Highlight:
KNOX3 gene is involved in symbiotic nodule development in Medicago truncatula and Pisum sativum, providing evidence that it may regulate cytokinin biosynthesis/activation upon nodulation.
Key words: Cytokinin, KNOX transcription factors, KNOX3, legume–rhizobium symbiosis, nodule development, plant meristem.
Abstract
KNOX transcription factors (TFs) regulate different aspects of plant development essentially through their effects on phytohormone metabolism. In particular, KNOX TF SHOOTMERISTEMLESS activates the cytokinin biosynthesis ISOPENTENYL TRANSFERASE (IPT) genes in the shoot apical meristem. However, the role of KNOX TFs in symbiotic nodule development and their possible effects on phytohormone metabolism during nodulation have not been studied to date. Cytokinin is a well-known regulator of nodule development, playing the key role in the regulation of cell division during nodule primordium formation. Recently, the activation of IPT genes was shown to take place during nodulation. Therefore, it was hypothesized that KNOX TFs may regulate nodule development and activate cytokinin biosynthesis upon nodulation. This study analysed the expression of different KNOX genes in Medicago truncatula Gaertn. and Pisum sativum L. Among them, the KNOX3 gene was upregulated in response to rhizobial inoculation in both species. pKNOX3::GUS activity was observed in developing nodule primordium. KNOX3 ectopic expression caused the formation of nodule-like structures on transgenic root without bacterial inoculation, a phenotype similar to one described previously for legumes with constitutive activation of the cytokinin receptor. Furthermore, in transgenic roots with MtKNOX3 knockdown, downregulation of A-type cytokinin response genes was found, as well as the MtIPT3 and LONELYGUY2 (MtLOG2) gene being involved in cytokinin activation. Taken together, these findings suggest that KNOX3 gene is involved in symbiotic nodule development and may regulate cytokinin biosynthesis/activation upon nodule development in legume plants.
Introduction
Interaction between legume plants and soil rhizobia leads to the development of new organs on roots, symbiotic nodules, where nitrogen fixation takes place. Bacterial signals Nod factors induce a complex of responses in epidermal cells and stimulate cell proliferation in the root pericycle and cortex, leading to nodule primordium formation (Schultze and Kondorosi, 1998). In legumes with indeterminate nodules, such as Medicago truncatula Gaertn. and Pisum sativum L., the nodule meristem is formed at the tip of primordium, providing the indeterminate growth of nodules (Rolfe and Gresshoff, 1988; Sprent and James, 2008).
A large number of studies on the model legumes have been carried out, demonstrating the important role of cytokinin and auxin in regulation of cell proliferation and differentiation during nodule development ((Libbenga et al. 1973; Gonzalez-Rizzo et al., 2006; Tirichine et al., 2007; Heckmann et al., 2011; Rightmyer and Long, 2011; Plet et al., 2011, reviewed by Ferguson and Mathesius, 2014). The positive role of cytokinin in nodule formation was first demonstrated in experiments with exogenous application of cytokinin, which promotes the formation of nodule-like structures on the roots of legume plants (Libbenga et al. 1973, Cooper and Long, 1994; Heckmann et al., 2011). Furthermore, a gain-of-function mutation in the Lotus histidine kinase1 (LjLHK1) gene encoding the cytokinin receptor in Lotus japonicus leads to spontaneous nodule formation (Murray et al. 2007; Tirichine et al., 2007). The lhk1-1 mutant carrying a loss-of-function mutation in the cytokinin receptor gene has reduced nodule formation, but it still develops a limited number of nodules at a later time point after rhizobial inoculation (Murray et al., 2007). Recently, it was shown that, along with LjLHK1, other cytokinin receptors such as LjLHK1A and LjLHK3 work partially redundantly promoting cell divisions during nodule primordium formation in L. japonicus (Held et al., 2014). Cytokinin-responsive genes, type-A response regulators, are activated in response to rhizobial inoculation, indicating that the cytokinin response is a part of a signalling cascade induced by rhizobia (Gonzalez-Rizzo et al., 2006; Op den Camp et al., 2011; Plet et al., 2011). In agreement with this, cytokinin activates the expression of genes encoding transcriptional regulators of nodule development, such as NSP1, NSP2, ERN1, and NIN (Plet et al., 2011). Recently, it was shown that the expression levels of genes involved in cytokinin biosynthesis were increased early in response to rhizobial signals. In L. japonicas, the expression level of the LjIPT3 gene encoding adenylate isopenthenyltransferase, the first and rate-limiting enzyme in cytokinin biosynthesis, was induced within 3h after Rhizobium inoculation. Its expression increased in roots during nodule development, reaching the highest level at the mature stage. Moreover, the expression of LjIPT1 was shown to be increased at 7 d after inoculation (Chen et al., 2013). In M. truncatula, the expression of MtIPT (Medtr2g022140) was induced as early as 1h after Nod factor treatment (van Zeijl et al., 2015). In addition, LONELYGUY (LOG) genes encoding cytokinin-activating enzymes were upregulated during nodulation in Medicago (Mortier et al., 2014, van Zeijl et al., 2015). Nod factor treatment was shown to induce accumulation of cytokinins, including trans-zeatin and isopentenyl adenine, in the root susceptible zone within 3h (van Zeijl et al., 2015). Taken together, these data suggest that cytokinin biosynthesis and activation are induced in legume roots in response to rhizobial signals.
Recently, it was shown that the cytokinin biosynthesis genes LjIPT3 and LjIPT1 were also induced in the shoot after rhizobial inoculation. Furthermore, LjIPT3 was shown to be activated in shoot phloem via the components of the AON (autoregulation of nodulation) system, thereby negatively affecting nodulation. Nodule number was increased in mutants defected in LjIPT3 and decreased in LjIPT3-overexpressing plants (Sasaki et al., 2014). In contrast to this, in previous study, Chen et al. (2013) found that RNA interference (RNAi) knockdown lines of LjIPT3 had decrease nodule number, suggesting that LjIPT3 has a positive effect on nodule development. Therefore, it was proposed that cytokinins have a dual role in nodulation depending on the time and place of their induction (Sasaki et al., 2014).
Along with cytokinin, auxin is also involved in nodule development. Exogenous application of polar auxin transport inhibitors stimulated the formation of nodule-like structures (Hirsch et al., 1989). There is evidence that polar auxin transport is regulated by cytokinin, as cytokinin negatively regulates the expression of PIN genes encoding auxin efflux carriers that mediate polar auxin transport (Plet et al., 2011).
The role of cytokinin and auxin as well as their crosstalk is well studied for plant meristems, specifically shoot and root apical meristems (SAM and RAM, respectively). In plant meristems, phytohormone metabolism, their transport and crosstalk are regulated by a set of transcription factors (TFs). A special place among them belongs to the TFs with a homeodomain of the WUSCHEL-RELATED HOMEOBOX (WOX) and KNOTTED-LIKE HOMEOBOX (KNOX) families (Mayer et al., 1998; Schoof et al., 2000; Hake et al., 2004; Sarkar et al., 2007).
Little is known about the role of meristem-specific TFs in nodulation. Previously, the TF WOX5, a member of the WOX family, was shown to be involved in nodule development (Osipova et al., 2012). However, the role of KNOX genes in the development of symbiotic nodules in legume plants remains unexplored.
The best-characterized member of the KNOX family, SHOOTMERISTEMLESS (STM), stimulates the expression of IPT genes in the SAM. Based on sequence similarity, gene structure, and expression pattern, KNOX TFs are grouped into two classes, class I KNOX (KNOXI) and class II KNOX (KNOXII) (Hay and Tsiantis, 2010). KNOXI genes, which are evolutionarily close to KNOTTED1 from maize, the first homeodomain-containing TF identified in plants (Vollbrecht et al., 1991), are expressed in the SAM and play crucial roles in the maintenance of the SAM (Long et al., 1996; Hake et al., 2004; Barkoulas et al., 2008; Hay and Tsiantis, 2010). KNOXII genes display diverse expression patterns and their function is not yet well understood (Serikawa et al., 1997; Byrne et al., 2002; Zhong et al., 2008).
KNOX genes have been characterized in legume plants. In total, 10 KNOX genes have been identified in M. truncatula (Di Giacomo et al., 2008, Peng et al., 2011; Zhou et al., 2014). In pea (P. sativum L.), two members of KNOX1 class, the KNOX1 and KNOX2 genes, have mainly been investigated and showed to be involved in the regulation of plant architecture (Hofer et al., 2001; Zhou et al., 2014).
This study questioned whether KNOX genes regulate nodule formation in legumes and whether they may regulate the expression of cytokinin biosynthesis genes upon nodulation in a manner similar to their action in the SAM. It was found that the KNOX3 gene was upregulated in response to rhizobial inoculation, and its promoter activity was observed in developing nodule primordium. Data from KNOX3 ectopic expression and RNAi suggested that the KNOX3 gene may regulate cytokinin biosynthesis and activation during nodule development.
Materials and methods
Plant material, bacterial strains, and growth conditions
M. truncatula Gaertn. Jemalong plants (wild-type A17) were grown in growth chambers (16h/8h day/night regime, 21 °C, and 75% relative humidity) and inoculated with Sinorhizobium meliloti strain Sm2011. The seeds were surface sterilized with concentrated sulphuric acid for 10min and washed five to six times with sterile water. Medicago seedlings were inoculated with 1ml of culture of per plant (OD600=0.7). For temporal expression during nodulation, M. truncatula plants inoculated with Sm2011 were grown in vermiculate-containing pots moistened with nitrogen-free Farhaeus medium (Fahraeus, 1957), and infected root tissue was harvested at different stages after inoculation with S. meliloti together with the uninoculated control plants. To avoid harvesting of lateral root primordia, only segments between emerged lateral roots were collected. Nodules were obtained at different stages after inoculation from the infected sites of roots.
P. sativum L. cv. Frisson seeds were surface sterilized with sulphuric acid for 5min, washed three times with water, transferred to 1% water agar plates and germinated at room temperature in the dark. After germination (for 4–5 d), plants were transferred into pots with vermiculite saturated with Jensen medium (van Brussel et al., 1982) and grown in a growth chamber at 21 °C in a 16h/8h light/dark cycle at 60% humidity. Pea seedlings were inoculated with 1ml of culture of Rhizobium leguminosarum bv. viciae CIAM1026 (ARRIAM, WDCM 966) per plant (OD600=0.5), and infected root tissue was harvested at different stages after inoculation together with the uninoculated control plants..
Molecular cloning
The MtKNOX3 promoter (2204bp) and the coding sequence of MtKNOX3 were first cloned into pDONR221 (Invitrogen, USA) and then into pBGFWS7.0 containing the β-glucuronidase (GUS) reporter gene and 35S terminator sequence and the pB7WG2D vector containing the 35S promoter and terminator sequences (VIB, Ghent, Belgium), respectively, with LR clonase enzyme (Invitrogen). For downregulation of the MtKNOX3 gene, a fragment of nt 1–151 in the coding sequence was amplified with the addition of the CACC sequence to the forward primer and cloned in the pENTR-D-TOPO vector (Invitrogen) and was cloned in the pK7GWIWG2D vector using LR clonase enzyme. The resulting vector contained a hairpin construct flanked by the 35S promoter and terminator sequences. The primers used for cloning are listed in Supplementary Table S1, available at JXB online. All the cloning vectors were sequenced.
The full-length coding sequence of the PsKNOX3 gene was amplified using cDNA as a matrix with corresponding primers (Supplementary Table S1). Amplification was done using Phusion Flash High-Fidelity PCR Master Mix (Thermo Scientific, USA). The amplified product was inserted in the pDONR221 vector with BP Clonase enzyme (Invitrogen) and finally into pB7WG2D with the LR clonase enzyme. The verified construct was transferred into Agrobacterium rhizogenes strain Arqua 1. The pB7WG2D and pK7GWIWG2D vectors used in this work contain the green fluorescent protein (GFP) marker gene for the selection of transgenic roots.
To identify putative KNOX genes in P. sativum, a BLASTN search was performed on the recently created Transcriptome Shotgun Assembly (TSA) [roots of pea line SGE inoculated with R. leguminosarum bv. viciae at 7 d post-inoculation (dpi); V. Zhukov, A. Zhernakov, O. Kulaeva, N. Ershov, A. Borisov and I. Tikhonovich, unpublished data) and other available TSAs of P. sativum L. [BioProject: PRJNA66035 (Franssen et al., 2011), BioProject: PRJNA81209 (Kaur et al., 2012), BioProject: PRJNA211622 (Duarte et al., 2014)] using full-length KNOX1–KNOX10 cDNAs of M. truncatula. This allowed the identification of five predicted full-length sequences that showed a high level of homology to MtKNOX3, MtKNOX4, MtKNOX5, MtKNOX9, and MtKNOX10, and these were named PsKNOX3, PsKNOX4, PsKNOX5, PsKNOX9, and PsKNOX10. The full-length coding sequences of PsKNOX3, PsKNOX4, PsKNOX5, PsKNOX9, and PsKNOX10 genes were verified by PCR amplification on cDNAs (synthesis was done on total RNA isolated from inoculated pea roots of the SGE line at 7 dpi) using the primer pairs listed in Supplementary Table S1, available at JXB online, and flanking predicted coding sequences.
In addition, partial transcripts of four predicted pea KNOX genes were identified and named PsKNOX1, PsKNOX2, PsKNOX6, and PsKNOX8. The RACE procedure was used to reveal the full-length coding sequences of the PsKNOX6 and PsKNOX8 genes using the primer pairs listed in Supplementary Table S1. Synthesis was done on total RNA isolated from inoculated pea roots of the SGE line for PsKNOX8 gene, but for PsKNOX6 total RNA was isolated from leaves, because PsKNOX6 was barely expressed in pea roots and nodules.
The full-length sequence of the PsKNOX3 gene was also identified by RACE on cDNA of P. sativum cv. Frisson as a template (Supplementary Table S1). Since experimental work was done on P. sativum cv. Frisson, the specificity of the designed primers for PCR and sequenced all amplicons was checked.
Quantitative reverse transcription PCR (qRT-PCR) analysis
Total RNA was isolated from roots with an RNeasy Plant Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. After a DNase (Thermo Scientific) treatment, the samples were extracted with an equal volume of chloroform, and RNA was precipitated from the aqueous phase with 3M sodium acetate and ethanol and subsequently quantified with a NanoDrop 2000c UV-Vvis Spectrophotometer (Thermo Scientific). RNA (from 200ng to 1 μg) was used for cDNA synthesis with RevertAid Reverse Transcriptase (Thermo Scientific). The efficacy of the DNase treatment was checked by using controls without reverse transcriptase for subsequent qRT-PCR analysis. The qRT-PCR experiments were done on a CFX-96 real-time PCR detection system with a C1000 thermal cycler (Bio-Rad Laboratories, USA), and SYBR Green and Eva Green intercalating dye were used for detection (Bio-Rad Laboratories). All reactions were done in triplicate and averaged. Cycle threshold (C t) values were obtained with the accompanying software and data were analysed with the 2–ΔΔ Ct method (Livak and Schmittgen, 2001). The relative expression was normalized against the constitutively expressed ubiquitin and actin genes in Medicago and pea. Medicago cDNA sequences were taken from the M. truncatula genome database Mt4.0v1. All primer pairs (Supplementary Table S2) were designed using the Vector NTI program and produced by Evrogen (http://www.evrogen.com). PCR amplification specificity was verified using a dissociation curve (55–95 °C). Each experiment was repeated at least three times with independent biological samples.
A. rhizogenes-mediated plant transformation
A. rhizogenes-mediated M. truncatula plants were transformed as described previously (Mortier et al., 2010). For pea transformation, the seeds were sterilized as described above. Seeds were germinated on 1% water agar in the darkness. Five-d-old seedlings were transferred in sterile plastic bags with Jensen medium to the light and incubated for 2–3 d. The pea roots were cut off in the area of the hypocotyl and transformed with freshly grown A. rhizogenes strain Arqua 1. Plants were put in special glass jars 120×60mm on Jensen agar (van Brussel et al., 1982), and the area of cut-off was covered with wet wool and foil. The seedlings were co-cultivated with A. rhizogenes for 10–12 d at 21 °C (16h/8h light/darkness) and subsequently transferred to Emergence medium (Limpens et al., 2004) containing 150mg ml–1 of cefotaxime. Plants were incubated on Emergence medium with antibiotic for 7 d and then transferred to fresh Emergence medium without antibiotic and incubated for 4–5 d until the new roots were formed (potentially co-transformed with the T-DNA of the binary vector). Emerging roots were analysed using an epifluorescent stereomicroscope (Stereo Discovery V8; Carl Zeiss). The plants were placed into vermiculite and kept under plastic film for 2–3 d. After inoculation with R. leguminosarum bv. viciae (OD600=0.1–0.2), the plants were checked for nodule formation after 14 –21 d.
Histochemical localization of GUS activity
GUS activity in transformed roots was analysed with 2mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid as substrate in NT buffer (100mm Tris/HCl, pH 7.5, 50mm NaCl) supplemented with 2mM ferricyanide (Van den Eede et al., 1992). The roots were vacuum infiltrated for 20min and subsequently incubated in GUS buffer [100mM Tris, 50mM NaCl, 1.9mM K3Fe(CN)6, 2.5mM X-Gluc (pre-diluted in DMSO)] at 37° C until sufficient blue staining had been developed (about 1h). After staining, the roots were infiltrated with fixative (3% paraformaldehyde, 0.25 % glutaraldehyde, 0.1 % Tween 20, 0.1 % Triton X-100 in 1/3× MTSB buffer) under vacuum (–1 atm), fixed overnight at 4 °C, and rinsed with 1/3× MTSB buffer (50mm PIPES, 5mm MgSO4, 5mm EGTA). The root segments were subsequently embedded in agarose (3%) and 50 µm sections were prepared with a microtome with a vibrating blade (VT-1200S; Leica). Photographs were taken with a fluorescent microscope AxioImager.Z1 (Carl Zeiss, Germany) equipped with an MRc5 digital camera (Carl Zeiss) and ZEN 2011 microscope software (Carl Zeiss).
Statistical methods and computer software
Multiple alignment of nucleotide sequences was performed using Clustal W (Thompson et al., 1994) using Vector NTI Advance 10 (InforMax, http://www.informaxinc.com). MEGA6 was used to generate graphic output of phylogenetic tree (Hall, 2013). One-way ANOVA and Student’s t-test were used to compare gene expression levels in transgenic roots.
Results
qRT-PCR analysis of KNOX gene expression upon nodulation in M. truncatula and P. sativum
Ten members of the KNOX family have been identified previously in M. truncatula genome (Di Giacomo et al., 2008; Peng et al., 2011; Zhou et al., 2014). In pea, only two genes of this family, KNOX1 (GenBank accession no. AF080104) and KNOX2 (AF080105), have been annotated in databases to date (Hofer et al., 2001). To identify other pea KNOX genes, the recently created TSA (V. Zhukov, A. Zhernakov, O. Kulaeva, N. Ershov, A. Borisov and I. Tikhonovich, unpublished data) and other available TSAs of P. sativum L. (Franssen et al., 2011; Kaur et al., 2012; Duarte et al., 2014) were searched using full-length KNOX1–KNOX10 cDNAs of M. truncatula. As a result, the full-length and partial transcripts of nine predicted pea KNOX genes were identified. Two of them, PsKNOX1 and PsKNOX2, corresponded to the previously characterized pea genes. The full-length coding sequences of PsKNOX genes were verified by PCR amplification of cDNAs using the primer pairs listed in Supplementary Table S1 and the flanking predicted coding sequences. Subsequent RACE analysis for partial transcripts allowed identification of other full-length pea cDNAs. Finally, identified genes were annotated as PsKNOX3–PsKNOX6, PsKNOX8–PsKNOX10 (GenBank accession nos KP296686/KP296687, KP296688, KP296689/KP296690, KT363851, KP296691, and KP296692).
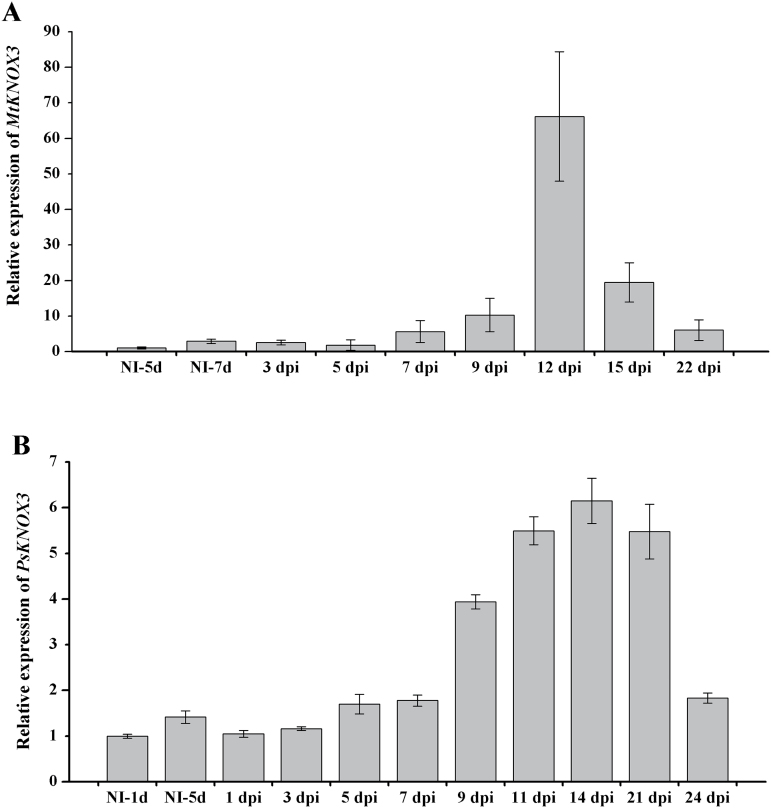
To study the expression dynamics of KNOX genes during nodule development, the relative transcript levels of KNOX genes were analysed at different stages after inoculation in M. truncatula and P. sativum using qRT-PCR. At 7 dpi, MtKNOX3 expression increased slightly in roots of M. truncatula inoculated with S. meliloti in compare with uninoculated control roots (Fig. 1A). At later stages, MtKNOX3 expression continued to increase up to a maximum at 12 dpi (on average more than 40-fold increase, compared with uninoculated control roots). Later at 15–22 dpi, MtKNOX3 expression decreased (Fig. 1A). The activation of the MtKNOX3 gene upon nodule development is consistent with data available in GENEEXPRESSION ATLAS (http://mtgea.noble.org/v3/). In addition to MtKNOX3, MtKNOX5 and MtKNOX9 were slightly upregulated during nodulation, although not as much as MtKNOX3 (Supplementary Fig. S1, available at JXB online). The expression levels of other MtKNOX genes in response to inoculation were not changed significantly (Supplementary Fig. S1).
Fig. 1.
qRT-PCR expression analysis of MtKNOX3 (A) and PsKNOX3 (B) in uninoculated plants (NI) and at different dpi. The relative expression was normalized against the constitutively expressed ubiquitin and actin genes. Results are means±SEM of three technical repeats.
In accordance with the data obtained for Medicago, PsKNOX3 expression levels significantly increased upon nodulation (Fig. 1B). The level of PsKNOX3 transcripts increased during nodule development starting from 7 dpi and reached the highest level at mature stages (14–21 dpi). PsKNOX3 expression then decreased at 24 dpi. At the same time, similarly to M. truncatula, the transcriptional levels of PsKNOX5 and PsKNOX9 genes were shown to increase upon nodulation (Supplementary Fig. S2, available at JXB online). In addition, in pea, the expression of PsKNOX10 was higher in developing nodules compared with uninoculated control roots. The expression levels of other PsKNOX genes did not change significantly in roots after inoculation (Supplementary Fig. S2). Since MtKNOX3 and its pea orthologue PsKNOX3 were the most strongly induced following inoculation, the KNOX3 gene was focused on for further functional studies.
Tissue-specific expression pattern of KNOX3 following nodulation by promoter:GUS analysis
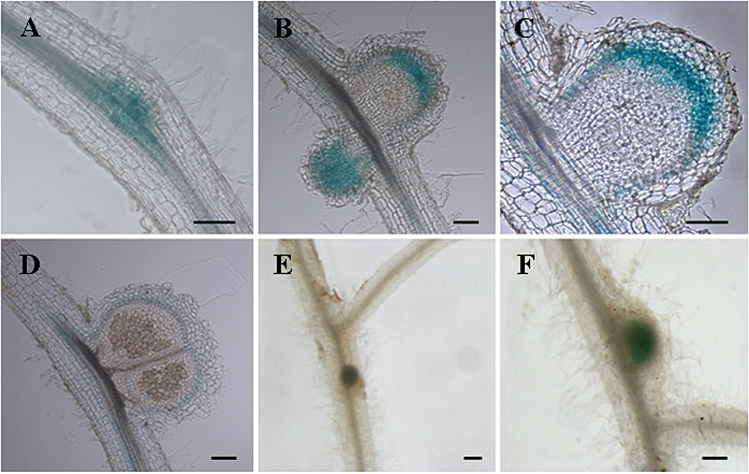
To localize KNOX3 expression during nodule development, a 2204bp MtKNOX3 putative promoter region was isolated and used to make a pMtKNOX3:GUS construct (see Materials and methods). This construct was introduced into M. truncatula roots via A. rhizogenes-mediated transformation. KNOX3 promoter activity was observed in developing nodule primordia at 7–10 dpi. At later stages (12–15 dpi), pKNOX3:GUS activity was restricted to provascular bundle tissues and to the apical part of nodule where apical meristem is formed (Fig. 2). At 18 dpi, MtKNOX3 promoter activity was detected at the periphery of mature nodules. The MtKNOX3 expression dynamic visualized with the MtKNOX3:GUS fusion was consistent with the qRT-PCR data. In addition to that in developing nodules, MtKNOX3 promoter activity was also found in lateral root primordia and root tips (Supplementary Fig. S3, available at JXB online).
Fig. 2.
Tissue-specific expression pattern of pMtKNOX3:GUS in M. truncatula. (A–D) Bright-field picture of a longitudinal section in nodule primordium (A), throughout proliferating cells in developing nodule (B), in the apical part of the nodule and in provascular bundles (C) and in late stages of nodule development (D). (E, F) pMtKNOX3:GUS expression in developing nodules. Bars, 100 μm. Thickness of sections, 50 μm.
Temporal dynamics of cytokinin response gene expression coincides with KNOX3 expression during nodulation
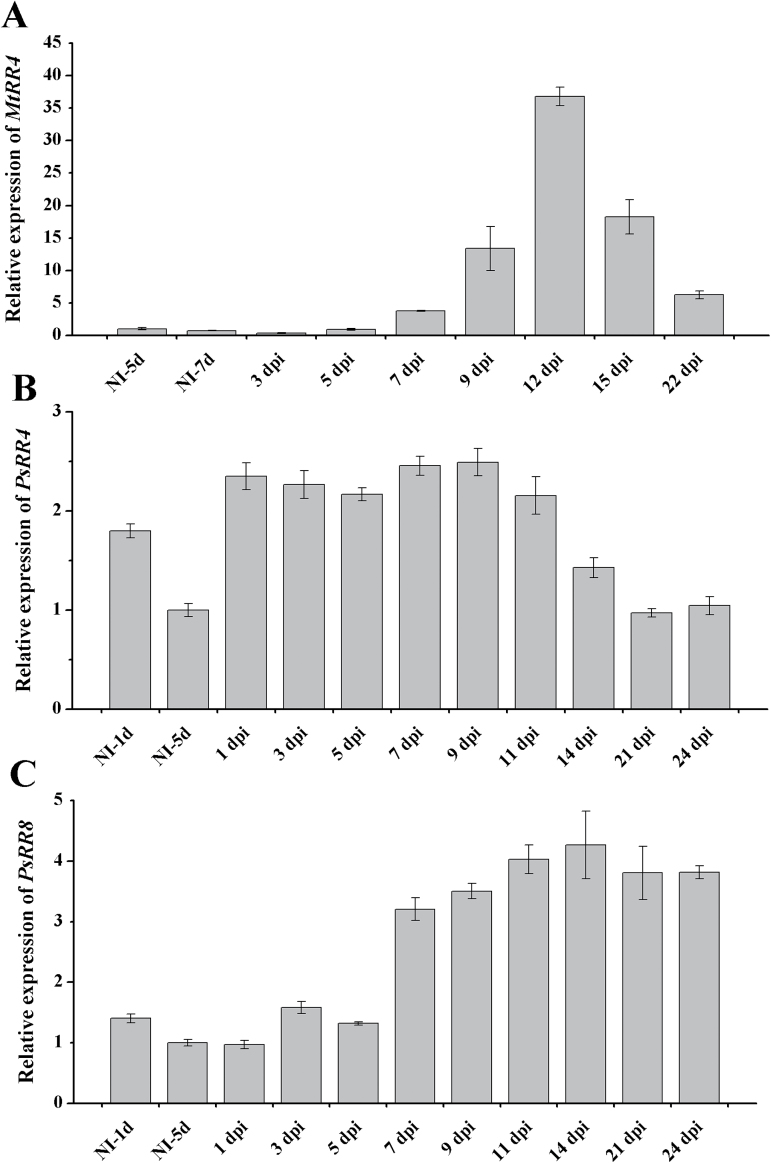
It is known that cytokinin response genes are activated upon nodule development (Lohar et al., 2004; Tirichine et al., 2007; Plet et al., 2011). It was shown previously that in M. truncatula the transcription level of five cytokinin response genes, MtRR4, MtRR5, MtRR8, MtRR9, and MtRR11, was markedly increased in response to inoculation with rhizobia in a CRE1-dependent manner (Gonzalez-Rizzo et al., 2006; Op den Camp et al., 2011). In our experiments, the expression of cytokinin response gene MtRR4 in M. truncatula reached its maximum at 12 dpi (Fig. 3A), i.e. coinciding with the maximum MtKNOX3 gene expression observed. In P. sativum, the expression of the PsRR4 (GenBank accession no. KP296699) and PsRR8 (KP296703) cytokinin response genes (closest homologuws of MtRR4 and MtRR8) were analysed at nodulation, which showed that PsRR8 expression reached a maximum at 14–21 dpi (Fig. 3B, C). The same expression dynamic was found for PsKNOX3. Thus, the KNOX3 expression maximum in both legumes was associated with the cytokinin response maximum upon nodule development, suggesting that KNOX3 may be involved in the cytokinin response activation.
Fig. 3.
Activation of cytokinin response regulator genes upon nodule development. Expression of MtRR4 (A) and PsRR4 (B), and PsRR8 (C) at the various stages of symbiosis development. NI, not inoculated. The relative expression was normalized against the constitutively expressed ubiquitin and actin genes. Results are means±SEM of three technical repeats.
Ectopic expression of KNOX3 causes the formation of nodule-like structures
To investigate the role of KNOX3 in root nodule development, transgenic roots overexpressing 35S:MtKNOX3 and 35S:PsKNOX3 were obtained and analysed by microscopy. qRT-PCR analysis confirmed the ectopic expression of MtKNOX3 and PsKNOX3 in individual transgenic roots (data not shown).
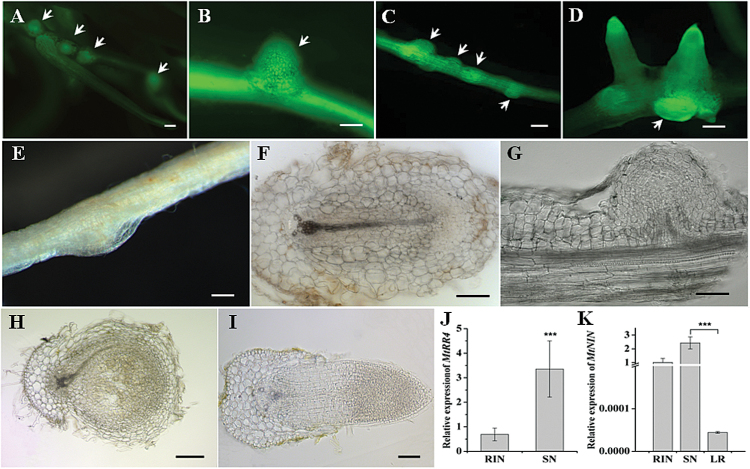
In uninoculated MtKNOX3-overexpressing and PsKNOX3-overexpressing roots, bumps were found resembling root nodule primordia (Fig. 4A–E). Such structures were not observed on the control roots with 35S:GUS overexpression. The frequency of such nodule-like structure formation was about 10–20 %: seven of 70 Medicago plants with GFP-positive transgenic roots formed these bumps (two to three per plant), whereas in pea, three of 15 plants with GFP-positive roots formed the bumps (four to five per plant). Microscopic analysis revealed that these nodule-like structures appeared as a result of cell divisions opposite to the protoxylem pole (Fig. 4F). In contrast to rhizobium-induced nodules, such structures did not develop a peripheral vascular system but instead formed one central vascular bundle similar to the lateral root primordial (Fig. 4F, G). In addition, expression of the nodulation-specific NIN gene was tested, which is expressed specifically in nodules and has a significantly lower expression level in lateral roots, as shown previously by Guan et al. (2013). NIN gene expression was significantly higher in bumps formed on KNOX3-overexpressing roots, compared with lateral root primordia. This suggested that the bumps observed were different from lateral root primordia and have nodule-like nature (Fig. 4K).
Fig. 4.
Phenotype of transgenic roots with overexpression of MtKNOX3 and PstKNOX3. The formation of structures resembling nodule primordia (spontaneous nodules) was found in the absence of rhizobia inoculation (A–G). (A–D) GFP fluorescence demonstrated expression of the transformation marker in M. truncatula (A, B) and P. sativum L. (C, D) Arrows indicate nodule-like structures. (E) Bright-field images of spontaneous nodules in M. truncatula. (F, G) Cross-section (F) and longitudinal section (G) of spontaneous nodules in M. truncatula with single vascular bundle. (H) Cross-section of wild-type nodule in M. truncatula with peripheral vascular bundles. (I) Cross-section of lateral root in M. truncatula. (J) Expression of MtRR4 in nodule-like structures in comparison with rhizobium-induced nodules (RIN) in M. truncatula. SN, spontaneous nodule. (K) Expression of nodule-specific genes MtNIN in SNs, RINs and lateral roots (LR). Error bars indicate the 95% confidence interval of three biological repeats. Asterisks indicate statistically significant differences compared with control (RIN): ***P<0.001. Bars, 100 μm. Thickness of sections, 50 μm.
Spontaneous nodules have been observed previously in legume plants with constitutive activity of cytokinin receptor gene. To check if nodule-like structures on KNOX3-overexpressing roots could be developed due to accumulation of cytokinin, the expression level of cytokinin response genes was analysed in these structures. Indeed, in Medicago, the expression of MtRR4 was significantly higher in nodule-like structures compared with rhizobium-induced nodules on GUS-overexpressing roots taken at 12 dpi (Fig. 4J).
Since these data supported the hypothesis about the possible role of KNOX3 gene in activation of cytokinin biosynthesis gene, the next step was to study the expression of cytokinin biosynthesis/activation genes in KNOX3-overexpressing and KNOX3-RNAi roots.
Expression of cytokinin biosynthesis/activation genes upon nodule development
To check our hypothesis that KNOX3 TF may activate expression of cytokinin biosynthesis/activation genes, the expression of IPT and LONELY GUY (LOG) genes, encoding cytokinin biosynthesis enzymes isopentenyl transferases and cytokinin-activating enzymes cytokinin riboside 5′-monophosphate phosphoribohydrolases, respectively, was analysed during nodule development in M. truncatula and P. sativum.
This study focused on the Medtr2g022140 gene (previously named MtIPT1 by van Zeijl et al., 2015), which was shown to be induced quite early after Nod factor treatment (1–3h) (van Zeijl et al., 2015), and the Medicago homologues of the LjIPT1 and LjIPT3 genes, which are induced during nodule development in L. japonicus (Chen et al., 2013). The M. truncatula genome database Mt4.0v1 was searched using LjIPT1 and LjIPT3 as reference sequences and closest homologues of these genes were found in M. truncatula. Phylogenetic analysis revealed that the closest homologue of LjIPT1 was Medtr1g110590, and the closest homologue of LjIPT3 was Medtr1g072540. Medtr2g022140 appeared to be the closest homologue of the LjIPT4 gene (Supplementary Fig. S4, available at JXB online). To avoid confusion, Medicago sequences are referred to according to the numbers given to corresponding IPT genes in L. japonicus, i.e. Medtr1g110590 is referred to as MtIPT1, Medtr1g072540 is referred to as MtIPT3, and Medtr2g022140 is referred to as MtIPT4.
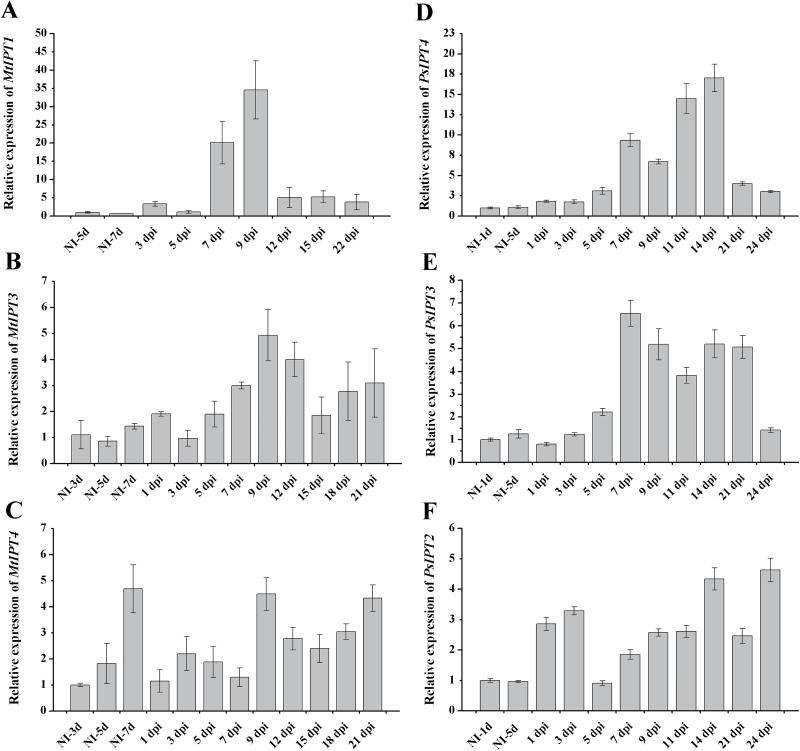
The expression dynamic of these MtIPTs was analysed in response to Rhizobium inoculation and it was found that Medtr1g110590 (MtIPT1) gene expression increased dramatically at 7 and 9 dpi (30- and 40-fold, respectively, compared with uninoculated control) (Fig. 5A). Moreover, the expression level of Medtr1g072540 (MtIPT3) was also increased during nodulation (Fig. 5B), whereas Medtr2g022140 (MtIPT4) expression was not changed significantly upon nodule development (Fig. 5C). Therefore, subsequent analyses focused on the Medtr1g110590 (MtIPT1) and Medtr1g072540 (MtIPT3) genes.
Fig. 5.
qRT-PCR expression analysis of MtIPT1 (A), MtIPT3 (B), MtIPT4 (C), PsIPT4 (D), PsIPT3 (E), and PsIPT2 (F) genes during nodule development in M. truncatula and P. sativum L. NI, not inoculated. Quantification was normalized using stable expressed reference genes ubiquitin and actin. Results are means±SEM of three technical repeats.
In P. sativum, two IPT genes (previously named PsIPT1 and PsIPT2) have been identified (Tanaka et al., 2006). PsIPT1 appeared to be the closest homologue of MtIPT2 (Medtr4g117330) and LjIPT2, whereas PsIPT2 seemed to be the closest homologue of MtIPT4 (Medtr2g022140) and LjIPT4 genes (Supplementary Fig. S4). To find P. sativum genes corresponding to M. truncatula and L. japonicus IPT1 and IPT3 genes, an in silico search was performed on TSA databases. As a result, two IPT genes designated PsIPT3 (GenBank accession no. KP296693) (the closest homologue of MtIPT3 (Medtr1g072540) and LjIPT3) and PsIPT4 (KP296694) (the closest homologue of MtIPT1 (Medtr1g110590) and LjIPT1) were identified.
Similarly to Medicago, in pea the transcription level of PsIPT4 (closest homologue of MtIPT1 and LjIPT1) was shown to be increased significantly during nodulation (Fig. 5D). The level of PsIPT4 expression increased at 5–7 dpi and reached a maximum at 11–14 dpi (on average a 10- to 15-fold increase compared with uninoculated control roots). The expression of PsIPT2 gene [closest homologue of MtIPT4 (Medtr2g022140)] was induced at early stages of nodule development (at 1–3 dpi) relative to uninoculated control and was slightly increased at mature stages of nodule development (Fig. 5F). The expression of the PsIPT3 gene was induced upon nodulation (on average 3- to 4-fold increase) but not as significantly as for PsIPT4 (Fig. 5E).
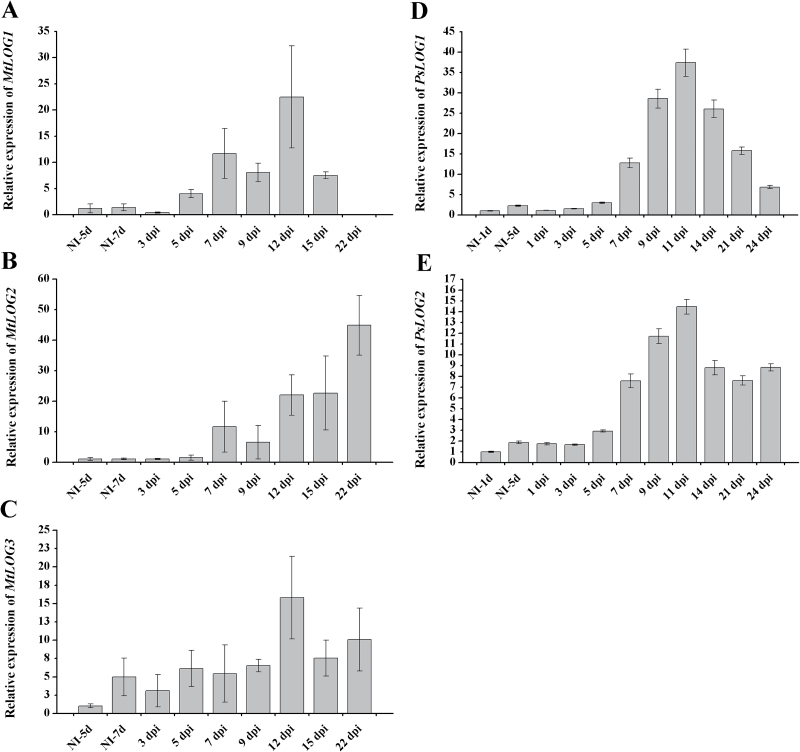
Among the MtLOG genes, MtLOG1 and MtLOG2 expression levels were increased upon nodule development (Fig. 6A, B). The highest level of MtLOG1 transcripts was observed at 12 dpi, whereas MtLOG2 expression level increased steadily with time after inoculation, reaching the highest level at 22 dpi. The data on MtLOG1/2 genes expression upon nodule development are consistent with data obtained by Mortier et al. (2014). Moreover, the accumulation of MtLOG3 gene transcripts at 12 dpi (Fig. 6C) was also observed.
Fig. 6.
Relative expression of MtLOG1 (A), MtLOG2 (B), MtLOG3 (C), PsLOG1 (D) and PsLOG2 (E) genes determined using qRT-PCR during nodule development in M. truncatula and P. sativum L. NI, not inoculated. The relative expression was normalized against the constitutively expressed ubiquitin and actin genes. Results are means±SEM of three technical repeats.
Similarly, in P. sativum, the closest homologues PsLOG1 (GenBank accession no. KP296697) and PsLOG2 (KP296698) showed significant upregulation in roots after rhizobial inoculation. The highest level of PsLOG1 and PsLOG2 transcripts was found at 11–14 dpi (Fig. 6D, E).
Effect of KNOX3 ectopic expression on symbiosis-inducible IPT and LOG genes
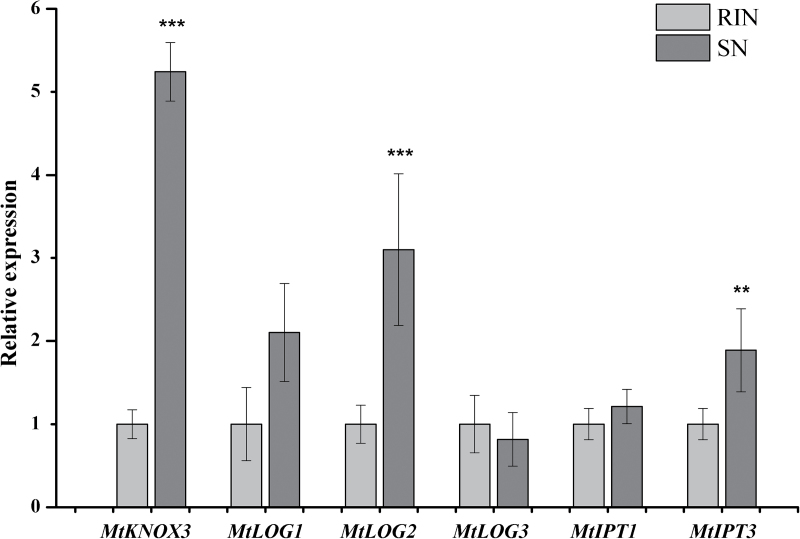
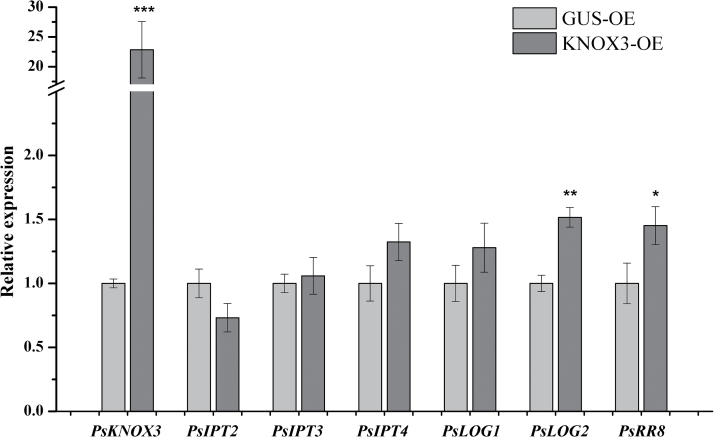
The expression was determined of symbiosis-inducible IPT and LOG genes in nodule-like structures formed on MtKNOX3-and PsKNOX3-overexpressing roots (Figs 7 and 8). As a control, the rhizobium-induced nodules on GUS-overexpressing roots were used. It was found that the expression of MtLOG2 and MtIPT3 was higher in nodule-like structures in comparison with the control (P<0.001 and P <0.01, respectively), while the expression of other analysed genes (MtLOG1, MtLOG3, and MtIPT1) did not show any statistically significant difference (Fig. 7). Likewise, it was found that expression of the PsLOG2 gene was significantly increased (P <0.01) in nodule-like structures on PsKNOX3-overexpressing roots (Fig. 8). Moreover, nodule-like structures on PsKNOX3-overexpressing roots demonstrated the increased level of cytokinin-responsive PsRR8 transcripts (P<0.05). However, the expression levels of PsLOG1, PsIPT3, and PsIPT4 were not significantly increased in the PsKNOX3 overexpression background (Fig. 8).
Fig. 7.
Expression of symbiosis-inducible IPT and LOG genes in nodule-like structures in comparison with rhizobium-induced nodules (RIN) in M. truncatula. SN, spontaneous nodule. Asterisks indicate statistically significant differences compared with control (RIN): ***P<0.001; **P<0.01; *P<0.05. Error bars indicate the 95% confidence interval of three biological repeats.
Fig. 8.
Effect of KNOX3 ectopic expression on symbiosis-inducible IPT and LOG genes in transgenic roots of P. sativum L. Asterisks indicate statistically significant differences compared with control [GUS-overexpressing (GUS-OE)]: ***P<0.001; **P<0.01; *P<0.05. Error bars indicate the 95% confidence interval of five biological repeats.
Effect of RNAi on nodule development and the expression of cytokinin biosynthesis/response genes
An RNAi construct was generated (KNOX3i) targeting MtKNOX3 mRNA (see Materials and methods). In KNOX3i transgenic roots and nodules, expression of MtKNOX3 gene decreased 4- to 5-fold compared with control roots (GUS-overexpressing control). No statistically significant difference was revealed between nodule number on MtKNOX3 RNAi roots and control roots (GUS-overexpressing roots) (Supplementary Fig. S5, available at JXB online). Moreover, nodules on MtKNOX3 RNAi roots demonstrated the wild-type phenotype (data not shown). The expression levels of other MtKNOX genes were not significantly changed by MtKNOX3 RNAi (data not shown).
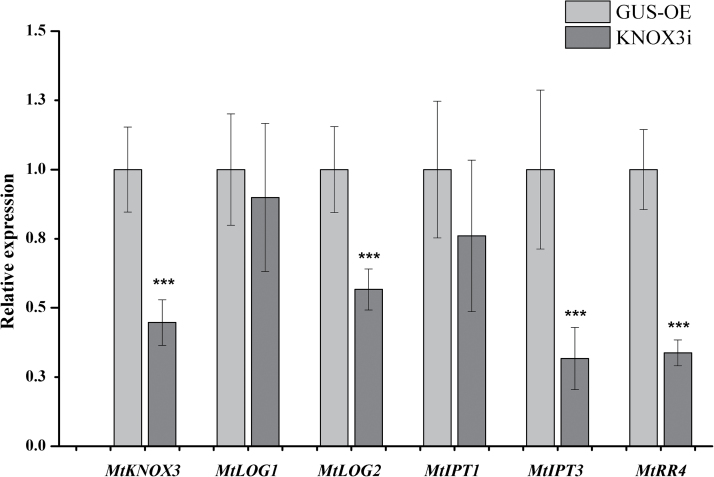
Next, the effect of MtKNOX3 RNAi on the expression of MtIPTs, MtLOGs and cytokinin-responsive gene MtRR4 in transgenic MtKNOX3 RNAi nodules was analysed with qRT-PCR. A statistically significant decrease was observed in MtLOG2, MtIPT3, and MtRR4 gene expression levels in transgenic nodules with MtKNOX3 RNAi in comparison with GUS-overexpressing control nodules (Fig. 9). The expression levels of Medtr1g110590 (MtIPT1) and MtLOG1 were not significantly changed in the MtKNOX3 RNAi background (Fig. 9). These data suggested that the MtKNOX3 TF may be involved in the activation of MtLOG2 and MtIPT3 genes. The decreased levels of cytokinin-responsive MtRR4 transcripts in MtKNOX3 RNAi roots are in line with the suggestion that MtKNOX3 may activate MtIPT and MtLOG genes and thereby increase cytokinin signalling during nodule development.
Fig. 9.
Reduction in gene expression level of MtLOG2, MtRR4, and MtIPT3 in MtKNOX3 RNAi transgenic M. truncatula plants. Asterisks indicate statistically significant differences compared with control [GUS-overexpressing (GUS-OE)]: *** P<0.001, ** P<0.01, * P<0.05. Error bars indicate the 95% confidence interval of eight biological repeats.
Discussion
Here, it was shown that the expression levels of the MtKNOX3 and PsKNOX3 genes encoding class II KNOTTED-like TFs increased significantly during nodulation in Medicago and pea. MtKNOX3 was shown to be closest homologue of the KNAT3 protein of Arabidopsis thaliana (Di Giacomo et al., 2008). The function of the AtKNAT3 protein is not well characterized. It was reported that AtKNAT3 is involved in embryo sac formation, germination, and early seedling development in Arabidopsis, where AtKNAT3 modulated the abscisic acid response via direct binding and activating the ABI3 promoter (Pagnussat et al., 2007; Kim et al., 2013). In addition, it was found that AtKNAT3 expression is repressed by moderate levels of cytokinin (Truernit et al., 2006). The KNAT3-like homeobox gene was also studied in black walnut (Juglans nigra L.), where it is expressed in pith meristem, roots, embryogenic callus, vascular cambium, leaves, and flowers. This gene is supposed to have a role during heartwood formation (Huang et al., 2009). The broad pattern of KNAT3-like gene expression in plants indicates that these genes may have diverse functions and mechanisms of action in plant development.
In this study, promoter:GUS analysis showed the KNOX3 expression pattern in M. truncatula nodule primordium cells and in the apical part of the nodule where the meristem is formed, as well as in provascular bundle tissues of developing nodules. KNOX3 promoter activity was also found in the lateral root primordium, in contrast to its putative orthologue in Arabisopsis AtKNAT3, for which expression was not observed in the lateral root primordium. Moreover, Di Giacomo et al. (2008) found that MtKNOX3 expression level increased rapidly in response to exogenous cytokinin by 30min after treatment. This also indicates a difference between MtKNOX3 and AtKNAT3, the latter being shown to be repressed by cytokinin in roots.
Activation of KNOX3 expression in response to cytokinin indicates a possible link between KNOX3 and cytokinin action during nodule development. In this study, it was hypothesized that KNOX3 may participate in the activation of cytokinin biosynthesis genes, as was shown for class I KNOX TFs in the SAM (Jasinski et al., 2005; Yanai et al., 2005).
According to the data, the MtKNOX3 expression maximum at 12 dpi in Medicago was associated with the cytokinin response gene MtRR4 maximum during nodulation. Similarly, the maximum of PsKNOX3 expression coincided with the expression maximum of the cytokinin response gene PsRR8. Moreover, the localization of KNOX3 expression visualized with promoter:GUS analysis coincided with the MtRR4 expression area in developing nodules studied using promoter:GUS analysis and RNA in situ hybridization (Vernié et al., 2008; Plet et al., 2011). In the current study, it was found that ectopic expression of the MtKNOX3 and PsKNOX3 genes resulted in the formation of bumps exhibiting increased expression of the cytokinin response genes MtRR4 and PsRR8, respectively. The presence of NIN gene expression in such structures suggested that they have a nodule-like nature. These data are in line with the hypothesis suggested in this study about the possible role of the KNOX3 gene in activation of cytokinin biosynthesis genes.
The formation of structures resembling nodule primordia on KNOX3-overexpressing roots in the absence of rhizobia inoculation (i.e. spontaneously) may indicate that the TF KNOX3 is involved in the regulation of cell proliferation upon nodule development. It was shown previously that a local increase in cytokinin concentration in Arabidopsis root pericycle cells due to overexpression of a cytokinin biosynthesis gene, IPT, affected lateral root development. Moreover, the addition of exogenous cytokinin altered the cell division plane of lateral root primordia, disrupting the structure and leading to the formation of ‘flat’ primordia (Laplaze et al., 2007). Lastly, the gain-of function mutation in cytokinin receptor gene LHK1 in L. japonicus (spontaneous nodule formation 2, snf2) resulted in nodule formation in the absence of rhizobia (spontaneous nodulation) (Tirichine et al., 2007). These data indicate that the increased cytokinin concentration/response in root tissues results in increased cell proliferation of root cortical cells leading to the formation of spontaneous nodules. Thus, here it was assumed that the nodule-like structures observed might also be associated with cytokinin activity, i.e. with the accumulation of active cytokinins in KNOX3-overexpressing roots. In contrast to rhizobium-induced nodules, nodule-like structures on KNOX3-overexpressing roots developed one central vascular bundle. Previously, such nodule-like structures with one central bundle were observed in transgenic roots with ectopic expression of the NIN gene and NF-Y subunit genes in L. japonicus, which act in the regulatory cascade induced by Nod factors (Soyano et al., 2013). Moreover, nodules with one central vascular bundle are formed in lin (lumpy infection) mutants defective for rhizobial infection (Kuppusamy et al., 2004; Guan et al., 2013). Xiao et al. (2014) suggested that such a phenotype could be explained by the higher cell proliferation in endodermis and pericycle cell layers. Thus, here it was assumed that KNOX3 ectopic expression changes the phytohormonal balance, which finally induces cell proliferation in the root pericycle, endodermis, and cortex, leading to nodule-like structure formation.
Previously, Chen et al. (2013) identified six IPT genes in the L. japonicus genome and found that LjIPT1 and LjIPT3 were activated in response to nodulation. In the current study, it was found that the expression level of MtIPT1 (Medtr1g110590) and its pea homologue, PsIPT4, were significantly increased upon nodulation. Changed expression levels were also observed for the MtIPT3 (Medtr1g072540) and PsIPT3 genes upon inoculation but were not as pronounced as for the MtIPT1 and PsIPT4 genes. These data are in agreement with the results obtained for L. japonicus by Chen et al. (2013). The expression level of MtIPT4 (Medtr2g022140), which was shown previously to be induced early by Nod factor treatment (van Zeijl et al., 2015), was not changed during nodule development in our experiment. It is suggested that the induction of this gene might be associated with the early steps of symbiosis establishment only, since it was induced by Nod factor as early as in 1h after treatment, whereas the later stages of nodule development in our experiment (1 dpi and later in Medicago) are unlikely to be regulated by the MtIPT4 (Medtr2g022140) gene. In agreement with this, in pea, induction of the PsIPT2 gene [MtIPT4 (Medtr2g022140) homologue] was also observed at the early stages of symbiosis development (1–3 dpi). This later response in gene expression following rhizobial inoculation in pea compared with Medicago is in line with previous observations that expression of early nodulin genes in pea reached a maximum in between 24 and 48h after rhizobial inoculation (Albrecht et al., 1998; Dolgikh et al., 2011).
The results of this study suggest that the MtIPT1 (Medtr 1g110590)/PsIPT4 gene is unlikely to be the target of the KNOX3 TF upon nodulation. No significant change in MtIPT1 (Medtr 1g110590)/PsIPT4 was found in the KNOX3-overexpressing background for both legumes. Again, MtIPT1 (Medtr1g110590) expression was not changed in transgenic nodules with KNOX3 RNAi in M. truncatula. At the same time, these data suggest that the MtIPT3 (Medtr1g072540) gene may be regulated by MtKNOX3 during nodulation, since its expression level was decreased in transgenic roots with MtKNOX3 RNAi and increased in the KNOX3-overexpressing background. The homologue of MtIPT3 (Medtr1g072540) in pea, PsIPT3, was not significantly upregulated in PsKNOX3-overexpressing nodules. Moreover, the PsIPT3 gene demonstrated a moderate increase throughout the nodule development stages in pea, and one may propose that the role and regulation of the IPT3 gene during nodule development could differ in both legumes; however, the additional analysis is needed to determine this.
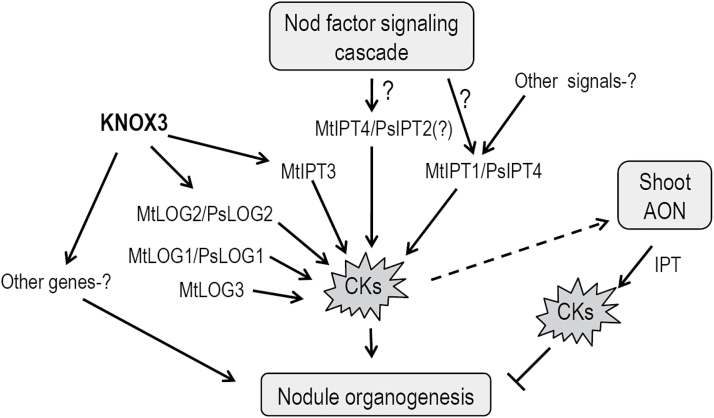
Lastly, our data suggest that the LOG2 gene may be regulated by KNOX3 during nodulation. This assumption is supported by the reduced level of MtLOG2 expression in transgenic roots with MtKNOX3 RNAi and the increased transcription level of the MtLOG2 and PsLOG2 genes in nodule-like structures with MtKNOX3 and PsKNOX3 overexpression. Based on these data, it is suggested that KNOX3 may activate LOG2 gene expression during nodulation, thereby stimulating the accumulation of cytokinin active forms (Fig. 10). This may account for the formation of nodule-like structures on KNOX3-overexpressing roots that demonstrated the increased cytokinin response. However, to prove the involvement of the TF KNOX3 in the activation of LOG2 gene expression upon nodule development, additional experiments are required, such as the measurement of active cytokinin content in transgenic roots with altered expression of KNOX3 and the identification of KNOX3 direct targets by promoter-binding studies.
Fig. 10.
Model of KNOX3 and cytokinin biosynthesis gene action in nodulation. The expression of three IPT genes (MtIPT1/PsIPT4, MtIPT3, and MtIPT4/PsIPT2) and three LOG genes (MtLOG1/PsLOG1, MtLOG2/PsLOG2, and MtLOG3) were induced in response to Rhizobium inoculation. The MtIPT4 gene (and presumably its pea orthologue PsIPT2) may be activated by the Nod factor signalling cascade at the early stages of nodulation, as shown by van Zeijl et al. (2015). It is proposed that the MtIPT3 and MtLOG2/PsLOG2 genes may be regulated by KNOX3 during nodulation since their expression levels were changed in roots with KNOX3-overexpression and RNAi. This may account for the increase in active cytokinin concentration and, as a consequence, the formation of nodule-like structures on KNOX3-overexpressing roots, which demonstrated the increased cytokinin response. Besides its positive effect on nodule formation, cytokinin also acts as negative regulator of nodulation, and its production is induced in shoots by the components of the AON system (Sasaki et al., 2014). CKs, cytokinins.
In plants, the functions of KNOX TFs are not limited to their effects on cytokinin metabolism genes. These transcriptional regulators are known to regulate the phytohormonal balance via multiple targets, including genes involved in metabolism and action of other hormones such as auxin, gibberellic acid, and abscisic acid (Sakamoto et al., 2001; Bolduc et al., 2012). It is quite possible that KNOX3 also affects multiple gene targets regulating hormonal balance in developing nodules, and future studies are required to unravel KNOX3 targets during nodulation.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Relative expression of MtKNOX genes in uninoculated plants (NI) and at different days post-inoculation (dpi).
Supplementary Fig. S2. Relative expression of PsKNOX genes in uninoculated plants (NI) and at different days post-inoculation (dpi).
Supplementary Fig. S3. Tissue-specific expression pattern of pMtKNOX3:GUS in root tips and lateral root primordia.
Supplementary Fig. S4. Phylogenetic tree of the MtIPT, PsIPT, and LjIPT gene families.
Supplementary Fig. S5. Effect of KNOX3 knockdown on nodulation.
Supplementary Table S1. List of primers used for cloning.
Supplementary Table S2. List of primers for qRT-PCR.
Acknowledgments
This work was supported at ARRIAM by the Russian Scientific Foundation (RSF project no. 14-24-00135) and at SPbSU by grants from Saint-Petersburg State University (1.38.676.2013, 1.38.229.2014, and 0.37.526.2013); the Ministry of Education and Science of the Russian Federation (State Contract 8045), grants RFBR 14-04-00591-а, 15-34-20071б, and 15-29-02737; and the President of Russia (HШ-5345.2012.4). The research was performed using the equipment of the Core Center ‘Genomic technologies, proteomics and cellular biology’ at ARRIAM. The authors acknowledge the Research Resource Center for Molecular and Cell Technologies of Saint-Petersburg State University for DNA sequencing and the equipment used in this study.
References
- Albrecht C, Geurts R, Lapeyrie F, Bisseling T. 1998. Endomycorrhizae and rhizobial Nod factors both require SYM8 to induce the expression of the early nodulin genes PsENOD5 and PsENOD12A . The Plant Journal 15, 605–614. [DOI] [PubMed] [Google Scholar]
- Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M. 2008. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta . Nature Genetics 40, 1136–1141. [DOI] [PubMed] [Google Scholar]
- Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O’Connor D, Grotewold E, Hake S. 2012. Unraveling the KNOTTED1 regulatory network in maize meristems. Genes & Development 26, 1685–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME, Simorowski J, Martienssen RA. 2002. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis . Development 129, 1957–1965. [DOI] [PubMed] [Google Scholar]
- Chen Y, Chen W, Li X, Jiang H, Wu P, Xia K, Yang Y, Wu G. 2013. Knockdown of LjIPT3 influences nodule development in Lotus japonicus . Plant and Cell Physiology 55, 183–193. [DOI] [PubMed] [Google Scholar]
- Cooper JB, Long SR. 1994. Morphogenetic rescue of Rhizobium meliloti nodulation mutants by trans-zeatin secretion. The Plant Cell Online 6, 215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Op den Camp RH, De Mita S, Lillo A, Cao Q, Limpens E, Bisseling T, Geurts R. 2011. A phylogenetic strategy based on a legume-specific whole genome duplication yields symbiotic cytokinin type-A response regulators. Plant Physiology 157, 2013–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giacomo E, Sestili F, Iannelli MA, Testone G, Mariotti D, Frugis G. 2008. Characterization of KNOX genes in Medicago truncatula . Plant Molecular Biology 67, 135–150. [DOI] [PubMed] [Google Scholar]
- Dolgikh E, Leppyanen I, Osipova M, Savelyeva N, Borisov A, Tsyganov V, Geurts R, Tikhonovich I. 2011. Genetic dissection of Rhizobium‐induced infection and nodule organogenesis in pea based on ENOD12A and ENOD5 expression analysis. Plant Biology 13, 285–296. [DOI] [PubMed] [Google Scholar]
- Duarte J, Rivière N, Baranger A, Aubert G, Burstin J, Cornet L, Lavaud C, Lejeune-Hénaut I, Martinant J-P, Pichon J-P. 2014. Transcriptome sequencing for high throughput SNP development and genetic mapping in pea. BMC Genomics 15, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahraeus G. 1957. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. Journal of General Microbiology 16: 374–381. [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Mathesius U. 2014. Phytohormone regulation of legume–rhizobia interactions. Journal of Chemical Ecology 40, 770–790. [DOI] [PubMed] [Google Scholar]
- Franssen SU, Shrestha RP, Bräutigam A, Bornberg-Bauer E, Weber AP. 2011. Comprehensive transcriptome analysis of the highly complex Pisum sativum genome using next generation sequencing. BMC Genomics 12, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F. 2006. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti . The Plant Cell Online 18, 2680–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, Stacey N, Liu C, Wen J, Mysore KS, Torres-Jerez I, Vernié T, Tadege M, Zhou C, Wang Z-Y. 2013. Rhizobial infection is associated with the development of peripheral vasculature in nodules of Medicago truncatula . Plant Physiology 162, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S, Smith HM, Holtan H, Magnani E, Mele G, Ramirez J. 2004. The role of KNOX genes in plant development. Annual Review of Cell and Developmental Biology 20, 125–151. [DOI] [PubMed] [Google Scholar]
- Hall BG. 2013. Building phylogenetic trees from molecular data with MEGA. Molecular Biology and Evolution 30, 1229–1235. [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. 2010. KNOX genes: versatile regulators of plant development and diversity. Development 137, 3153–3165. [DOI] [PubMed] [Google Scholar]
- Heckmann AB, Sandal N, Bek AS, Madsen LH, Jurkiewicz A, Nielsen MW, Tirichine L, Stougaard J. 2011. Cytokinin induction of root nodule primordia in Lotus japonicus is regulated by a mechanism operating in the root cortex. Molecular Plant–Microbe Interactions 24, 1385–1395. [DOI] [PubMed] [Google Scholar]
- Held M, Hou H, Miri M, Huynh C, Ross L, Hossain MS, Sato S, Tabata S, Perry J, Wang TL. 2014. Lotus japonicus cytokinin receptors work partially redundantly to mediate nodule formation. The Plant Cell Online 26, 678–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch AM, Bhuvaneswari TV, Torrey JG, Bisseling T. 1989. Early nodulin genes are induced in alfalfa root outgrowths elicited by auxin transport inhibitors. Proceedings of the National Academy of Sciences, USA 86, 1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer J, Gourlay C, Michael A, Ellis TN. 2001. Expression of a class 1 knotted1-like homeobox gene is down-regulated in pea compound leaf primordia. Plant Molecular Biology 45, 387–398. [DOI] [PubMed] [Google Scholar]
- Huang Z, Meilan R, Woeste K. 2009. A KNAT3-like homeobox gene from Juglans nigra L., JnKNAT3-like, highly expressed during heartwood formation. Plant Cell Reports 28, 1717–1724. [DOI] [PubMed] [Google Scholar]
- Jasinski S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M. 2005. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Current Biology 15, 1560–1565. [DOI] [PubMed] [Google Scholar]
- Kaur S, Pembleton LW, Cogan NO, Savin KW, Leonforte T, Paull J, Materne M, Forster JW. 2012. Transcriptome sequencing of field pea and faba bean for discovery and validation of SSR genetic markers. BMC Genomics 13, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Cho Yh, Ryu H, Kim Y, Kim TH, Hwang I. 2013. BLH1 and KNAT3 modulate ABA responses during germination and early seedling development in Arabidopsis . The Plant Journal 75, 755–766. [DOI] [PubMed] [Google Scholar]
- Kuppusamy KT, Endre G, Prabhu R, Penmetsa RV, Veereshlingam H, Cook DR, Dickstein R, VandenBosch KA. 2004. LIN, a Medicago truncatula gene required for nodule differentiation and persistence of rhizobial infections. Plant Physiology 136, 3682–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, Maes L, Vanneste S, Swarup R, Weijers D, Calvo V, Parizot B, Herrera-Rodriguez MB. 2007. Cytokinins act directly on lateral root founder cells to inhibit root initiation. The Plant Cell Online 19, 3889–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libbenga K, Van Iren F, Bogers R, Schraag-Lamers M. 1973. The role of hormones and gradients in the initiation of cortex proliferation and nodule formation in Pisum sativum L. Planta 114, 29–39. [DOI] [PubMed] [Google Scholar]
- Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R. 2004. RNA interference in Agrobacterium rhizogenes‐transformed roots of Arabidopsis and Medicago truncatula . Journal of Experimental Botany 55, 983–992. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lohar DP, Schaff JE, Laskey JG, Kieber JJ, Bilyeu KD, Bird DM. 2004. Cytokinins play opposite roles in lateral root formation, and nematode and rhizobial symbioses. The Plant Journal 38, 203–214. [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. 1996. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis . Nature 379, 66–69. [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. [DOI] [PubMed] [Google Scholar]
- Mortier V, Den Herder G, Whitford R, Van de Velde W, Rombauts S, D’haeseleer K, Holsters M, Goormachtig S. 2010. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiology 153, 222–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier V, Wasson A, Jaworek P, De Keyser A, Decroos M, Holsters M, Tarkowski P, Mathesius U, Goormachtig S. 2014. Role of LONELY GUY genes in indeterminate nodulation on Medicago truncatula . New Phytologist 202, 582–593. [DOI] [PubMed] [Google Scholar]
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K. 2007. A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315, 101–104. [DOI] [PubMed] [Google Scholar]
- Osipova MA, Mortier V, Demchenko KN, Tsyganov VE, Tikhonovich IA, Lutova LA, Dolgikh EA, Goormachtig S. 2012. WUSCHEL-RELATED HOMEOBOX5 gene expression and interaction of CLE peptides with components of the systemic control add two pieces to the puzzle of autoregulation of nodulation. Plant Physiology 158, 1329–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu H-J, Sundaresan V. 2007. Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1 . The Plant Cell Online 19, 3578–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Yu J, Wang H, Guo Y, Li G, Bai G, Chen R. 2011. Regulation of compound leaf development in Medicago truncatula by fused compound leaf1, a class M KNOX gene. The Plant Cell Online 23, 3929–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plet J, Wasson A, Ariel F, Le Signor C, Baker D, Mathesius U, Crespi M, Frugier F. 2011. MtCRE1‐dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula . The Plant Journal 65, 622–633. [DOI] [PubMed] [Google Scholar]
- Rightmyer AP, Long SR. 2011. Pseudonodule formation by wild-type and symbiotic mutant Medicago truncatula in response to auxin transport inhibitors. Molecular Plant–Microbe Interactions 24, 1372–1384. [DOI] [PubMed] [Google Scholar]
- Rolfe BG, Gresshoff PM. 1988. Genetic analysis of legume nodule initiation. Annual Review of Plant Physiology and Plant Molecular Biology 39, 297–319. [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. 2007. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Suzaki T, Soyano T, Kojima M, Sakakibara H, Kawaguchi M. 2014. Shoot-derived cytokinins systemically regulate root nodulation. Nature Communications 5, 4983. [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T. 2000. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. [DOI] [PubMed] [Google Scholar]
- Schultze M, Kondorosi A. 1998. Regulation of symbiotic root nodule development. Annual Review of Genetics 32, 33–57. [DOI] [PubMed] [Google Scholar]
- Serikawa KA, Martinez‐Laborda A, Kim HS, Zambryski PC. 1997. Localization of expression of KNAT3, a class 2 knotted1‐like gene. The Plant Journal 11, 853–861. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Kamiya N, Ueguchi-Tanaka M, Iwahori S, Matsuoka M. 2001. KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes & Development 15, 581–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Kouchi H, Hirota A, Hayashi M. 2013. Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus . PLoS Genetics 9, e1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent JI and James EK. 2008. Legume–rhizobial symbiosis: an anorexic model? New Phytologist 179, 3–5. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H. 2006. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. The Plant Journal 45, 1028–1036. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L, Sandal N, Madsen LH, Radutoiu S, Albrektsen AS, Sato S, Asamizu E, Tabata S, Stougaard J. 2007. A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315, 104–107. [DOI] [PubMed] [Google Scholar]
- Truernit E Siemering KR Hodge S Grbic V and Haseloff1 J. 2006. A map of KNAT gene expression in the Arabidopsis root. Plant Molecular Biology 60, 1–20. [DOI] [PubMed] [Google Scholar]
- van Brussel AAN, Tak T, Wetselaar A, Pees E, Wijffelman CA. 1982. Small leguminosae as test plants for nodulation of Rhizobium leguminosarum and other rhizobia and agrobacteria harbouring a leguminosarum sym-plasmid. Plant Science Letters 27, 317–325. [Google Scholar]
- Van den Eede G, Deblaere R, Goethals K, Van Montagu M, Holsters M. 1992. Broad host range and promoter selection vectors for bacteria that interact with plants. Molecular Plant–Microbe Interactions 5, 228–234. [DOI] [PubMed] [Google Scholar]
- van Zeijl A, den Camp RHO, Deinum EE, Charnikhova T, Franssen H, den Camp HJO, Bouwmeester H, Kohlen W, Bisseling T, Geurts R. 2015. Rhizobium lipo-chitooligosaccharide signaling triggers accumulation of cytokinins in Medicago truncatula roots. Molecular Plant 8, 1213–1226. [DOI] [PubMed] [Google Scholar]
- Vernié T, Moreau S, De Billy F, Plet J, Combier J-P, Rogers C, Oldroyd G, Frugier F, Niebel A, Gamas P. 2008. EFD is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula . The Plant Cell Online 20, 2696–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S. 1991. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350, 241–243. [DOI] [PubMed] [Google Scholar]
- Xiao TT, Schilderink S, Moling S, Deinum EE, Kondorosi E, Franssen H, Kulikova O, Niebel A, Bisseling T. 2014 Fate map of Medicago truncatula root nodules. Development 141, 3517–3528. [DOI] [PubMed] [Google Scholar]
- Yanai O, Shani E, Dolezal K, Tarkowski P, Sablowski R, Sandberg G, Samach A, Ori N. 2005. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Current Biology 15, 1566–1571. [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye Z-H. 2008. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis . The Plant Cell Online 20, 2763–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Han L, Li G, Chai M, Fu C, Cheng X, Wen J, Tang Y, Wang Z-Y. 2014. STM/BP-like KNOXI is uncoupled from ARP in the regulation of compound leaf development in Medicago truncatula . The Plant Cell Online 26, 1464–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.