the female flower, the expression domain of CsHAN1 is mostly in the developing ovary (Fig. 2H), ovules (Fig. 2I), and the base of the embryo sac (Fig. 2J). CsHAN1 is also strongly expressed in vascular tissues in all of the examined samples (arrows in Fig. 2I, J). No signal is detected upon hybridization with the sense CsHAN1 probe (Fig. 2K).
Ectopic expression of CsHAN1 in Arabidopsis
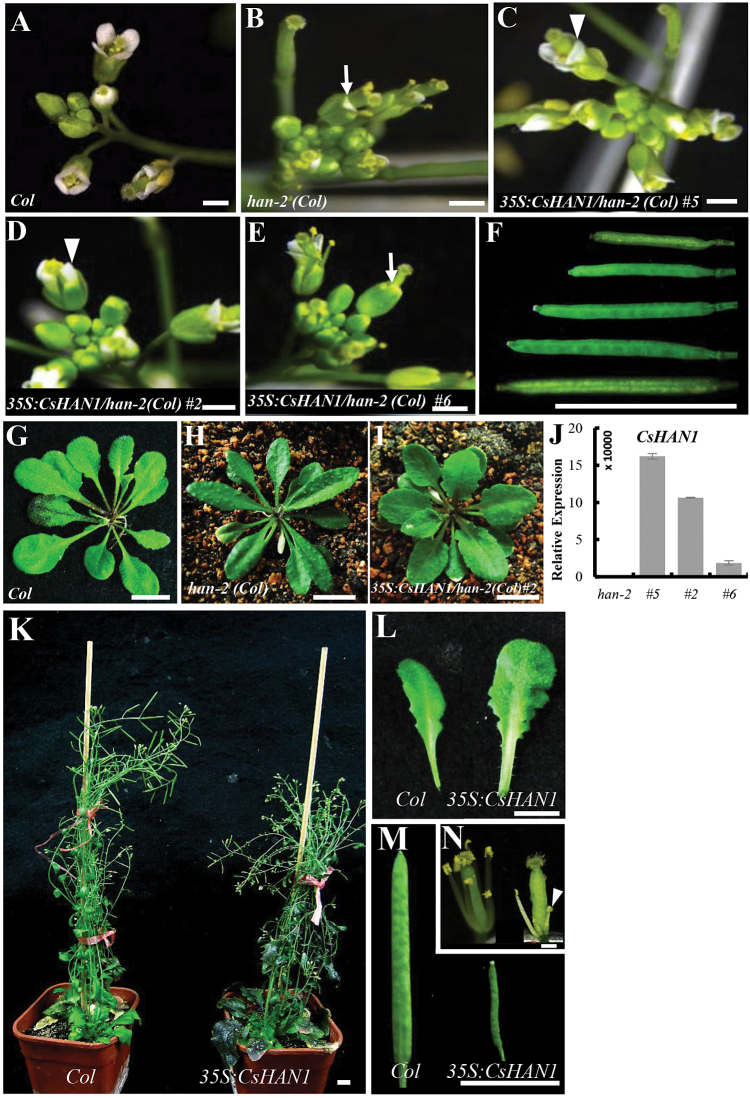
To investigate the function of CsHAN1, CsHAN1 was first ectopically introduced under the Cauliflower mosaic virus (CaMV) 35S promoter into the han-2 mutant in the Ler background. However, only two transgenic plants were produced, screened from ~3ml (1.1×104) of seeds, and died without producing any seeds (Supplementary Fig. S1A, B at JXB online). This is similar to the overexpression of AtHAN itself in Arabidopsis (Zhao et al., 2004). Next, CsHAN1 was ectopically expressed in the han-2 mutants in the Col background which displays similar reduced floral organs and decreased silique length to those in the Ler background (Zhang et al., 2013). Fortunately, 17 independent transgenic lines were produced, and the degree of rescue of the han-2 mutant phenotype positively correlates with the ectopic CsHAN1 expression (Fig. 3A–J) (investigation of the phenotype was performed in the T2 transgenic lines). For example, the number of petals rescued ranged from 1.9±1.1 in han-2 to 3.6±0.5 in the strongest transgenic line 5, and 3.1±0.9 in the weakest line 6 (Fig. 3C–E; Table 1). Similarly, the length of the silique in the three CsHAN1 transgenic lines was also increased, and line 5 recovered almost to the length of the WT (Fig. 3F). In addition, in contrast to the slightly serrated margin in Col, it was noticed that the rosette leaves of han-2(Col) are spindly with smooth margins and short petioles (Fig. 3G, H), and CsHAN1 can rescue the smooth leaves to serrated upon ectopic expression in Arabidopsis (Fig. 3I). These results suggest that CsHAN1 may play a role in regulation of flower organ and leaf shape development.
Fig. 3.
Ectopic expression of CsHAN1 in han-2 mutant and wild-type Arabidopsis. (A–E) The inflorescences of Col (A), han-2(Col) (B), 35S:CsHAN1/han-2(Col) line 5 (C), 35S:CsHAN1/han-2(Col) line 2 (D), and 35S:CsHAN1/han-2(Col) line 6 (E). The arrows indicate the flowers with 1–2 petals, and the arrowheads show the flowers with 3–4 petals. (F) The siliques of WT, 35S:CsHAN1/han-2(Col) line 5, 35S:CsHAN1/han-2(Col) line 2, 35S:CsHAN1/han-2(Col) line 6, and the han-2 mutant at the same developmental stage (from bottom to top). (G–I) Rosette leaves of Col (G), han-2(Col) (H), and 35S:CsHAN1/han-2(Col) line 2 (I) show the partially rescued leaf shape. (J) qRT–PCR analyses of CsHAN1 in the three transgenic lines in the han-2(Col) background. Arabidopsis ACTIN2 was used as an internal standard to normalize the templates. (K–N) The whole plants (K), rosette leaves (L), fruits (M), and flowers (N) of Col (left) and the CsHAN1 overexpression line (right) in the Col background. Sepals and petals were removed in (N), and the white arrowhead shows the retarded stamen in the overexpression line. Bar=1mm. (This figure is available in colour at JXB online.)
Table 1.
CsHAN1 can partly rescue the number of floral organs in the han-2 mutant in Arabidopsis
| Genotype | Sepal | Petal | Stamen | Carpal |
|---|---|---|---|---|
| Col | 4.0±0.0 | 4.0±0.0 | 5.5±0.3 | 2.0±0.0 |
| han-2(Col) | 3.6±0.6 | 1.9±1.1 | 4.5±0.6 | 2.0±0.0 |
| 35S:CsHAN1/han-2 #5 | 3.8±0.5 | 3.6±0.5 | 4.5±0.5 | 2.0±0.0 |
| 35S:CsHAN1/han-2 #2 | 3.6±0.5 | 3.2±0.8 | 4.7±0.6 | 2.0±0.0 |
| 35S:CsHAN1/han-2 #6 | 3.5±0.8 | 3.1±0.9 | 4.4±0.5 | 2.0±0.0 |
The values shown are the means ±SE, n=30.
To explore further the function of CsHAN1, transgenic lines overexpressing CsHAN1 in Arabidopsis WT Col were also generated. A total of 12 independent transgenic lines were obtained. Overexpression of CsHAN1 leads to serrated leaves in both rosette leaves and cauline leaves, and produces short siliques that may partially result from the 28% short stamens that are not long enough to reach the stigma and/or immature anthers (Fig. 3K–N; Supplementary Fig. S1C at JXB online). However, the number of floral organs is unchanged in the CsHAN1 overexpression lines.
CsHAN1 may be involved in shoot apical meristem development in cucumber
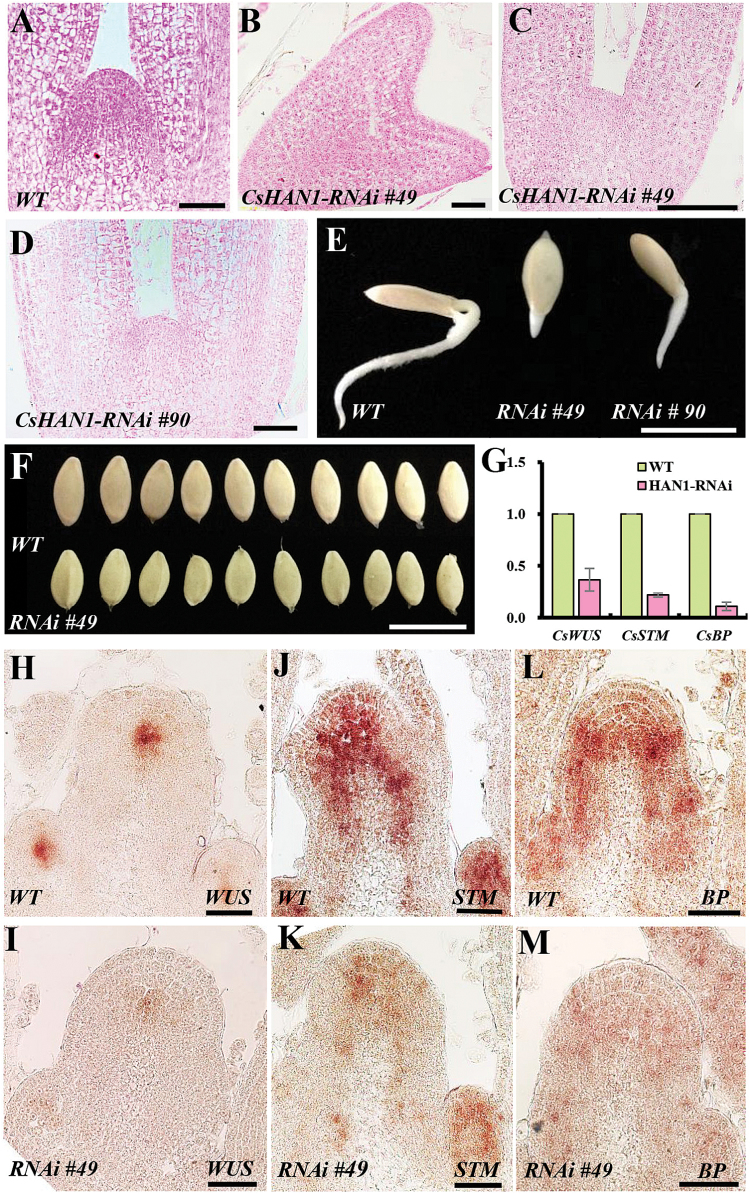
To understand further the function of CsHAN1 in cucumber, the 35S promoter followed by the CsHAN1 coding sequence (CsHAN1-OE) or the double-stranded RNAi construct containing the specific sequence of CsHAN1 (CsHAN1-RNAi) was introduced into the cucumber inbred line R1407 through Agrobacterium-mediated cotyledon transformation, and positive transplants were selected based on antibiotic selection as well as PCR analyses using primers from the vector (Y. Zhang et al., 2014; Cheng et al., 2015). Nine CsHAN1-OE and 11 CsHAN1-RNAi independent T0 transgenic lines were obtained. Surprisingly, the expression of CsHAN1 was down-regulated in the CsHAN1-OE lines whereas it was up-regulated in the CsHAN1-RNAi lines (Fig. 4A; Supplementary Fig. S2 at JXB online), which can be explained by co-suppression in the CsHAN1-OE lines and negative autoregulation of HAN in the CsHAN1-RNAi lines (Baudry et al., 2006; Zhang et al., 2013). Next three representative transgenic lines for each construct were selected for further characterization (Fig. 4A). In the three T0 CsHAN1-OE lines, transcripts of CsHAN1 declined to 30, 31, and 40% in lines 9, 12, and 15, respectively, as compared with those in the empty vector (WT). In the T0 CsHAN1-RNAi lines, the expression of CsHAN1 is up-regulated 1.6- to 3-fold (Fig. 4A). Line 9 of CsHAN1-OE grew slowly, with very few flower buds and lobed leaves (Fig. 4B; Supplementary Fig. S3), and it died after 3 months without generating any seeds. Despite CsHAN1-OE lines 12 and 15 producing several seeds, the resulting T1 plants display severely retarded growth and appear to be sterile (no seeds produced after pollination). Similarly, the T0 CsHAN1-RNAi line also grew slowly (Fig. 4B; Supplementary Fig. S3). The stunted transgenic lines suggest that CsHAN1 may function in SAM development. To confirm this notion, embryo development was characterized in the T2 CsHAN1-RNAi lines (Fig. 5). In the WT, the cucumber embryo developed to the torpedo stage and the meristem protruded upward, forming a dome at 16 d after pollination (Fig. 5A) (Atsmon and Galun, 1960; Sun et al., 2010). In the CsHAN1-RNAi line 49, ~30% of embryos remained at the heart stage (Fig. 5B), and 60% embryos were in the torpedo stage with a flat meristem (Fig. 5C). In the CsHAN1-RNAi line 90, although 75% of embryos were at the torpedo stage, the meristem was small or did not fully protrude (Fig. 5D). Next, the rate of seed germination was compared for 36h; the root of WT cucumbers was ~3cm long (Fig. 5E), while the root in the CsHAN1-RNAi lines just began to emerge or was <2cm long (Fig. 5E). The seed morphology was also affected in the CsHAN1-RNAi lines, with 39% of seeds obviously crapy and smaller than those in the WT (Fig. 5F). Therefore, CsHAN1 can retard plant growth early after embryogenesis.
Fig. 4.
Phenotypes of transgenic cucumber. (A) qRT–PCR analyses of CsHAN1 expression in transgenic overexpression and RNAi lines in cucumber. (B) Plant phenotypes of a transgenic CsHAN1 overexpression line (left), the wild type (middle), and a CsHAN1-RNAi line (right) which are 50 days old. Bar=5cm. (This figure is available in colour at JXB online.)
Fig. 5.
CsHAN1 is required for shoot apical meristem development. (A–D) Embryo phenotypes in the normal torpedo stage of the wild type (A), the heart stage in the CsHAN1-RNAi line 49 (B), the retarded torpedo embryo in the CsHAN1-RNAi line 49 (C), and in the CsHAN1-RNAi line 90 (D) at 16 d after fertilization. Bar=50 μm. (E) Phenotypes of wild-type (left), CsHAN1-RNAi line 49 (middle), and CsHAN1-RNAi line 90 (right) seeds at 36h after germination. Bar=1cm. (F) Seed morphology in the WT and CsHAN1-RNAi line 49. Bar=1cm. (G) qRT–PCR analyses of CsWUS, CsSTM, and CsBP in the shoot apexes of the wild type and the CsHAN1-RNAi line. The UBI-ep gene was used as an internal reference to normalize the expression data. (H–M) The expression of CsWUS (H, I), CsSTM (J, K), and CsBP (L, M) in the wild type (H, J, L) and CsHAN1-RNAi line 49 (I, K, M) in the apex of 6-day-old seedlings as detected by in situ hybridization. Bar=50μm. (This figure is available in colour at JXB online.)
Given that WUS was shown to be a classical meristem marker that functions in specifying the stem cell identity in the shoot meristem, and STM and BP are KNOX family genes that promote meristem maintenance (Long et al., 1996; Byrne et al., 2002; Douglas et al., 2002; Lenhard et al., 2002), it was next explored whether HAN suppresses SAM development through these genes. qRT–PCR analyses showed that the expression of CsWUS, CsSTM, and CsBP was greatly decreased in the shoot apexes of CsHAN1-RNAi lines (Fig. 5G). In situ hybridization of CsWUS, CsSTM, and CsBP was also performed in the shoot apexes of CsHAN1-RNAi lines. The CsWUS signal was detected in the organizing centre of the SAM, IM, and FM in the WT, consistent with findings in other species (Fig. 5H), while the expression of CsWUS was significantly reduced in the CsHAN1-RNAi line (Fig. 5I). CsSTM is expressed throughout the SAM and FM but not in the organ primordia in the WT (Fig. 5J). In the CsHAN1-RNAi line, the CsSTM signal is also greatly decreased (Fig. 5K). Similarly, CsBP mRNA is detected at the base of the SAM and the cortex of the stem in the WT (Fig. 5L), and the transcript level of CsBP is greatly reduced in the CsHAN1-RNAi line (Fig. 5M). These data suggested that CsHAN1 may regulate meristem development by mediating the expression of CsWUS, CsSTM, and CsBP in cucumber.
CsHAN1 regulates leaf shape development in cucumber
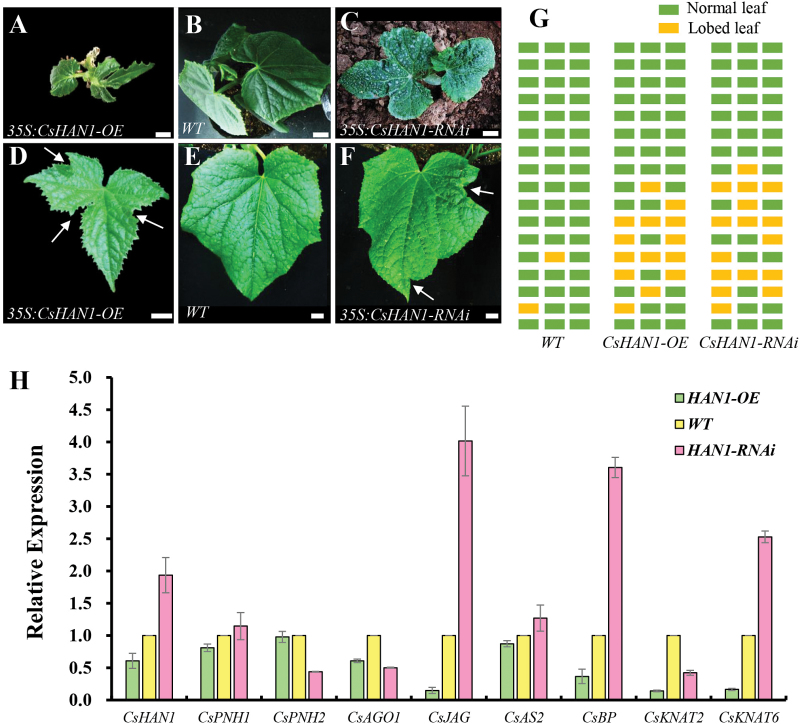
In addition to the retarded growth, another obvious phenotype in the CsHAN1 transgenic cucumber was the lobed leaves (Fig. 6). In contrast to the palmate entire leaves in the WT, a high proportion of the leaves in both CsHAN1-OE and CsHAN1-RNAi lines were highly lobed (Fig. 6A–F), especially in leaves at the first 10 nodes, probably due to different penetrance and developmental cues at different nodes (Fig. 6G) (Weigel et al., 1992; Ji et al., 2011). To explore the mechanism by which CsHAN1 regulates leaf shape development in cucumber, the known leaf developmental genes in cucumber were first isolated using a BLAST search, and then the expression in the fourth young leaves was examined by qRT–PCR in the T2 plants. The expression of CsJAG, CsBP, and CsKNAT6 was down-regulated in the CsHAN1-OE lines and up-regulated in the CsHAN1-RNAi lines, whereas the expression of CsAGO1 and CsKNAT2 was reduced in both the CsHAN1-OE lines and CsHAN1-RNAi lines (Fig. 6H). The expression of CsPNH2 was reduced >2-fold in the CsHAN1-RNAi lines, but was unchanged in the CsHAN1-OE lines (Fig. 6H). The expression of CsPNH1 and CsAS2 appears to be unaffected in both transgenic lines (Fig. 6H), suggesting that CsHAN1 regulates leaf shape development through a complicated gene regulatory network in cucumber.
Fig. 6.
CsHAN1 regulates leaf shape development. (A–F) Transgenic cucumber plants (A–C) and representative leaves (D–F) of a CsHAN1 overexpression line (A, D), the wild type (B, E), and a CsHAN1-RNAi line (C, F) which are 20 days old. Arrows showed the notches. Bar=1cm. (G) Diagrammatic data show the position of lobed leaves in the WT and CsHAN1 transgenic lines. Each column represents an individual plant, and each rectangle represents a node. (H) qRT–PCR analyses of leaf developmental genes in CsHAN1 overexpression and RNAi lines. The cucumber UBI-ep gene was used as an internal reference to normalize the expression data, and the experiments were repeated in triplicate independent samples. Error bars represent the SE. (This figure is available in colour at JXB online.)
Discussion
CsHAN1 may regulate shoot meristem development through regulating WUS and STM pathways in cucumber
In Arabidopsis, WUS and STM function in independent pathways and play essential roles for SAM establishment and maintenance (Lenhard et al., 2002). Here it was found that both CsHAN1-OE and CsHAN1-RNAi lines displayed retarded growth, but CsHAN1-OE lines displayed a more severe phenotype than the CsHAN1-RNAi lines, probably due to the huge reduction caused by co-suppression in the CsHAN1-OE lines (Figs 4–6). In situ hybridization showed that the expression of CsWUS was significantly reduced in the CsHAN1-RNAi lines, despite the fact that the expression domain remained unchanged (Fig. 5H, I). However, the expression of AtWUS was diffused and shifted to the L2 or L1 layer in the han-1 mutant plants in Arabidopsis (Zhao et al., 2004), implying that CsHAN1 and AtHAN may regulate WUS in a different way. In addition, embryo development in the han-1 mutant was uncoordinated in Arabidopsis, resulting in misshapen embryos (Zhao et al., 2004; Nawy et al., 2010), whereas the embryo development in the CsHAN1-RNAi line was delayed, with no obvious change of embryo shape. There are two possibilities to explain this difference: one is that CsHAN1 and AtHAN regulate embryo development through a distinct mechanism, and the other possibility is that the embryo defects in the CsHAN1-RNAi line were covered by CsHAN1 autoregulation; the 1.6- to 3-fold increase of CsHAN1 expression in the CsHAN1-RNAi line was within the buffer threshold that fails to produce any morphological defects in the embryo. A clean loss of function of CsHAN1 like that in the han-1 null allele would better elucidate the function of CsHAN1 in embryo development in cucumber.
Further, it was found that the expression of CsSTM and CsBP was greatly reduced in the CsHAN1-RNAi lines (Fig. 5J–M). STM and BP both belong to the class 1 KNOX genes, and were shown to function redundantly in meristem maintenance in Arabidopsis (Byrne et al., 2002; Douglas et al., 2002; Lenhard et al., 2002). Given that the expression domain of CsHAN1 overlaps with that of CsWUS, CsSTM, and CsBP (Figs 2, 5), CsHAN1 may regulate meristem development through physical interactions with CsWUS and CsSTM, bridging the previously speculated two parallel pathways in cucumber. Further studies using inducible CsHAN1 lines and ChIP assay will be helpful to test the above hypothesis.
Elaborate expression of CsHAN1 is required for normal leaf shape development
Previous studies of HAN emphasized its role in flower and embryo development (Zhao et al., 2004; Nawy et al., 2010), while the function of HAN in leaf development was largely neglected. Here it was found that leaves of the han-2 mutant in the Col background changed from serrated into smooth margins (Fig. 3G, H), Together with the finding that ectopic expression of AtHAN led to lobed leaves in Arabidopsis (Zhao et al., 2004), a function for AtHAN in leaf shape development is hypothesized.
In this study, it was found that ectopic expression of CsHAN1 can rescue the smooth margin phenotype in the han-2 mutant and resulted in lobed leaves in WT Arabidopsis (Fig. 3). More importantly, both CsHAN1-OE and CsHAN1-RNAi lines produced highly lobed leaves in cucumber (Fig. 6A–F), especially in the leaves at the first 10 nodes (Fig. 6G). The CUC boundary genes have been shown to play a role in leaf development (Aida et al., 1999; Nikovics et al., 2006; Hasson et al., 2011). In tomato, both reduction and overexpression of the CUC homologous gene GOBLET (GOB) led to a change from complex leaves in the WT into simpler leaves with no sharp leaf margin (Blein et al., 2008; Berger et al., 2009). These data suggest that the elaborate expression of the boundary genes HAN and CUC is essential for leaf shape development, with increased or reduced expression resulting in a change in leaf margins. However, molecular and genetic studies are required to establish whether HAN and CUC may be part of the same pathway or act independently in leaf development.
Further, the present data showed that despite the CsHAN1-OE lines and CsHAN1-RNAi lines displaying similar leaf phenotype, the underlying gene expression was different (Fig. 6). As a co-suppression event may be involved in the CsHAN1-OE lines and negative autoregulation of AtHAN1 has been well documented (Zhang et al., 2013), the final phenotypes of transgenic plants might derive from different levels of HAN proteins. The expression of CsJAG, CsBP, CsKNAT2, and CsKNAT6 was down-regulated in the CsHAN1-OE lines (Fig. 6). Interestingly, JAG, BP, KNAT2, and KNAT4 were also shown to be down-regulated upon AtHAN induction in Arabidopsis (Zhang et al., 2013), suggesting that a similar regulatory mechanism may be involved between HAN, JAG, BP, and KNOX genes in cucumber and Arabidopsis. CsBP was found to be down-regulated in the meristem but up-regulated in the leaves of CsHAN1-RNAi cucumber plants, implying that different regulatory networks exist in different tissues and/or developmental stages.
Partially conserved function of CsHAN1 in flower development
The most obvious phenotype in the han mutant was the reduced floral organs (especially petals and stamens) and fused sepals in Arabidopsis (Zhao et al., 2004). Despite the fact that ectopic expression of CsHAN1 can partially rescue the floral organ and silique length phenotype of the han-2 mutant (Fig. 3), no obvious flower organ defects were observed in either CsHAN1-OE or CsHAN1-RNAi lines in cucumber (data not shown), suggesting that CsHAN1 may possess both conserved and divergent functions in flower development. Morphologically, the flowers in Arabidopsis and cucumber are quite different. Arabidopsis has bisexual flowers with separate sepals, petals, and stamens, and fused carpels in the innermost whorl (Supplementary Fig. S4A, B at JXB online). In cucumber, flowers are unisexual (male and female flower) with a tubular structure consisting of fused sepals, petals, stamens, or pistils at the base of a flower (Supplementary Fig. S4C–F). In Arabidopsis, AtHAN is transcribed at the boundaries between the meristem and its newly initiated organ primordia and at the boundaries between different floral whorls (Zhao et al., 2004). Such boundary expression, especially in the boundaries between floral meristem and sepal primordia, or between sepal and petal primordia, was not observed for CsHAN1 in cucumber (Fig. 2), suggesting that the boundary expression of HAN may be essential for floral organ separation and therefore affects organ numbers. Considering that there are two HAN homologues in cucumber (Fig. 1), lack of flower phenotype in the CsHAN1 transgenic lines may due to the redundant role of CsHAN2 in cucumber, or the function of the remaining CsHAN1 in the knock-down lines (CsHAN1-OE). Future studies using the knock-out lines of both CsHAN1 and CsHAN2 through the CRISPR/Cas9 system (H. Zhang et al., 2014) would be a promising way to dissect the specific function of CsHAN genes during the unisexual flower development in cucumber.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. CsHAN1 overexpression in WT Arabidopsis.
Figure S2. PCR identification and qRT-PCR analyses of transgenic cucumber.
Figure S3. Leaf phenotype in the CsHAN1 transgenic cucumber.
Figure S4. Flower morphology in Arabidopsis and cucumber.
Table S1. Primers used in this study.
Acknowledgements
The authors thank members of the Zhang lab for technical assistance and stimulating discussions, Dr Jinsheng Lai for providing the machine during real-time qRT–PCR analysis, and Dr Xuexian Li for critical reading and comments on the manuscript. This study was supported by the National Basic Research of China 973 program [2012CB113900], the National Natural Science Foundation of China [31171399], the Chinese Universities Scientific Fund [2013RC030], and Earmarked Fund for Beijing Leaf Vegetables Innovation Team of Modern Agro-industry Technology Research System [BLVT-08].
Glossary
Abbreviations:
- AGO1
ARGONAUTE1
- AS1/2
ASYMMETRIC LEAVES1/2
- BP
KNAT1/BREVIPEDICELLUS
- CaMV
Cauliflower mosaic virus
- CDS
coding sequence
- CLV1/2/3
CLAVATA1/2/3
- CUC1/2/3
CUP-SHAPED COTYLEDON1/2/3
- FM
flower meristem
- HAN
HANABA TARANU
- IM
inflorescence meristem
- JAG
JAGGGED
- KNAT1/2/6
KNOTTED1-LIKE HOMEOBOX GENE1/2 /6
- KNOX
KNOTTED1-LIKE HOMEOBOX
- NJ
Neighbor–Joining
- NL1
NECK LEAF1
- OE
overexpression
- PNH
AGO10/PINHEAD
- SAM
shoot apical meristem
- STM
SHOOT MERISTEMLESS
- TSH1
TASSEL SHEATH1
- WT
wild type
- WUS
WUSCHEL
References
- Aida M, Ishida T, Tasaka M. 1999. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126, 1563–1570. [DOI] [PubMed] [Google Scholar]
- Aida M, Tasaka M. 2006. Genetic control of shoot organ boundaries. Current Opinion in Plant Biology 9, 72–77. [DOI] [PubMed] [Google Scholar]
- Atsmon D, Galun E. 1960. A morphogenetic study of staminate, pistillate and hermaphrodite flowers in Cucumis sativus L. Phytomorphology 10, 110–115. [Google Scholar]
- Bai S-L, Peng Y-B, Cui J-X, Gu H-T, Xu L-Y, Li Y-Q, Xu Z-H, Bai S-N. 2004. Developmental analyses reveal early arrests of the spore-bearing parts of reproductive organs in unisexual flowers of cucumber (Cucumis sativus L.). Planta 220, 230–240. [DOI] [PubMed] [Google Scholar]
- Bar M, Ori N. 2015. Compound leaf development in model plant species. Current Opinion in Plant Biology 23, 61–69. [DOI] [PubMed] [Google Scholar]
- Baudry A, Caboche M, Lepiniec L. 2006. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. The Plant Journal 46, 768–779. [DOI] [PubMed] [Google Scholar]
- Belles-Boix E, Hamant O, Witiak SM, Morin H, Traas J, Pautot V. 2006. KNAT6: an Arabidopsis homeobox gene involved in meristem activity and organ separation. The Plant Cell 18, 1900–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger Y, Harpaz-Saad S, Brand A, Melnik H, Sirding N, Alvarez JP, Zinder M, Samach A, Eshed Y, Ori N. 2009. The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136, 823–832. [DOI] [PubMed] [Google Scholar]
- Bilsborough GD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C, Laufs P, Hay A, Prusinkiewicz P, Tsiantis M. 2011. Model for the regulation of Arabidopsis thaliana leaf margin development. Proceedings of the National Academy of Sciences, USA 108, 3424–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckmann A, Weidtkamp-Peters S, Seidel CA, Simon R. 2010. Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiology 152, 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blein T, Pautot V, Laufs P. 2013. Combinations of mutations sufficient to alter Arabidopsis leaf dissection. Plants 2, 230–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blein T, Pulido A, Vialette-Guiraud A, Nikovics K, Morin H, Hay A, Johansen IE, Tsiantis M, Laufs P. 2008. A conserved molecular framework for compound leaf development. Science 322, 1835–1839. [DOI] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. 1998. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO Journal 17, 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. 2000. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. [DOI] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. 2000. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. [DOI] [PubMed] [Google Scholar]
- Byrne ME, Simorowski J, Martienssen RA. 2002. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129, 1957–1965. [DOI] [PubMed] [Google Scholar]
- Cheng J, Wang Z, Yao F, Gao L, Ma S, Sui X. 2015. Down-regulating CsHT1, a cucumber pollen-specific hexose transporter, inhibits pollen germination, tube growth and seed development. Plant Physiology 168, 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Lincoln C, Hake S. 1996. KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. The Plant Cell 8, 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Di Giacomo E, Iannelli MA, Frugis G. 2013. TALE and shape: how to make a leaf different. Plants 2, 317–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Yadegari R, Fischer RL, Yanofsky MF, Weigel D. 2004. The role of JAGGED in shaping lateral organs. Development 131, 1101–1110. [DOI] [PubMed] [Google Scholar]
- Douglas SJ, Chuck G, Dengler RE, Pelecanda L, Riggs CD. 2002. KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. The Plant Cell 14, 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endrizzi K, Moussian B, Haecker A, Levin JZ, Laux T. 1996. The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. The Plant Journal 10, 967–979. [DOI] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu CM. 2005. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. The Plant Cell 17, 2542–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC. 2002. Shoot and floral meristem maintenance in Arabidopsis. Annual Review of Plant Biology 53, 45–66. [DOI] [PubMed] [Google Scholar]
- Guo M, Thomas J, Collins G, Timmermans MC. 2008. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. The Plant Cell 20, 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S, Smith HM, Holtan H, Magnani E, Mele G, Ramirez J. 2004. The role of knox genes in plant development. Annual Review of Cell and Developmental Biology 20, 125–151. [DOI] [PubMed] [Google Scholar]
- Hao Y-J, Wang D-H, Peng Y-B, Bai S-L, Xu L-Y, Li Y-Q, Xu Z-H, Bai S-N. 2003. DNA damage in the early primordial anther is closely correlated with stamen arrest in the female flower of cucumber (Cucumis sativus L.). Planta 217, 888–895. [DOI] [PubMed] [Google Scholar]
- Hasson A, Plessis A, Blein T, Adroher B, Grigg S, Tsiantis M, Boudaoud A, Damerval C, Laufs P. 2011. Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. The Plant Cell 23, 54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. 2010. KNOX genes: versatile regulators of plant development and diversity. Development 137, 3153–3165. [DOI] [PubMed] [Google Scholar]
- Huang S, Li R, Zhang Z, et al. 2009. The genome of the cucumber, Cucumis sativus L. Nature Genetics 41, 1275–1281. [DOI] [PubMed] [Google Scholar]
- Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H. 2006. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313, 842–845. [DOI] [PubMed] [Google Scholar]
- Iwakawa H, Iwasaki M, Kojima S, Ueno Y, Soma T, Tanaka H, Semiarti E, Machida Y, Machida C. 2007. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. The Plant Journal 51, 173–184. [DOI] [PubMed] [Google Scholar]
- Iwakawa H, Ueno Y, Semiarti E, et al. 2002. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant and Cell Physiology 43, 467–478. [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Lund L, Sinha N. 1998. Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiology 117, 771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L, Liu X, Yan J, Wang W, Yumul RE, Kim YJ, Dinh TT, Liu J, Cui X, Zheng B. 2011. ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genetics 7, e1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Ding L, He B, Shen J, Xu Z, Yin M, Zhang X. 2014. Systemic gene silencing in plants triggered by fluorescent nanoparticle-delivered double-stranded RNA. Nanoscale 6, 9965–9969. [DOI] [PubMed] [Google Scholar]
- Kater MM, Franken J, Carney KJ, Colombo L, Angenent GC. 2001. Sex determination in the monoecious species cucumber is confined to specific floral whorls. The Plant Cell 13, 481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. 2006. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313, 845–848. [DOI] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jurgens G. 1996. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96. [DOI] [PubMed] [Google Scholar]
- Lenhard M, Jurgens G, Laux T. 2002. The WUSCHEL and SHOOTMERISTEMLESS genes fulfil complementary roles in Arabidopsis shoot meristem regulation. Development 129, 3195–3206. [DOI] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. 1994. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. The Plant Cell 6, 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Yao X, Pi L, Wang H, Cui X, Huang H. 2009. The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. The Plant Journal 58, 27–40. [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. 1996. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69. [DOI] [PubMed] [Google Scholar]
- Malepszy S, Niemirowicz-Szczytt K. 1991. Sex determination in cucumber (Cucumis sativus) as a model system for molecular biology. Plant Science 80, 39–47. [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T. 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. [DOI] [PubMed] [Google Scholar]
- Morel J-B, Godon C, Mourrain P, Béclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. 2002. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. The Plant Cell 14, 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T, Bayer M, Mravec J, Friml J, Birnbaum KD, Lukowitz W. 2010. The GATA factor HANABA TARANU is required to position the proembryo boundary in the early Arabidopsis embryo. Developmental Cell 19, 103–113. [DOI] [PubMed] [Google Scholar]
- Nicotra A, Cosgrove M, Cowling A, Schlichting C, Jones C. 2008. Leaf shape linked to photosynthetic rates and temperature optima in South African Pelargonium species. Oecologia 154, 625–635. [DOI] [PubMed] [Google Scholar]
- Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P. 2006. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. The Plant Cell 18, 2929–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. 2008. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319, 294. [DOI] [PubMed] [Google Scholar]
- Ohno CK, Reddy GV, Heisler MG, Meyerowitz EM. 2004. The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 131, 1111–1122. [DOI] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. 2000. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127, 5523–5532. [DOI] [PubMed] [Google Scholar]
- Papp I, Dulai S, Koncz C. 2004. A mutation in the Cap Binding Protein 20 gene confers drought. Plant Molecular Biology 55, 679–686. [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Wagner DR. 2001. The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. The Plant Cell 13, 1263–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor–joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T. 2000. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. [DOI] [PubMed] [Google Scholar]
- Spinelli SV, Martin AP, Viola IL, Gonzalez DH, Palatnik JF. 2011. A mechanistic link between STM and CUC1 during Arabidopsis development. Plant Physiology 156, 1894–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM. 1989. Patterns in plant development . Cambridge: Cambridge University Press. [Google Scholar]
- Sun X-l, Liu W-x, Cao Q-w, Zhang W-h. 2010. Anatomical observation of the female gametophyte development and embryogenesis of cucumber. China Cucurbits and Vegetables 2, 010. [Google Scholar]
- Tsukaya H. 2006. Mechanism of leaf-shape determination. Annual Review of Plant Biology 57, 477–496. [DOI] [PubMed] [Google Scholar]
- Tucker MR, Laux T. 2007. Connecting the paths in plant stem cell regulation. Trends in Cell Biology 17, 403–410. [DOI] [PubMed] [Google Scholar]
- Wang H, Sui X, Guo J, Wang Z, Cheng J, Ma S, Li X, Zhang Z. 2014. Antisense suppression of cucumber (Cucumis sativus L.) sucrose synthase 3 (CsSUS3) reduces hypoxic stress tolerance. Plant, Cell and Environment 37, 795–810. [DOI] [PubMed] [Google Scholar]
- Wang L, Yin H, Qian Q, Yang J, Huang C, Hu X, Luo D. 2009. NECK LEAF 1, a GATA type transcription factor, modulates organogenesis by regulating the expression of multiple regulatory genes during reproductive development in rice. Cell Research 19, 598–611. [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. 1992. LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. [DOI] [PubMed] [Google Scholar]
- Whipple CJ, Hall DH, DeBlasio S, Taguchi-Shiobara F, Schmidt RJ, Jackson DP. 2010. A conserved mechanism of bract suppression in the grass family. The Plant Cell 22, 565–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Girke T, Jonsson H, Reddy GV. 2011. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes and Development 25, 2025–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N. 2014. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnology Journal 12, 797–807. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhou Y, Ding L, Wu Z, Liu R, Meyerowitz EM. 2013. Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. The Plant Cell 25, 83–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang X, Liu B, Wang W, Liu X, Chen C, Liu X, Yang S, Ren H. 2014. A GAMYB homologue CsGAMYB1 regulates sex expression of cucumber via an ethylene-independent pathway. Journal of Experimental Botany 65, 3201–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Medrano L, Ohashi K, Fletcher JC, Yu H, Sakai H, Meyerowitz EM. 2004. HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. The Plant Cell 16, 2586–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.