Highlight
An anthocyanin transporter gene that contains a high frequency of non-transposable element-related mutations is responsible for variegated colouration of flowers in peach.
Key words: Anthocyanin, chimera, GST, indel, peach, variegation.
Abstract
The ornamental peach cultivar ‘Hongbaihuatao (HBH)’ can simultaneously bear pink, red, and variegated flowers on a single tree. Anthocyanin content in pink flowers is extremely low, being only 10% that of a red flower. Surprisingly, the expression of anthocyanin structural and potential regulatory genes in white flowers was not significantly lower than that in both pink and red flowers. However, proteomic analysis revealed a GST encoded by a gene—regulator involved in anthocyanin transport (Riant)—which is expressed in the red flower, but almost undetectable in the variegated flower. The Riant gene contains an insertion-deletion (indel) polymorphism in exon 3. In white flowers, the Riant gene is interrupted by a 2-bp insertion in the last exon, which causes a frameshift and a premature stop codon. In contrast, both pink and red flowers that arise from bud sports are heterozygous for the Riant locus, with one functional allele due to the 2-bp deletion or a novel 1-bp insertion. Southern blot analysis indicated that the Riant gene occurs in a single copy in the peach genome and it is not interrupted by a transposon. The function of the Riant gene was confirmed by its ectopic expression in the Arabidopsis tt19 mutant, where it complements the anthocyanin phenotype, but not the proanthocyanidin pigmentation in seed coat. Collectively,these results indicate that a small indel mutation in the Riant gene, which is not the result of a transposon insertion or excision, causes variegated colouration of peach flowers.
Introduction
Colouration is an important agronomic trait that contributes to the ornamental value of plants, which is determined by three major classes of plant pigments: anthocyanin, chlorophyll, and carotenoid. In flowers, anthocyanin is the major pigment and confers red, violet, or blue colours (Tanaka et al., 2008). The anthocyanin biosynthetic pathway has been well studied in a variety of plants (Winkel-Shirley, 2001; Grotewold, 2006). This pathway starts with a condensation of malonyl-CoA and 4-coumaroyl CoA, and is catalysed by multiple enzymes including chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), flavonoid 3′5′-hydroxylase (F3′5′H), dihydroflavonol 4-reductase (DFR), and UDPG-flavonoid glucosyltransferase (UFGT), to generate anthocyanins. Subsequently, water-soluble anthocyanin is transported into the vacuole.
Two mechanisms have been proposed for anthocyanin transport from the site of synthesis to the vacuole: vesicle-mediated transport and transporter-mediated transport (Grotewold and Davis, 2008; Zhao and Dixon, 2010). The evidence for the former mechanism comes from microscopy observations that anthocyanin accumulates initially in vesicle-like structures alongside the tonoplast that merge with the central vacuole (Zhang et al., 2006; Poustka et al., 2007; Conn et al., 2010). The other mechanism is supported by transporter proteins located in the tonoplast, including multidrug resistance-associated proteins (MRP) and multidrug and toxic compound extrusion (MATE) transporters, shown to mediate anthocyanin transport (Debeaujon et al., 2001; Goodman et al., 2004; Grotewold, 2004; Gomez et al., 2009; Zhao and Dixon, 2009; Francisco et al., 2013). In addition, GSTs also have an essential role in transport of anthocyanins from the ER to the vacuole (Larsen et al., 2003; Conn et al., 2008; Gomez et al., 2011). Initially, GST was proposed to conjugate anthocyanins with glutathione to form stable water-soluble conjugates, which were then transported into vacuoles by ABC transmembrane transporters (Marrs et al., 1995). However, no anthocyanin–glutathione conjugates have been found in vivo. Instead, evidence suggests that GST functions as an anthocyanin carrier that may escort anthocyanins from the ER to the tonoplast (Mueller et al., 2000; Sun et al., 2012).
Variegation (variable colouration with patches of different colours) is one of the quality parameters sought after by the plant ornamental industry. Variegation is a common phenomenon, with the mechanism of variegation being studied in numerous plant species. In maize, insertion and excision of transposable elements located in the promoter region of the bronze gene encoding GST causes unstable pigmentation in kernels (Schiefelbein et al., 1988). Transposable elements are also responsible for the unstable pigmentation phenotype in ornamental plants such as petunia (Quattrocchio et al., 1999; Spelt et al., 2000), snapdragon (Noda et al., 1994), carnation (Itoh et al., 2002; Nishizaki et al., 2011), and morning glory (Habu et al., 1998; Inagaki et al., 1994). Besides transposable elements, RNA interference (RNAi) and DNA methylation are involved in unstable pigmentation in plants. For example, silencing of the CHS gene results in flower colour variegation in petunia (Koseki et al., 2005). High methylation levels in the promoter region of MYB10 inhibit gene expression, which causes variable colour patterns in the peel of apple (Telias et al., 2011).
Peach [Prunus persica L. (Batsch)] is one of the most popular fruit trees in the world. Peach belongs to the Rosaceae family and serves as a model species of woody perennial angiosperms due to its small genome size of about 230Mb/haploid (The International Peach Genome Initiative, 2013). Peach trees are primarily grown for their fruit, but some cultivars are selected for their ornamental value. In China, the cultivation of ornamental peach has a long history. The ancient book Luoyanghuamuji written by Shihou Zhou in 1082 records ornamental peach cultivars such as Ersetao (variegated flower) and Ziyetao (red-leaved); and the variegated flower was highly praised in an ancient poem entitled ‘Er Se Tao’ written by Yong Shao in Song Dynasty, approximately 1000 years ago. Modern varieties have been bred for flower variegation such as Hongbaihuatao (HBH), which produces red, pink, and variegated flowers. Several studies have been conducted to investigate the genetic basis for peach flower colour. Chen et al. (2014) found that anthocyanin pathway genes such as CHS, CHI, and F3H show higher level of expression in red flowers than in white flower in ornamental peach (Prunus persica f. versicolor [Sieb.] Voss). The expression of a MYB-like gene (Peace) controls the pigmentation of flowers in the flowering peach ‘Genpei’. However, the mechanism underlying variegation in peach remains unclear.
In this study, cv. HBH was selected to investigate the molecular basis for peach flower variegation. A GST gene—regulator involved in anthocyanin transport (Riant)—was found to be associated with variegation. Levels of the Riant protein were high in red flower, but barely detectable in variegated flowers. This was due to small insertions and deletions (indels) in the last exon, resulting in a frameshift mutation. Anthocyanin accumulation in the flower of cv. HBH therefore appears to be regulated at the post-transcriptional level, unlike previous reports of transcriptional regulation in flowers of other peach varieties. This study demonstrates that the gene encoding GST is critical for anthocyanin accumulation in peach, and is helpful in understanding the mechanism underling the variegation in peach flowers.
Materials and methods
Plant material
Peach (P. persica) cultivars used in this study, including HBH, ‘Mantianhong’, ‘Hongcuizhi’, and ‘Sahongtao’, are maintained at Wuhan Botanical Garden of the Chinese Academy of Sciences (Hubei Province, China). Cv. HBH has variegation in flower colouration, while the other three cultivars bear only red flowers. Young leaves and flower buds with a diameter ranging from 0.5 to 0.8cm were collected in spring. Petals and sepalswere removed from flower samples and individually put into separate aluminium bags. All samples were immediately frozen in liquid nitrogen, and then stored at −80 °C until use.
HPLC analysis of anthocyanin in peach flower
Anthocyanin was extracted according to a previously reported protocol (Cheng et al., 2014). Approximately 0.5g of tissue was ground in liquid nitrogen and then added to 25ml of extraction solution (80:20 v/v methanol/water mixture containing 1.18mM HCl). The mixture was centrifuged at 10000 g for 10min. An aliquot of 10ml of supernatant was collected and evaporated under vacuum at 30 °C using a rotary evaporator. The residual was resuspended in acidified water (1.18mM HCl).
Anthocyanin was dissolved in 1ml methanol, filtered through a 0.22 μm Millipore membrane, and analysed using an HPLC-ESI-MS/MS system (ThermoFisher Scientific, Pittsburgh, PA, USA). The analytical column was a ZORBAX Extend C18, 4.6×250mm, with a particle size of 5 μm (Agilent Technologies, Waldbronn, Germany). The analytical column was sequentially eluted using mobile phase A (formic acid:water, 5:95, v/v) and mobile phase B (methanol) with a flow rate of 0.8ml/min. The linear gradient of phase B was as follows: 0–2min, 5%; 2–7min, 5–15%; 7–20min, 15–20%; 20–25min, 20–27%; 25–32min, 27%; 32–41min, 27–35%; 41.01–43min, 5%. UV-visible light detector wavelength was set at 520nm.
Analysis of gene expression using quantitative real-time PCR
Total RNA was extracted using ZP401 kit (Zoman, Beijing, China) according to the manufacturer’s instructions. Total RNA was treated with DNase I (Takara, Dalian, China) to remove any contamination of genomic DNA. Approximately 3 μg of total RNA was used for cDNA synthesis using PrimeScriptTM RT-PCR Kit (Takara, Dalian, China). An SYBR green-based real-time PCR assay was carried out in a total volume of 20 µl reaction mixture containing 10.0 µl of 2× SYBR Green I Master Mix (Takara, Dalian, China), 0.2 µM of each primer, and 100ng of template cDNA. A peach actin gene PpGAPDH (ppa008812m) was used as a constitutive control. Primer sequences of genes involved in anthocyanin biosynthesis and transport are listed in Supplementary Table S1, available at JXB online.
Amplification was conducted using StepOnePlus Real-Time PCR System (Applied Biosystems, Foster, CA, USA). The amplification programme consisted of an initial denaturing step at 95°C for 30 s, followed by 40 cycles of 95°C for 30 s, and 60°C for 34 s. The fluorescent product was detected at the second step of each cycle. Melt curve analysis was performed at the end of 40 cycles to ensure the proper amplification of target fragments. Fluorescence readings were consecutively collected during the melting process from 60 to 90 °C at the heating rate of 0.5°C/sec. All analyses were repeated three times using biological replicates.
Genomic DNA blot analysis
For cv. HBH, total DNA was separately extracted from red and variegated flowers at balloon stage, while total DNA of three cultivars used as controls (Mantianhong, Hongcuizhi, and Sahongtao), was extracted from young leaves. DNA extraction was performed using a cetyltrimethylammonium bromide (CTAB) method. Approximately 5 μg of genomic DNA was digested with HindIII, SpeI, and XbaI, separated on a 1.0% agarose gel, and transferred onto nylon membranes (Hybond-N, Amersham, UK) using the capillary transfer method. A pair of primers (5′-CTCAGTTCCTCTCCCGTCAG-3′/5′-CCAGCCAGATAGCTGCTCTT-3′) was designed to synthesize DNA probes using cDNA from leaves of cv. HBH as a template. The probe consisted of the last 17bp in exon 1, a complete exon 2, and a partial segment of exon 3 of Riant. Hybridization was carried out using the DIG Easy Hyb kit (Roche Applied Science, Indianapolis, IN, USA) according to the manufacturer’s instructions. Blots were exposed to a Lumi-Film X-ray film (Hyperfilm, Amersham) at room temperature for 25min.
Phylogenetic analysis
The amino acid sequences of genes encoding GST from different plants were used for phylogenetic analysis. Sequence alignment was performed using CLUSTAL X, and the resulting data matrix was analysed using equally weighted neighbour joining (NJ). An NJ tree was generated using MEGA (version 5.0). Bootstrap values were calculated from 1000 replicate analyses.
2D electrophoresis (2-DE)
The protein extraction protocol was based on the phenol method (Hurkman and Tanaka, 1986; Ahsan et al., 2008). Briefly, 2g of flower petal was ground in liquid nitrogen and added to 10ml extraction buffer containing 0.5M Tris-HCl (pH 8.3), 0.1M KCl,50mM EDTA, 2% (v/v) 2-mercaptoethanol, and 0.7M sucrose. An equal volume of Tris-HCl-saturated phenol (pH 8.0) was subsequently added and mixed by vigorous vortexing for 2min followed by centrifugation at 3500g for 15min. After centrifugation, the top phenol phase was collected and proteins were precipitated by adding four volumes of cold methanol containing 0.1M ammonium acetate at −20 °C for 2h. The precipitated proteins were recovered by centrifugation at 3500 g for 10min followed by three washes with cold methanol containing 0.1M ammonium acetate. The protein pellet was dried at room temperatureuntil needed for solubilization.
Protein concentration was quantified according to the Bradford method (Bradford, 1976). A total of 1.2mg of protein was loaded into the IPG strips (pH4–7, 24cm, Bio-Rad, USA) through rehydration for 12h at room temperature. Isoelectric focusing (IEF) electrophoresis was conducted using the following procedure: 200V for 1h (step and hold); 500V for 1.5h (step and hold); 1000V for 1h (gradient), 8000V for 2h (gradient), and 8000V for 6h (step and hold) for a total of 42000 volt-hoursusing a Protean IEF Cell (Bio-Rad). Then, the IPG strips were equilibrated for 15min in the equilibration buffer (50mM Tris-HCl pH 6.8, 6M urea, 10% v/v glycerol, 2.5% w/v SDS, and 5% 2-mercaptoethanol) containing 0.5% DTT, followed by 15min in the equilibration buffer containing 25mg/ml iodoacetamide. The second dimensional electrophoresis was run on 12% SDS-PAGE and conducted using the following procedure: 100V, 30min; 250V, 5h. The 2-DE gels were stained with Coomassie Brilliant Blue (CBB R-250).
Trypsin digestion and MALDI-TOF-MS analysis
The differentially expressed protein spots were excised from the gel manually and washed with double-distilled water twice. The gel slices were destained, dehydrated, and digested with trypsin. The digested protein peptides were analysed over a mass range of 800–4,000Da using an Autoflex speed™ MALDI-TOF-TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). Subsequently, the obtained PMF data were searched against the NCBI nr database and Swiss-Port database using MASCOT software (Mascot Wizard 1.2.0, Matrix Science Ltd., www.matrixscience.com). The parameters were set as follows: carbamidomethylation of cysteine and oxidation of methionine; peptide charge state of +1 and peptide mass tolerance of 0.5Da; a maximum of one for missed cleavages and monoisotopic.
PAGE
PCR products were mixed with an equal volume of formamide loading buffer (98% formamide, 10mM EDTA pH 8.0, 0.025% Bromophenol Blue and Xylene Cyanol). The mixture was denatured at 94°C for 3min, and then immediately chilled on ice. An aliquot of 2 μl mixture was loaded on a 6% polyacrylamide gel, and electrophoresed for 1.5h at 1200V. Bands were visualized after silver staining, and recorded on a ScanMaker 3830 (Microtek, Shanghai, China).
Expression vector construction and plant transformation
A pair of primers, 5′-GAATTCATGGTTGTGAAAGTGTAT GGTCC-3′/5′-CTCGAGTGGGGGTATCTCATATCTAGTAGTC-3′, was designed to amplify the whole coding region of the Riant gene using cDNA synthesized from flowers of cv. HBH as templates. The forward and reverse primers contained EcoRI and XhoI sites at the 5′ end, respectively. The PCR product was digested with EcoRI and XhoI, and inserted into EcoRI/XhoI-digested pSAK277 (Hellens et al., 2005). The Arabidopsis tt19 mutant (CS60000) with the Columbia genetic background was obtained from the Arabidopsis Biological Resource Center (Ohio State University, OH, USA). Arabidopsis transformation was performed according to the floral dip method (Clough and Bent, 1998). For transgenic plant selection, T0 seeds were sterilized and germinated on Murashige and Skoog (MS) medium containing 12 μg ml−1 kanamycin and 3% (w/v) Suc. Following 1 week of selection, kanamycin-resistant plants with red hypocotyls were transplanted to soil and placed in a growth chamber at 25°C and 50–80% relative humidity.
Results
Colouration and anthocyanin composition in red, pink, and variegated petals of cv. HBH
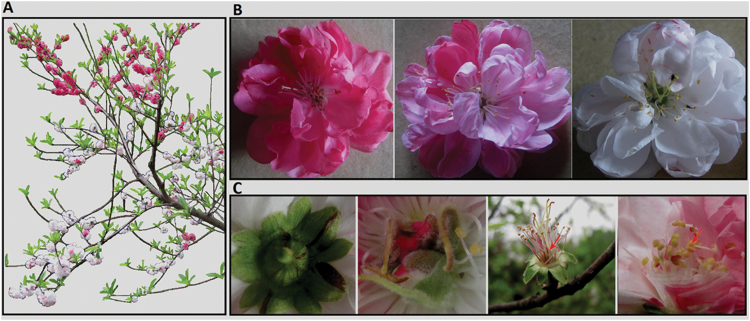
Flowering peach cv. HBH produces red, pink, and variegated flowers on a single tree (Fig. 1A). The variegated flowers show a great variation in petal colouration (Fig. 1B), and can be classified into four types: white and red/pink spotted, white and pink somatic sectors, white and red somatic sectors, and pink and red somatic sectors. Most variegated flowers belong to type 1, while the other three types of variegated flowers are occasionally produced. Besides the petal tissue, variegation also appears in the colouration of sepal, stamen, and pistil tissues (Fig. 1C). Based on the flower colouration, the branches of cv. HBH can be grouped into types: red-, pink-, and white-flower branches. Red-flower branches produce exclusively red flowers with no variegated flowers. Pink-flower branches bear predominantly pink flowers, and occasionally produce pink flowers with red somatic sectors. White-flower branches bear predominantly white flowers with red/pink spotted, and occasionally produce white flowers with pink or red somatic sectors, pink flowers, and red flowers. Since the white-flower branch is the principal branch in cv. HBH, the pink- and red-flower branches are deemed to arise from bud sports.
Fig. 1.
Flower colouration of ornamental cv.HBH.(A) Peach tree cv. HBH bears pigmented and variegated flowers at bloom stage. (B) Three kinds of coloured flowers within a single tree.(C) The colouration in sepal, pistil, and stamen. The arrow indicates the stamen. (A colour version of this figure is available at JXB online.)
HPLC analysis was conducted to investigate differences in anthocyanin content among red petal, pink petal, and white petal with red/pink spotted (termed variegated petalhereinafter). The red petal consists of six peaks (Supplementary Fig. S1, available at JXB online). Based the authors’ previous study (Cheng et al. 2014), peaks 1–6 correspond to cyanidin 3-galactoside, cyanidin 3-glucoside, cyanidin 3-rutinoside, peonidin 3-glucoside, cyanidin 3-rhamnoside, and peonidin 3-rutinoside, respectively. The main component in peach flower is cyanidin 3-glucoside, and its content varies greatly among red, pink, and variegated petals. The content of cyanidin 3-glucoside in red and pink petals is 12.1 and 1.26 μg/100g fresh weight (FW), respectively. However, cyanidin 3-glucoside is almost undetectable in variegated petals. In addition, the flavonol content was also determined. Overall, there is no striking difference in flavonol content among red, pink, and variegated petals. The amount of flavonol was slightly higher in variegated petals (4.8 μg/100g FW) than in red petals (4.1 μg/100g FW) and pink petals (4.6 μg/100g FW). Taken together, these results show that anthocyanin is responsible for the red and pink pigmentation in petals.
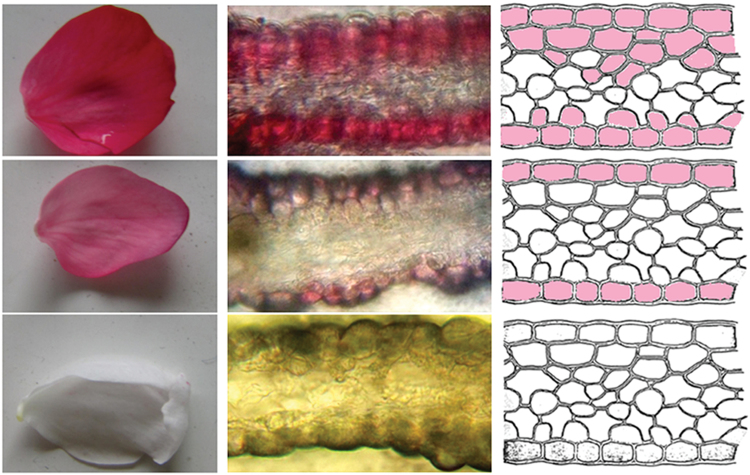
Microscopic analysis of fresh hand-cut sections of flower petals showed that red petals had several layers of coloured cells, with anthocyanin accumulation in both epidermal and sub-epidermal layers (Fig. 2). Pink petals only accumulated anthocyanins in the upper and lower epidermal layers. However, white petals accumulated no anthocyanins in either epidermal or sub-epidermal layers.
Fig. 2.
Examination of anthocyanin accumulation in petal cell layers. The middle column represents cross-sections of red, pink, and white petals (photo taken with microscope), while the right column is a diagrammatic representation of the cross-sections of red, pink, and white petals. (A colour version of this figure is available at JXB online.)
Expression profiling of genes involved in anthocyanin biosynthesis and transport
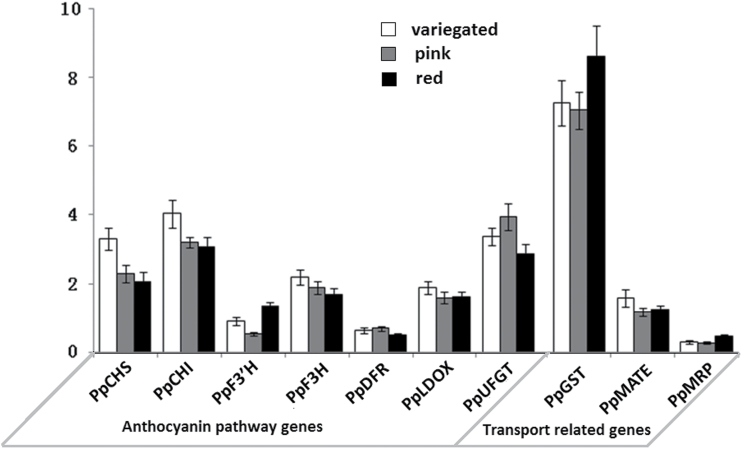
Red, pink, and variegated petals of flowers at balloon stage were collected to investigate the expression profile of anthocyanin biosynthesis genes using real-time PCR, including PpCHS, PpCHI, PpF3′H, PpF3H, PpDFR, PpLDOX, and PpUFGT. No striking difference in expression level was observed for any of these anthocyanin biosynthesis genes when comparing red, pink, and variegated petals (Fig. 3). Surprisingly, two genes—PpCHS and PpCHI—showed a significantly higher level of expression in variegated petals than in red and pink petals (P<0.05). Similarly, all the anthocyanin regulatory genes of MYBs also showed no significant difference in expression levels among pink, red, and white petals (Supplementary Fig. S2, available at JXB online).
Fig. 3.
Expression level of genes involved in biosynthesis and transport of anthocyanin in petals of cv. HBH.
Subsequently, the expression level of three genes involved in anthocyanin transport, e.g. PpMRP, PpMATE, and PpGST, was also measured. All these anthocyanin transport-related genes, like the anthocyanin biosynthesis genes, showed no significant difference in expression level among red, pink, and variegated petals (Fig. 3). These results suggested that anthocyanin accumulation is not regulated at the transcriptional level in the flower of cv. HBH.
Proteomic analysis reveals a candidate GST correlated with flower colour variegation in cv. HBH
To determinate whether anthocyanin accumulation in the flower of cv. HBH is controlled at the post-transcriptional level, 2-DE protein analysis was performed. Petals from red and variegated flowers at balloon stage were chosen for 2-DE analysis. As a result, a total of 84 protein spots displaying differential abundance (>1.6-fold change) were identified (Supplementary Fig. S3). Of these proteins, 40 and 44 were up- and down-regulated in red petals, respectively.
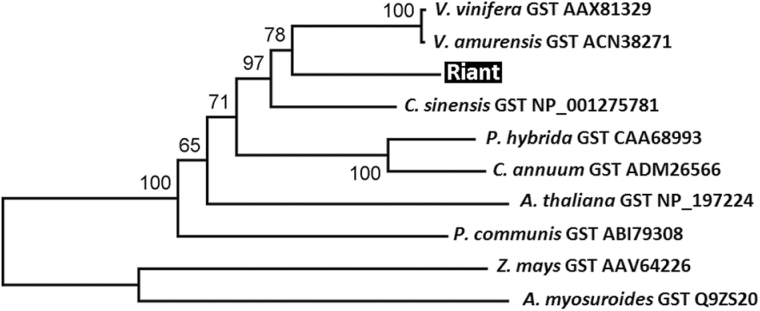
These differentially abundant proteins were digested with trypsin and analysed using MALDI TOF/TOF MS/MS. Of the 84 proteins surveyed, 53 were successfully identified. Among these identified proteins (Table 1), two, PpCHS-like and PpGST with spot numbers B41 and B54, respectively, are potentially related to anthocyanin pigmentation. The PpCHS-like protein was highly expressed in red petals but undetectable in variegated petals. Coding sequences of the PpCHS-like gene were cloned from both red and variegated petals, but no difference was identified. Phylogenetic analysis indicated that the PpCHS-like gene is closely related to genes encoding a biphenyl synthase in Malus (Supplementary Fig. S4, available at JXB online). These results suggest that the PpCHS-like gene is unlikely involved in regulation of anthocyanin pigmentation. In contrast, the PpGST protein was found in both red and variegated petals, but its level was much higher in red petals than in variegated petals (Supplementary Fig. S4). Phylogenetic analysis showed that the PpGST gene is closely related to VvGST (Fig. 4). Since the VvGST gene is known for anthocyanin transport in grapevine (Gomez et al., 2011), the PpGST gene, Riant, is a strong candidate responsible for variegation of flower colouration.
Table 1.
Proteins differentially expressed in red and white flower from P. persica cv. HBH*
| No. | Spot no. | Protein name | MW(Da) | pI | PSC(%) |
|---|---|---|---|---|---|
| 1 | A27 | Geranylgeranyl pyrophosphate synthase family protein (Populus trichocarpa) | 37305 | 5.06 | 23% |
| 2 | A42 | Coatomer subunit epsilon-2-like (Fragaria vesca subsp. vesca) | 32395 | 5.16 | 9% |
| 3 | A49 | Triose phosphate isomerase cytosolic isoform-like protein (Capsicum annuum) | 27433 | 6.00 | 37% |
| 4 | A68 | PREDICTED: peroxiredoxin-2E, chloroplastic-like (F. vesca) | 24230 | 8.96 | 20% |
| 5 | A48 | S-locus lectin protein kinase family protein (Theobroma cacao) | 87288 | 6.64 | 1% |
| 6 | A22 | Calcium-binding EF hand family protein (T. cacao) | 31298 | 4.8 | 18% |
| 7 | A62 | Adenine nucleotide hydrolases-like superfamily protein (T. cacao) | 17992 | 6.2 | 48% |
| 8 | A64 | Hypothetical protein | 17685 | 4.77 | 14% |
| 9 | A33 | PREDICTED: 14-3-3-like protein-like (F. vesca subsp. vesca) | 29687 | 4.77 | 29% |
| 10 | A55 | Chalcone-flavanone isomerase family protein (T. cacao) | 32276 | 7.77 | 35% |
| 11 | A26 | Fructose-bisphosphate aldolase 4 (Camellia oleifera) | 42688 | 8.15 | 16% |
| 12 | A38 | EF hand family protein, expressed isoform 1 (T. cacao) | 30276 | 6.44 | 13% |
| 13 | A28 | Temperature-induced lipocalin (P. persica) | 21450 | 5.60 | 38% |
| 14 | A44 | ATP-dependent Clp protease proteolytic subunit 4 (T. cacao) | 32028 | 5.97 | 19% |
| 15 | A13 | Actin 7 (Arabidopsis thaliana) | 41954 | 5.31 | 50% |
| 16 | A58 | Cyclophilin peptidyl-prolyl cis-trans isomerase family (T. cacao) | 15270 | 5.61 | 45% |
| 17 | A23 | Annexin-like protein RJ4 (P. trichocarpa) | 35923 | 6.19 | 49% |
| 18 | A24 | RNA-binding protein Nova-1-like (Vitis vinifera) | 30618 | 6.01 | 38% |
| 19 | B15 | Full=UDP-sugar pyrophosphorylase | 67671 | 5.71 | 21% |
| 20 | B29 | 26S proteasome non-ATPase regulatory subunit 4-like (F. vesca) | 43042 | 4.48 | 19% |
| 21 | B35 | Monodehydroascorbate reductase (Malus domestica) | 47111 | 6.51 | 26% |
| 22 | B40 | Isovaleryl-CoA dehydrogenase 1, mitochondrial-like (F. vesca) | 43644 | 6.20 | 23% |
| 23 | B41 | Chalcone synthase 1-like (F. vesca subsp. vesca) | 43390 | 5.97 | 32% |
| 24 | B54 | GST-like protein (M. domestica) | 24389 | 5.34 | 37% |
| 25 | B28 | TCP domain class transcription factor (M. domestica) | 57477 | 5.72 | 40% |
| 26 | B23 | RNA-binding KH domain-containing protein isoform 1 (T. cacao) | 59072 | 6.12 | 35% |
| 27 | B09 | Lipoxygenase (M. domestica) | 90278 | 5.40 | 20% |
| 28 | B21 | Starch synthase isoform I (Manihot esculenta) | 71556 | 5.38 | 22% |
| 29 | B48 | Papain family cysteine protease (T. cacao) | 40868 | 5.86 | 17% |
| 30 | B17 | Mediator of RNA polymerase II transcription subunit 37e-like (F. vesca) | 69534 | 5.25 | 20% |
| 31 | B03 | Patellin-3-like (F. vesca subsp. vesca) | 65404 | 4.85 | 6% |
| 32 | B01 | Heat shock protein 70 (Hsp 70) family protein isoform 1 (T. cacao) | 100326 | 5.40 | 20% |
| 33 | B27 | TCP domain class transcription factor (M. domestica) | 57477 | 5.72 | 44% |
| 34 | B05 | Patellin-3-like (F. vesca subsp. vesca) | 65404 | 4.85 | 10% |
| 35 | B16 | NADP-malic protein (Prunus armeniaca) | 65358 | 5.73 | 24% |
| 36 | B33 | 3-Ketoacyl-acyl carrier protein synthase I (T. cacao) | 52446 | 6.38 | 23% |
| 37 | B19 | DC1 domain-containing protein (T. cacao) | 65260 | 4.80 | 31% |
| 38 | B06 | Cytosolic aconitase (Pyrus pyrifolia) | 108637 | 6.98 | 10% |
| 39 | B58 | PREDICTED: allene oxide cyclase 4, chloroplastic-like (F. vesca) | 20527 | 5.41 | 19% |
| 40 | B55 | Chaperonin 20 isoform 1 (T. cacao) | 26399 | 7.79 | 11% |
| 41 | B44 | Caffeic acid 3-O-methyltransferase 1 (T. cacao) | 42020 | 5.40 | 21% |
| 42 | B25 | TCP domain class transcription factor (M. domestica) | 45351 | 5.26 | 26% |
| 43 | B18 | UDP-sugar pyrophosphorylase (T. cacao) | 67671 | 5.71 | 14% |
| 44 | B31 | Hypothetical protein | 39546 | 6.77 | 7% |
| 45 | B20 | Nucleoporin nup211-like (Glycine max) | 54774 | 5.99 | 16% |
| 46 | B24 | Glucose-6-phosphate 1-dehydrogenase | 59358 | 6.13 | 24% |
| 47 | B50 | Prolyl 4-hydroxylase alpha subunit, putative (Ricinus communis) | 33559 | 6.26 | 36% |
| 48 | B13 | Oligopeptidase A-like (F. vesca subsp. vesca) | 90582 | 6.42 | 12% |
| 49 | B49 | Probable rhamnose biosynthetic enzyme 1-like (Citrus sinensis) | 33636 | 6.17 | 21% |
| 50 | B57 | Peroxiredoxin-2B-like (F. vesca subsp. vesca) | 17480 | 5.70 | 57% |
| 51 | B60 | Regulator of ribonuclease-like protein 2-like (F. vesca subsp. vesca) | 18008 | 5.69 | 64% |
| 52 | B51 | Protein PPLZ12, putative (R. communis) | 31730 | 5.27 | 41% |
| 53 | B65 | Ribonuclease UK114-like (F. vesca subsp. vesca) | 19977 | 8.99 | 44% |
* The protein associated with the variegated colouration of the peach flower is highlighted in bold.
Fig. 4.
A phylogenetic tree derived from amino acid sequences of GST genes from plants. GenBank accession numbers are listed after the gene name. The Riant gene isolated in this study is highlighted. The numbers indicate bootstrap values calculated from 1000 replicate analyses.
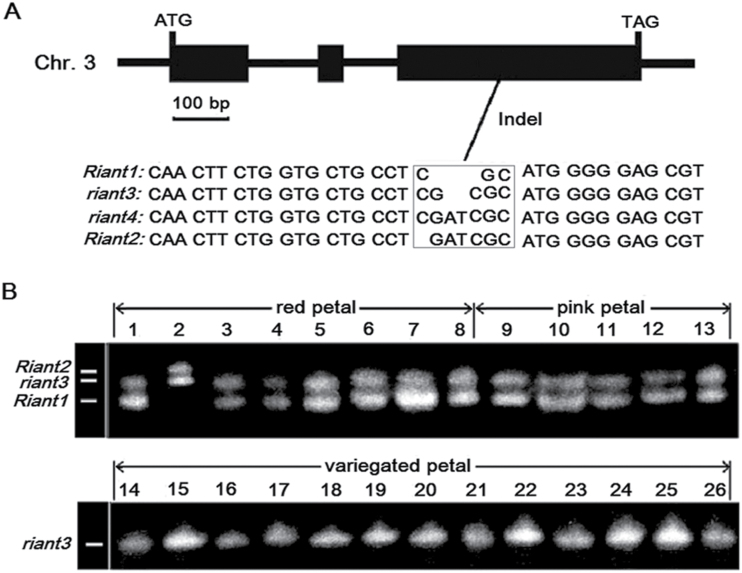
Small indels in the Riant gene and their association with variegation in petal colouration
To confirm that the Riant gene is responsible for petal variegation, its coding sequences from red, pink, and variegated petals were cloned and sequenced. Comparison of the coding sequences revealed a 2-bp insertion in the third exon of Riant (Fig. 5A). The 2-bp insertion causes a frameshift and a premature stop codon. Subsequently, a pair of primers flanking the 2-bp insertion, 5′-CTCTGGTGGATCAGTGGCT-3′ (GIF) and 5′-TATCCCTGGAAGATGGCTC-3′ (GIR), was designed to amplify red, pink, and variegated petals from different clones of cv. HBH (Fig. 5B). Interestingly, all the red or pink petals contained two bands, suggesting they are heterozygous at the Riant locus. Three alleles, designated Riant1, Riant2, and riant3, were detected among the red and pink petals. Of the 13 pigmented petals tested, 12 have a Riant1/riant3 genotype. One (the second sample in Fig. 5B) has a Riant2/riant3 genotype. Sequencing of the PCR products revealed that both Riant1 and Riant2 have an intact ORF, while the riant3 allele has a frameshift mutation due to a 2-bp insertion in the third exon (Fig. 5A). It is worth noting that Riant2 contains a 3-bp insertion in the third exon, so should not induce a frame shift. All the variegated petals amplified only one band that corresponds to the riant3 allele, suggesting they are homozygous at the Riant locus.
Fig. 5.
The Riant gene isolated from peach cv. HBH. (A) Genomic structure and genetic variation highlighted in a square box. (B) Genotyping of red, pink, and variegated flowers based on the indel in the last exon of the Riant gene; the detected alleles are indicated.
To determine whether there are other alleles of the Riant gene, 15 more flowers (five with red petals, five with pink petals, and five with variegated petals) were randomly collected from different clones of cv. HBH. The petals of these flowers were individually subjected to genomic DNA extraction, and the extracted DNA was subsequently amplified using the primers GIF/GIR as mentioned above. Cloning and sequencing of the PCR products revealed one more frameshift mutant allele from the variegated flower, designated riant4, which contains a 4-bp insertion (Fig. 5A). All four alleles—Riant1 to Riant4—were deposited in GenBank under accession nos. KT312847 to KT312850, respectively.
Copy number of the Riant gene in the peach genome
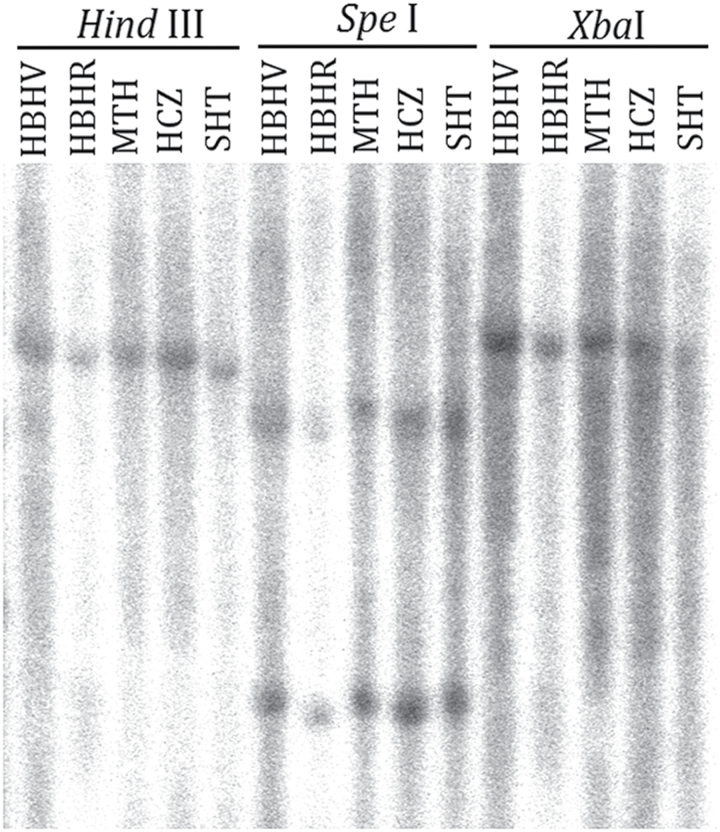
In many species, variegation in flower colouration is caused by transposon activity. To determine whether the small indels in the third exon of the Riant gene have also arisen from a transposon or other DNA fragment that was not amplified by the primers GIF and GIR, aDNA gel blot analysis was performed. Three cultivars (Mantianhong, Hongcuizhi, and Sahongtao) that bear red flowers were used as controls. Genomic DNA was digested with SpeI, HindIII, and XbaI. The digested DNA was hybridized with a probe covering the indel site in the third exon of the Riant gene. Both HindIII and XbaI digestions yielded a single hybridizing DNA band in all the tested samples, while there were two hybridizing bands for SpeI digestion in all the tested samples (Fig. 6). This result indicates that only one copy of the Riant gene is present in the peach genome, and it is not interrupted by a transposon. In other words, the small indel mutation in the Riant gene is not due to insertion and excision of a transposable element.
Fig. 6.
Southern blot analysis of peach genomic DNA. The Riant-specific probe consists of a partial sequence of the first exon, whole fragment of the second exon, and a partial sequence of the third exon that covers the indel site. HBHV and HBHR represent variegated and red flowers from cv. HBH, respectively. MTH, Mantianhong; HCZ, Hongcuizhi; SHT, Sahongtao.
Functional analysis of the Riant gene in the Arabidopsis transparent testa19 mutant
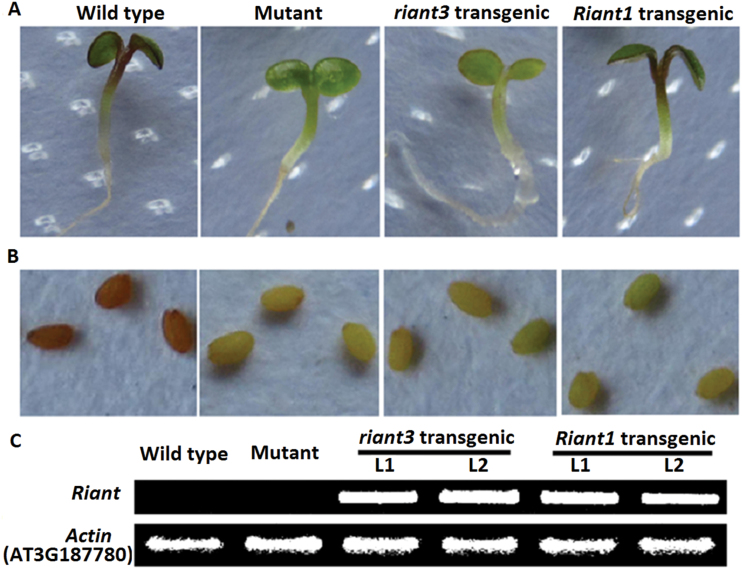
The Arabidopsis transparent testa19 (tt19) mutant, lacking GST, was selected to investigate the functionality of the Riant gene. The coding sequences of both Riant1 and riant3 alleles were separately transferred into the Arabidopsis tt19 mutant under the control of the cauliflower mosaic virus 35S promoter, and several transgenic lines were generated for each construct. Seeds of the Arabidopsis tt19 mutant, T1 transgenic lines, and wild-type Arabidopsis were germinated and grown on MS medium. Germinating seedlings of wild-type plants and transgenic lines expressing Riant1 had red hypocotyls, whereas hypocotyls of the Arabidopsis tt19 mutant and transgenic lines expressing riant3 were green (Fig. 7A). Moreover, seeds collected from kanamycin-resistant T1 plants and the Arabidopsis tt19 mutant were pale brown in colour, while seeds of wild-type plants were dark brown in colour (Fig. 7B). This suggests that Riant, like petunia AN9, complements the anthocyanin accumulation in vegetative tissues, but not the brown pigmentation in the seed coat (Kitamura et al., 2004). In addition, reverse transcription (RT)-PCR analysis showed that both Riant1 and riant3 were highly expressed in transgenic lines (Fig. 7C). Taken together, these results demonstrated that, the Riant1 allele is involved in the transport of anthocyanins from cytosol to vacuole, but the riant3 allele is nonfunctional.
Fig. 7.
Complementation of the pigmentation of Arabidopsis tt19 mutant seedlings of the ecotype Columbia with the Riant gene.(A) Phenotypes of wild-type, mutant, and transgenic Arabidopsis seedlings. (B) Phenotypes of wild-type, mutant, and transgenic Arabidopsis seeds. (C)Expression level of the Riant gene in wild-type, mutant, and transgenic Arabidopsis seedlings. Two transgenic lines each of Riant1 and riant3 were analysed, and these exhibited similar phenotypes, as shown. (A colour version of this figure is available at JXB online.)
Discussion
The Riant gene is involved in anthocyanin transport and is critical for flower colouration in peach cv. HBH
In plants, flower pigmentation is mainly attributed to anthocyanin accumulation (Bogs et al., 2005; Cutanda-Perez et al., 2009). In this study, HPLC analysis revealed that cyanidin 3-glucoside is the main component of anthocyanin in flowers of cv. HBH, which is consistent with previous reports (Chaparro et al., 1995a ; Cheng et al., 2014; Uematsu et al., 2014). Both red and pink petals accumulate cyanidin 3-glucoside, and its content is approximately 10-fold higher in red petals than in pink petals. In contrast, cyanidin 3-glucoside is almost undetectable in white sectors of the variegated petal. This suggests that anthocyanin accumulation contributes to flower colouration in cv. HBH and its flower colour variegation is related to a change in anthocyanin accumulation.
In contrast to anthocyanin, flavonol shows the highest level of accumulation in the variegated petal, followed by pink and red petals. This is consistent with previous findings that blocking anthocyanin accumulation strengthens the metabolic flux towards flavonols (Gou et al., 2011; Sun et al., 2012). Real-time PCR analysis reveals that early biosynthetic genes such as CHS, CHI, and F3H show higher levels of expression in variegated petals than in red and pink petals, whereas, the expression levels of late biosynthetic genes such as DFR, LDOX, and UFGT are not significantly different among red, pink, and variegated petals. Thus, it seems that anthocyanin and flavonol biosynthesis is coordinately regulated by anthocyanin pathway genes (Owens et al., 2008; Han et al., 2010). However, all the potential MYB regulatory genes tested in this study showed no significant difference in expression level among red, pink, and white petals of cv. HBH.
The vacuole is the cellular compartment where anthocyanins accumulate. Increasing evidence shows that GST is indispensable for the transport of anthocyanins from the ER to the vacuole (Marrs et al., 1995; Alfenito et al., 1998; Zhao and Dixon, 2010; Gomez et al., 2011; Gou et al., 2011). This study also demonstrates that the Riant gene encoding GST is essential for anthocyanin pigmentation in peach cv. HBH. In variegated petals, two alleles of the Riant gene were identified, but both of them encode truncated nonfunctional proteins due to 2- or 4-bp insertions in the third exon that result in frameshift mutations. Interestingly, the mutation of the Riant gene does not alter the expression level of genes involved in anthocyanin biosynthesis. Similar results have been reported in the Arabidopsis tt19 mutant (Sun et al., 2012). In Arabidopsis, anthocyanin accumulates first in vesicles and then is transported to the vacuole via fusion with the tonoplast (Poustka et al., 2007). Knockout of the GST gene results in weak accumulation of anthocyanins in vesicles, but not in the vacuole (Goodman et al., 2004; Gomez et al., 2011; Li et al., 2011; Sun et al., 2012). In peach,no anthocyanin was foundin white flowers, but some was present in variegated petals. Anthocyanins may be temporarily accumulated in vesicles, but subsequently degraded in variegated flowers of cv. HBH. In addition, the PpMATE gene shows a higher level of expression in variegated petals than in red and pink petals. This suggests a relationship between the PpMATE and Riant genes, to coordinately transport anthocyanins from the ER to the vacuole. MATE is involved in anthocyanin transport via the transporter-mediated mechanism, suggesting that both vesicle-mediated trafficking and MATE transporter-mediated mechanisms are involved in the sequestration of anthocyanins to vacuoles in peach.
Genetic mapping reveals that two loci, B and Fc, are responsible for flower colour in peach. The B locus has been mapped to an interval flanked by two markers, Pr1-12 and BPPCT028, on the bottom of chromosome 1 (Martínez-Gómez et al., 2007), while the Fc locus is anchored to an interval flanked by two markers, OPJ01 and MA039a, on the upper region of chromosome 3 (Yamamoto et al., 2005). The Peace gene that regulates petal pigmentation in peach ‘Genpei’ is located at the bottom of chromosome 1 (Uematsu et al., 2014). The peach reference genome (The International Peach Genome Initiative, 2013) has been searched, and the Riant gene was found to be located at the top of chromosome 3. Thus, it is worthy of further study to clarify whether the Peace and Riant genes are actually candidates of the B and Fc loci, respectively.
In Arabidopsis, TT19 participates in both anthocyanin accumulation in vegetative tissues and proanthocyanidin (PA) accumulation in seed coats (Kitamura et al., 2004). The accumulation of PA pigments is responsible for brown colouration in Arabidopsis seed coats. Petunia anthocyanin 9 (AN9) is an orthologue of Arabidopsis TT19. However, its ectopic expression in the Arabidopsis tt19 mutant complements the anthocyanin accumulation in vegetative tissues, but not the brown pigmentation in the seed coat (Kitamura et al., 2004). Like AN9, Riant also complements the anthocyanin phenotype, but not the PA defect of the tt19 mutant.
Potential mechanism underlying the mutation of the Riant gene in flowering peach cv. HBH
Transposable elements are often responsible for the phenotype of colour variegation in plants (Habu et al., 1998; van Houwelingen et al., 1998; Pooma et al., 2002; Schwinn et al., 2006; Nishizaki et al., 2011; Lazarow et al., 2012). In this study, variegation in peach was shown to be associated with small indels in the last exon of the Riant gene. Both red and pink petals are heterozygous at the Riant locus, with a functional allele and a frameshift mutant allele, whereas variegated petals contain two nonfunctional alleles (homozygous, Fig. 5). Moreover, DNA blot analysis shows that there is no polymorphism between red and variegated petals. These results strongly suggest that there are no transposable elements in the Riant locus in cv. HBH.
Flowering peach cv. HBH is quite similar in variegation to the previously reported peach cultivar Pillar, which bears dark pink, light pink, and white flowers on the same tree (Chaparro et al., 1995a ). The phenotype in cv. Pillar is assumed to be controlled by an active transposable element in the W locus (Chaparro et al., 1995b ). If the dark pink flowers carry a functional W allele that is reverted by excision of the transposable element, its self-pollinated progeny are expected to segregate for flower colouration. However, self-pollinated seeds of the dark pink flowers on Pillar trees yield only anthocyanin-deficient progeny (Chaparro et al., 1995b ). This suggests that the unstable phenotype in cv. Pillar cannot be ascribed to an active transposable element. Similarly, no transposable element is identified in the Peace gene responsible for the variegated phenotype in flowering peach ‘Genpei’ that produces pink and variegated flowers on the same tree (Uematsu et al., 2014). Thus, it appears that flower colour variegation is not due to transposable elements. DNA methylation and RNAi can also cause colour variegation in plants (Koseki et al., 2005; Telias et al., 2011). These two mechanisms lead to down-regulation of the anthocyanin pathway. However, expression levels of genes were similar among red, pink, and variegated flowers. This suggests colour variegation in cv. HBH is controlled at the translational level, and is unlikely to be related to DNA methylation or RNAi.
A total of four alleles were identified at the Riant locus. Of these alleles, Riant1 has the same coding sequence as the GST gene (ppa011307m) retrieved from the peach reference genome of cv. Lovell (The International Peach Genome Initiative 2013). All the tested flowers contain the riant3 allele. Thus, the riant3 allele probably represents an original allele in cv. HBH, whereas, Riant1, Riant2, and riant4 are mutants of the riant3 allele. These small indels appear responsible for the DNA variation at the Riant locus.
Strand slippage during DNA replication is a well-understood mechanism of the small indel mutagenesis (Garcia-Diaz and Kunkel 2006; Montgomery et al. 2013), and the G–C pairing surrounding the indels contributes to the rate of small indels (Tanay and Siggia, 2008). The Riant1 allele has a 2-bp (GC) deletion compared with the riant3 allele. Interestingly, the indel polymorphic locus of the riant3 allele contains a (GC)2 sequence, which has the potential to induce strand slippage. Thus, strand slippage may be responsible for the DNA variation at the Riant locus. Besides strand slippage, DNA single- or double-stranded breaks are also required for the generation of small indels as specialized translesion synthesis polymerases are capable of bypassing DNA lesions without repairing them, which allows replication on damaged DNA substrates and, in some cases, to promote mutagenic DNA synthesis (Waters et al., 2009; De and Babu, 2010; Roerink et al., 2012). For example, Sulfolobus solfataricus DNA polymerase IV (Dpo4), a member of the Y family of DNA polymerases, can generate deletions and mismatches at an unusually high average rate and preferentially at cytosine flanked by 5′-template guanine (Goodman, 2002). In this study, small indels also occur at the site with GC nucleotide sequences. Therefore, it cannot be excluded that the DNA variation of the Riant gene may be caused by similar mechanisms.
Relationship between somatic chimerism and flower colour variegation in cv. HBH
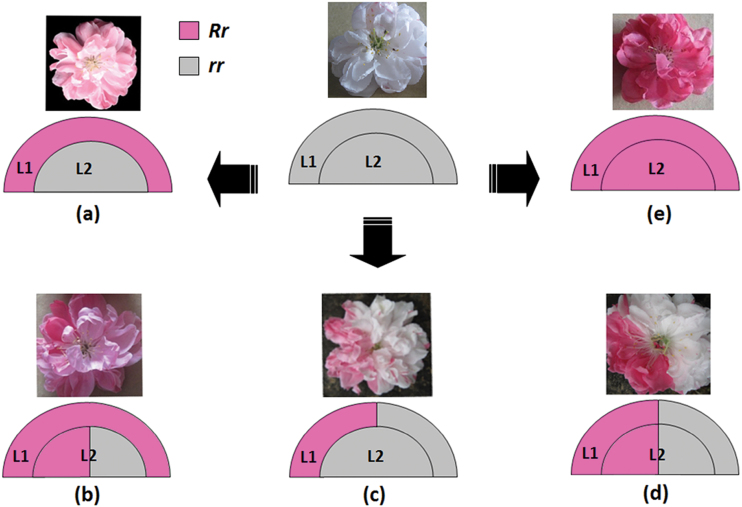
Chimerism is one of the factors that causes variegated colouration in plants (Stewart and Dermen, 1979; Mandal et al., 2000; Walker et al., 2006; Pelsy, 2010). Dicotyledonous plants usually have stratified apical meristems containing three layers of dividing cells, L1, L2, and L3, and each layer contributes to different tissues of the developing organs (Carles and Fletcher, 2003). The word chimera indicates that one or more layers consist of genetically distinct cells, and chimerascan be classified as periclinal, mericlinal, or sectorial chimeras. For floral meristems, layers L1 and L2 contribute to epidermis and internal tissues, respectively, while layer L3 forms the innermost tissues such as vascular tissues (Brand et al., 2001; Filippis et al., 2013). Thus, layers L1 and L2 play an important role in determining flower colouration (Stewart and Dermen, 1979; Carles and Fletcher, 2003).
Anthocyanin content in pink-coloured petals is extremely low—only approximately 10% of that in red-coloured petals. A similar result is also observed in grapevine. ‘Cabernet Sauvignon’ and its bud sport ‘Malian’ bears dark red and pink berries, respectively, with Malian berry containing only 10% of the anthocyanin content of the Cabernet Sauvignon berry (Boss et al., 1996). In Cabernet Sauvignon, both the L1 and L2 layers carry one red and one white allele, giving rise to a coloured epidermis derived from the L1 layer and several sub-epidermal coloured layers in the skin derived from L2 (Walker et al., 2006). In contrast, Malian is a periclinal chimera with the L1 layer carrying one red and one white allele of the berry colour locus while two white alleles for the L2 layer, resulting in only the epidermis containing anthocyanin. The pink flower of peach cv. HBH is a periclinal chimera with the L1 layer capable of accumulating anthocyanin while the L2 layer cannot. Since both pink and red flowers belong to bud sports and are heterozygous for the Riant gene responsible for anthocyanin accumulation, the L1 layer in the pink flower should be heterozygous with one functional allele such as Riant1 or Riant2, whereas, both the L1 and L2 layers in the red flower are heterozygous. The variegated flowers, including pink and red somatic sectors (type 1), pink and white somatic sectors (type 2), and red and white somatic sectors (type 3), can be attributed to the existence of genetically distinct cells within the same layer. Thus, a model is proposedfor the variegated phenotype in flower colouration of peach cv. HBH (Fig. 8).
Fig. 8.
A proposed model for the variegated phenotype in flower colouration of peach cv. HBH. L1 and L2 indicate different layers of floral meristems, and R and r represent functional and nonfunctional alleles of the Riant gene, respectively. White flower with red/pink spots carrying two nonfunctional alleles of the Riant gene. (a) Pink flower derived from periclinal chimera, (b) pink flower with red somatic sectors derived from mericlinal chimera, (c) white flower with pink somatic sectors derived from mericlinal chimera, (d) white flower with red somatic sectors derived from sectorial chimera, (e) red flower carrying one functional and one nonfunctional allele of the Riant gene. (A colour version of this figure is available at JXB online.)
The type 1 variegated flower results from a sectorial chimera, whereas, both the type 2 and type 3 variegated flowers have arisen from a mericlinal chimera. These three types of variegated flowers occur at low frequency, with no branches that produce predominantly one of the three types of variegated flowers. This is consistent with a previous finding that sectorial and mericlinal chimeras are unstable (Pelsy, 2010; Filippis et al., 2013). In contrast, periclinal chimeras are very stable giving rise to the pink-flower branch that bears predominantly pink flowers.
No pure white flowers are found on the trees of cv. HBH, and the petals carrying no functional Riant allele still have red- and pink-coloured spots. This is consistent with previous reports that knockout of the GST gene cannot completely inhibit anthocyanin accumulation in maize (Goodman et al., 2004) and Arabidopsis (Li et al., 2011; Sun et al., 2012). The development of white with red/pink spotted flowers could be explained by the following reasons. First, mutated cells capable of accumulating anthocyanin appear at late stages of floral meristem development, and they are distributed within the L1 and/or L2 layers. An invasion by cells from the inner L2 layers into the outer L1 layer, termed ‘displacement’, has been reported in grapevine (Hocquigny et al., 2004; Walker et al., 2006; Pelsy, 2010). Thus, these mutated cells will be mixed with the wild-type cells incapable of accumulating anthocyanin in petal tissue, resulting in white and red/pink spotted flowers. Second, functional redundancy in the GST gene family has been reported in grapevine (Conn et al., 2008). Thus, it is unclear whether other anthocyanin carrier(s) could complement anthocyanin accumulation, resulting in variegated colouration.
This study reveals that the Riant gene encoding GST is essential for flower colouration in peach. Mutations involving small indels frequently occur in the last exon of the Riant gene, which causes the variegated flower in peach cv. HBH. However, the mechanism underlying the small indel formation requires further study.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primers for real-time PCR.
Fig. S1. HPLC analysis of anthocyanin composition in red, pink, and variegated flowers of cv. HBH.
Fig. S2. Expression level of potential regulatory genes involved in anthocyanin biosynthesis in petals of cv. HBH.
Fig. S3. Gel image of proteins separated by 2D PAGE.
Fig. S4. A phylogenetic tree of CHS genes from different plant species.
Acknowledgements
This project was supported by funds received from the National High Technology Research and Development Program of China (Grant No. 2011AA100206) and the National Natural Science Foundation of China. We would also like to thank Dr Andrew Charles Allan for his critical review of the manuscript.
References
- Ahsan N, Lee DG, Alam I, et al. 2008. Comparative proteomic study of arsenic-induced differentially expressed proteins in rice roots reveals glutathione plays a central role during As stress. Proteomics 8, 3561–3576. [DOI] [PubMed] [Google Scholar]
- Alfenito MR, Souer E, Goodman CD, Buell R, Mol J, Koes R, Walbot V. 1998. Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell 10, 1135–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J, Downey MO, Harvey JS, Ashton AR, Tanner GJ, Robinson SP. 2005. Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiology 139, 652–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP. 1996. Anthocyanin composition and anthocyanin pathway gene expression in grapevine sports differing in berry skin colour. Australian Journal of Grape and Wine Research 2, 163–170. [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brand U, Hobe M, Simon R. 2001. Functional domains in plant shoot meristems. Bioessays 23, 134–141. [DOI] [PubMed] [Google Scholar]
- Carles CC, Fletcher JC. 2003. Shoot apical meristem maintenance, the art of a dynamic balance. Trends in Plant Science 8, 394–401. [DOI] [PubMed] [Google Scholar]
- Chaparro JX, Werner DJ, Whetten RW, O’Malley DM. 1995. a Inheritance, genetic interaction, and biochemical characterization of anthocyanin phenotypes in peach. Journal of Heredity 86, 32–37. [Google Scholar]
- Chaparro JX, Werner DJ, Whetten RW, O’Malley DM. 1995. b Characterization of an unstable anthocyanin phenotype and estimation of somatic mutation rates in peach. Journal of Heredity 86, 186–193. [Google Scholar]
- Chen Y, Mao Y, Liu H, Yu F, Li S, Yin T. 2014. Transcriptome analysis of differentially expressed genes relevant to variegation in peach flowers. PLoS One 9, e90842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Wei G, Zhou H, Gu C, Vimolmangkang S, Liao L, Han Y. 2014. Unraveling the mechanism underlying the glycosylation and methylation of anthocyanins in peach. Plant Physiology 166, 1044–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip, a simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana. Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Conn S, Curtin C, Bezier A, Franco C, Zhang W. 2008. Purification, molecular cloning, and characterization of glutathione S-transferases.(GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. Journal of Experimental Botany 59, 3621–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn S, Franco C, Zhang W. 2010. Characterization of anthocyanic vacuolar inclusions in Vitis vinifera L. cell suspension cultures. Planta 231, 1343–1360. [DOI] [PubMed] [Google Scholar]
- Cutanda-Perez MC, Ageorges A, Gomez C, Vialet S, Terrier N, Romieu C, Torregrosa L. 2009. Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport. Plant Molecular Biology 69, 633–648. [DOI] [PubMed] [Google Scholar]
- De S, Babu MM. 2010. A time-invariant principle of genome evolution. Proceedings of the National Academy of Sciences 107, 13004–13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Peeters AJ, Léon-Kloosterziel KM, Koornneef M. 2001. The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13, 853–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippis I, Lopez-Cobollo R, Abbott J, Butcher S, Bishop GJ. 2013. Using a periclinal chimera to unravel layer-specific gene expression in plants. Plant Journal 75, 1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco RM, Regalado A, Ageorges A, et al. 2013. ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-glucosides. Plant Cell 25, 1840–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Diaz M, Kunkel TA. 2006. Mechanism of a genetic glissando: structural biology of indel mutations. Trends in Biochemical Sciences 31, 206–214. [DOI] [PubMed] [Google Scholar]
- Gomez C, Terrier N, Torregrosa L, et al. 2009. Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiology 150, 402–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez C, Conejero G, Torregrosa L, Cheynier V, Terrier N, Ageorges A. 2011. In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST. Plant Journal 67, 960–970. [DOI] [PubMed] [Google Scholar]
- Goodman CD, Casati P, Walbot V. 2004. A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. Plant Cell 16, 1812–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MF. 2002. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annual review of biochemistry 71, 17–50. [DOI] [PubMed] [Google Scholar]
- Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW. 2011. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23, 1512–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E. 2004. The challenges of moving chemicals within and out of cells: insights into the transport of plant natural products. Planta 219, 906–909. [DOI] [PubMed] [Google Scholar]
- Grotewold E. 2006. The genetics and biochemistry of floral pigments. Annual Review of Plant Biology , 57, 761–780. [DOI] [PubMed] [Google Scholar]
- Grotewold E, Davis K. 2008. Trafficking and sequestration of anthocyanins. Natural Product Communications 3, 1251–1258. [Google Scholar]
- Habu Y, Hisatomi Y, Iida S. 1998. Molecular characterization of the mutable flaked allele for flower variegation in the common morning glory. Plant Journal 16, 371–376. [DOI] [PubMed] [Google Scholar]
- Han Y, Vimolmangkang S, Soria-Guerra RE, Rosales-Mendoza S, Zheng D, Lygin AV, Korban SS. 2010. Ectopic expression of apple F3’H genes contributes to anthocyanin accumulation in the Arabidopsis tt7 mutant grown under nitrogen stress. Plant Physiology 153, 806–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. 2005. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocquigny S, Pelsy F, Dumas V, Kindt S, Heloir MC, Merdinoglu D. 2004. Diversification within grapevine cultivars goes through chimeric states. Genome 47, 579–589. [DOI] [PubMed] [Google Scholar]
- Hurkman WJ, Tanaka CK. 1986. Solubilization of plant membrane proteins for analysis of two-dimensional gel electrophoresis. Plant Physiology 81, 802–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki Y, Hisatomi Y, Suzuki T, Kasahara K, Lida S. 1994. Isolation of a suppressor-mutator / enhancer-like Transposable element, Tpnl, from Japanese bearing variegated flowers morning glory. Plant Cell 6, 375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Higeta D, Suzuki A, Yoshida H, Ozeki Y. 2002. Excision of transposable elements from the chalcone isomerase and dihydroflavonol 4-reductase genes may contribute to the variegation of the yellow-flowered carnation (Dianthus caryophyllus). Plant and Cell Physiology 43, 578–585. [DOI] [PubMed] [Google Scholar]
- Kitamura S, Shikazono N, Tanaka A. 2004. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant Journal 37, 104–114. [DOI] [PubMed] [Google Scholar]
- Koseki M, Goto K, Masuta C, Kanazawa A. 2005. The star-type color pattern in petunia hybrida ‘Red Star’ flowers is induced by sequence-specific degradation of chalcone synthase RNA. Plant and Cell Physiology 46, 1879–1883. [DOI] [PubMed] [Google Scholar]
- Larsen ES, Alfenito MR, Briggs WR, Walbot V. 2003. A carnation anthocyanin mutant is complemented by the glutathione S-transferases encoded by maize Bz2 and petunia An9. Plant Cell Reports 21, 900–904. [DOI] [PubMed] [Google Scholar]
- Lazarow K, Du M, Weimer R, Kunze R. 2012. A hyperactive transposase of the maize transposable element Activator(Ac). Genetics 191, 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gao P, Cui D, Wu L, Parkin I, Saberianfar R, Menassa R, Pan H, Westcott N, Gruber MY. 2011. The Arabidopsis tt19-4 mutant differentially accumulates proanthocyanidin and anthocyanin through a 3’ amino acid substitution in glutathione S-transferase. Plant Cell and Environment 34, 374–388. [DOI] [PubMed] [Google Scholar]
- Mandal AKA, Chakrabarty D, Datta SK. 2000. In vitro isolation of solid novel flower colour mutants from induced chimeric ray florets of chrysanthemum. Euphytica 114, 9–12. [Google Scholar]
- Marrs KA, Alfenito MR, Lloyd AM, Walbot V. 1995. A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 375, 397–400. [DOI] [PubMed] [Google Scholar]
- Martínez-Gómez P, Sánchez-Pérez R, Dicenta F, Howad W, Arús P, Gradziel TM. 2007. Almond. In: Kole C, ed. Genome Mapping and Molecular Breeding in Plants. Fruits and Nuts . Berlin Heidelberg: Springer-Verlag, 229–242. [Google Scholar]
- Montgomery SB, Goode DL, Kvikstad E, et al. 2013. The origin, evolution, and functional impact of short insertion-deletion variants identified in 179 human genomes. Genome Research 23, 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LA, Goodman CD, Silady RA, Walbot V. 2000. AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiology 123, 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizaki Y, Matsuba Y, Okamoto E, Okamura M, Ozeki Y, Sasaki N. 2011. Structure of the acyl-glucose-dependent anthocyanin 5-O-glucosyltransferase gene in carnations and its disruption by transposable elements in some varieties. Molecular Genetics and Genomics 286, 383–394. [DOI] [PubMed] [Google Scholar]
- Noda K, Glover BJ, Linstead P, Martin C. 1994. Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature 369, 661–664. [DOI] [PubMed] [Google Scholar]
- Owens DK, Alerding AB, Crosby KC, Bandara AB, Westwood JH, Winkel BS. 2008. Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiology 147, 1046–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelsy F. 2010. Molecular and cellular mechanisms of diversity within grapevine varieties. Heredity 104, 331–340. [DOI] [PubMed] [Google Scholar]
- Pooma W, Gersos C, Grotewold E. 2002. Transposon insertions in the promoter of the Zea mays a1 gene differentially affect transcription by the Myb factors P and C1. Genetics 161, 793–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poustka F, Irani NG, Feller A, Lu Y, Pourcel L, Frame K, Grotewold E. 2007. A trafficking pathway for anthocyanins overlaps with the endoplasmic reticulum-to-vacuole protein-sorting route in Arabidopsis and contributes to the formation of vacuolar inclusions. Plant Physiology 45, 1323–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing J, Woude K, Souer E, Vetten N, Mol J, Koes R. 1999. Molecular analysis of the anthocyanin2 gene of Petunia and its role in the evolution of flower color. Plant Cell 11, 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roerink SF, Koole W, Stapel LC, Romeijn RJ, Tijsterman M. 2012. A broad requirement for TLS polymerases η and κ, and interacting sumoylation and nuclear pore proteins, in lesion bypass during C. elegans embryogenesis. PLoS Genetics 8, e1002800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein JW, Furtek DB, Dooner HK, Nelson OE. 1988. Two mutations in a maize bronze-1 allele caused by transposable elements of the Ac-Ds family alter the quantity and quality of the gene product. Genetics 120, 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E., Oyama R, Bailey P, Davies K, Martin C. 2006. A Small Family of MYB-Regulatory Genes Controls Floral Pigmentation Intensity and Patterning in the Genus Antirrhinum. Plant Cell 18, 831–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelt C, Quattrocchio F, Mol J, Koes R. 2000. anthocyanin1 of petunia encodes a basic Helix-Loop-Helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12, 1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RN, Dermen H. 1979. Ontogeny in monocotyledons as revealed by studies of the developmental anatomy of periclinal chloroplast chimeras. American Journal of Botany 66, 47–58. [Google Scholar]
- Sun Y, Li H, Huang JR. 2012. Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Molecular Plant 5, 387–400. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A. 2008. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant Journal 54, 733–749. [DOI] [PubMed] [Google Scholar]
- Tanay A, Siggia ED. 2008. Sequence context affects the rate of short insertions and deletions in flies and primates. Genome Biology 9, R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telias A, Lin-Wang K, Stevenson DE, Cooney JM, Hellens RP, Allan AC, Hoover EE, Bradeen JM. 2011. Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biology 11, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Peach Genome Initiative. 2013. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nature Genetics 45, 487–494. [DOI] [PubMed] [Google Scholar]
- Uematsu C, Katayama H, Makino I, Inagaki A, Arakawa O, Martin C. 2014. Peace, a MYB-like transcription factor, regulates petal pigmentation in flowering peach ‘Genpei’ bearing variegated and fully pigmented flowers. Journal of Experimental Botany 65, 1081–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houwelingen A, Souer E, Spelt K, Kloos D, Mol J, Koes R. 1998. Analysis of flower pigmentation mutants generated by random transposon mutagenesis in Petunia hybrida. Plant Journal 13, 39–50. [DOI] [PubMed] [Google Scholar]
- Walker AR, Lee E, Robinson SP. 2006. Two new grape cultivars, bud sports of Cabernet Sauvignon bearing pale-coloured berries, are the result of deletion of two regulatory genes of the berry colour locus. Plant Molecular Biology 62, 623–635. [DOI] [PubMed] [Google Scholar]
- Waters LS, Minesinger BK, Wiltrout ME, D’Souza S, Woodruff RV, Walker GC. 2009. Eukaryotic Translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiology and Molecular Biology Reviews 73, 134–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. 2001. Flavonoid biosynthesis.A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology 126 (2), 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Yamaguchi M, Hayashi T. 2005. An Integrated Genetic Linkage Map of Peach by SSR, STS, AFLP and RAPD. Journal of The Japanese Society for Horticultural Science 74, 204–213. [Google Scholar]
- Zhang H, Wang L, Deroles S, Bennett R, Davies K. 2006. New insight into the structures and formation of anthocyanic vacuolar inclusions in flower petals. BMC Plant Biology 17, 6–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Dixon RA. 2009. MATE transporters facilitate vacuolar uptake of epicatechin 3’-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 21, 2323–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Dixon RA. 2010. The ‘ins’ and ‘outs’ of flavonoid transport. Trends in Plant Science 15, 72–80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.