Abstract
The recently published genome of Brassica napus offers for the first time the opportunity to gain insights into the genomic organization and the evolution of miRNAs in oilseed rape. In this study, 12 small RNA libraries from two B. napus cultivars (Tapidor and Ningyou7) and their four double-haploid lines were sequenced, employing the newly sequenced B. napus genome, together with genomes of its progenitors Brassica rapa and Brassica oleracea. A total of 645 miRNAs including 280 conserved and 365 novel miRNAs were identified. Comparative analysis revealed a high level of genomic conservation of MIRNAs (75.9%) between the subgenomes of B. napus and its two progenitors’ genomes, and MIRNA lost/gain events (133) occurred in B. napus after its speciation. Furthermore, significant partitioning of miRNA expressions between the two subgenomes in B. napus was detected. The data of degradome sequencing, miRNA-mediated cleavage, and expression analyses support specific interactions between miRNAs and their targets in the modulation of diverse physiological processes in roots and leaves, as well as in biosynthesis of, for example, glucosinolates and lipids in oilseed rape. These data provide a first genome-wide view on the origin, evolution, and genomic organization of B. napus MIRNAs.
Key words: allopolyploid evolution, Brassica napus, expression partitioning, microRNA.
Introduction
Small RNAs include two main types of small non-coding RNAs, microRNAs (miRNAs) and small interfering RNAs (siRNAs). miRNAs are endogenous 20–24 nt RNAs that can regulate gene expression via complementary binding to specific mRNAs and achieve their functions via post-transcriptional gene silencing in animals and plants (Voinnet, 2009; Rogers and Chen, 2013). The MIRNA gene is first transcribed into primary miRNA (pri-miRNA) by RNA polymerase II (Pol II) and then catalysed by endoribonuclease Dicer-like 1 (DCL1) in association with HYPONASTICLEAVES 1 (HYL1) and SERRATE (SE) proteins leading to the formation of a hairpin structure, or precursor miRNA (pre-miRNA). The pre-miRNA hairpin structure is further processed into a miRNA/miRNA* duplex, of which the mature miRNA is loaded onto the Argonaute (AGO) protein complex to execute its function (Vazquez et al., 2010). miRNAs are involved in regulating many plant physiological processes including plant development and biotic and abiotic stress responses (Chen, 2009; Khraiwesh et al., 2012; Sunkar et al., 2012; Shen et al., 2014; Zhang 2015).
Brassica napus, known as oilseed rape, is second only to soybean as an oil crop with a world production of over 60 million t (Bancroft et al., 2011). B. napus is an allopolyploid (AnAnCnCn) species that evolved from the spontaneous hybridization of Brassica rapa (ArAr) and Brassica oleracea (CoCo) about 7500–12 500 years ago (Nagaharu, 1935; Chalhoub et al., 2014). The lack of the genomic sequence has severely impeded research on B. napus miRNAs over the last years. Nevertheless, several studies, mainly based on EST/GSS sequences of B. napus or genomes of B. rapa and B. oleracea, have reported several hundred miRNAs and demonstrated their possible functions in regulating diverse physiological processes (He et al., 2008; Korbes et al., 2012; Xu et al., 2012; Yu et al., 2012; Zhao et al., 2012; Zhou et al., 2012; Huang et al., 2013; Lukasik et al., 2013; Wang et al., 2013; Shen et al., 2014). Recently, the genomes of B. napus and its two progenitors (ArAr and CoCo) have been sequenced (Wang et al., 2011; Chalhoub et al., 2014; Liu et al., 2014), providing for the first time an opportunity to identify and characterize B. napus miRNAs at the whole-genome level.
This work presents the results of identification and characterization of B. napus miRNAs by analysis of small RNA populations from two B. napus cultivars (one European winter cultivar,, Tapidor and one Chinese semi-winter cultivar, Ningyou7) and four double-haploid (DH) lines derived from a cross between Tapidor and Ningyou7. These data provide a first genome-wide view on the origin, evolution, and genomic organization of B. napus MIRNAs. Potential roles of miRNAs and their targets in the modulation of diverse physiological processes, e.g. for biosynthesis of glucosinolates and lipids, in oilseed rape are discussed.
Materials and methods
Plant materials
Two B. napus cultivars (Tapidor and Ningyou7) and their four DH lines (TN151, TN156, TN177, and TN186) (Qiu et al., 2006) were used for small RNA population investigation. Tapidor and Ningyou7 have significant differences in seed erucic/glucosinolate content and flowering time. Tapidor was used to generate degradome data. The cultivar Express 617 (Shen et al., 2014) was included in miRNA expression analyses. The plants were grown in a growth chamber at 24 °C and a 14h daytime photoperiod. Leaves and roots were harvested at the six-leaf stage for RNA collection. Total RNA was isolated using Trizol reagent (Invitrogen, USA). Each RNA sample was generated from leaves or roots of three different plants. The tissue samples used in RNA extraction and small RNA sequencing were collected from two replicated experiments.
Small RNA and degradome sequencing
Small RNA libraries were constructed using the standard Illumina protocol and sequenced by Illumina HiSeq 2000 (BIOMARKER, China). Twelve libraries (two cultivars and four DH lines from two replicated experiments) were sequenced (Supplementary Table S1, available at JXB online). Before miRNA prediction, the adaptors and low-quality reads were removed. The clean reads were further compared with the annotated non-coding RNA sequences, including plant snoRNA (version 1.2; http://bioinf.scri.sari.ac.uk/cgi-bin/plant_snorna/home), tRNA (http://gtrnadb.ucsc.edu/), rRNA (V11.0; http://rfam.xfam.org/) and rasiRNA (release 09-02-2014; http://www.girinst.org/server/RepBase/). Two degradome libraries from the leaf and root of the six-leaf stage of Tapidor were sequenced by Illumina HiSeq 2000 (BIOMARKER) resulting in a total of 47 million raw tags. All sequences have been submitted to the GenBank/EMBL data libraries (accession no. PRJNA272953).
Prediction of miRNA and targets
The clean reads were aligned with the genome of B. napus (Chalhoub et al., 2014; version 5.0) for identification of miRNAs. Mireap (http://sourceforge.net/projects/mireap/) was applied to predict secondary hairpin structures of MIRNAs with following parameters: the hairpin structure with free energy lower than –18 kcal mol–1; the space between miRNA and miRNA* is less than 300 nt; and with more than 16 matched nucleotides and fewer than four nucleotide bulges between miRNA and miRNA*. Only small RNAs with at least two reads in a library were used for miRNA prediction. By comparison with the miRBase (http://www.mirbase.org, release 21), a miRNA was considered as conserved if its mature sequence had two nucleotide or fewer than two nucleotide mismatches to the known miRNA or as a novel miRNA when there are more than two nucleotide mismatches (Meyers et al., 2008). For the conserved miRNAs, the same miRNA/family names as in miRBase were assigned, but with new serial numbers (such as b, c, d) in some cases. For the novel miRNAs, the names bna_novel_miRX1 to bna_novel_miRX214 were given (Meyers et al., 2008).
The targets of miRNAs in B. napus were predicted via the online sever psRNATarget (Dai and Zhao, 2011). Using the function ‘User-submitted small RNAs/ user-submitted transcripts’, the query of transcripts was generated from the B. napus genome. Default parameters were used to filter candidates. The software PatMan was used to map the degradomic reads to the targets, and custom perl scripts were employed to identify candidate degradative targets. According to previous results (Jones-Rhoades and Bartel, 2004; Shen et al., 2014; Song et al., 2010a, b ; Zhao et al., 2012), the ends of degradomic reads were considered when they were within the 5 nt region of the cleavage site.
Lipid biosynthesis-related genes
As reference, the Arabidopsis lipid-related gene database was downloaded at http://aralip.plantbiology.msu.edu/ (Li-Beisson et al., 2013). The annotation of lipid-related genes in B. napus was carried out using the method of Wang et al. (2014) with minor modifications, i.e. BLASTp under the E-value <1e–5 and sequence identify >50% for the search of orthologous genes.
Genomic synteny of MIRNAs
The synteny or co-linearity of MIRNAs among the three Brassica species (B. napus, B. rapa, and B. oleracea) was detected by MCScanX (Wang et al., 2012). The genomes of B. oleracea (http://ocri-genomics.org/bolbase/, version 1.0), B. rapa (Phytozome 10.2) and B. napus (http://www.genoscope.cns.fr/brassicanapus/data/, version 5.0), and their annotated gene sets were used for the genomic synteny analysis. BLASTp was employed to determine the synteny by pairwise comparison with the parameters of E-vlaue <1e–5 and max_target_seqs < 6. MIRNA orthologues in B. napus, B. rapa, and B. oleracea were analysed using a microsynteny-based method (Ma et al., 2010; Shen et al., 2014). For each MIRNA, its 10 flanking protein-coding loci were retrieved from the B. rapa and B. oleracea genomes, respectively. Homology tests of MIRNA and flanking genes were performed by BLASTn and the top five hits of each MIRNA were chosen for flanking loci tests. A syntenic MIRNA pair among B. napus, B. rapa, and B. oleracea was defined with at least one identical upstream or downstream flanking protein-coding gene. Syntenic MIRNAs were divided into four sets as described by Shen et al. (2014). The first three sets were taken as syntenic MIRNAs for construction of the Circos map (Krzywinski et al., 2009).
Transcript analysis of miRNAs and target genes
Low-molecular-weight RNA was enriched by 5% polyethylene glycol (M r 8000) and 0.5M NaCl precipitation (Lu et al., 2007). Mature miRNA expression analysis was performed by stem–loop reverse transcription quantitative PCR (RT-qPCR) (Shen et al., 2014). For cDNA synthesis, 200ng of low-molecular-weight RNA from each sample was reverse transcribed using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) with U6 snRNA as the internal control. For cDNA synthesis of predicted targets, 2 µg of total RNA from each sample was used. qPCR reactions were performed in a CFX96™ Real-Time System/C1000 Touch™ Thermal Cycler (BioRad) with MAXIMA® SYBR Green Master Mix (Fermentas) and actin as the reference gene. Invariant expression of the actin gene in seedling roots and leaves was assured beforehand. The relative expression fold change of miRNAs and related genes were calculated using the comparative C T method. All reactions were performed in triplicate with three independent experiments. The primer pairs were listed in Supplementary Table S2, available at JXB online.
Expression levels of miRNAs in parental and DH lines
The expression levels of miRNAs were measured by the small RNA abundance. The amount of small RNA reads was transformed into reads per million (RPM) with the formula RPM=reads count/clean reads×106). PatMan was employed to map the small RNA reads to mature miRNA sequences. Small RNA reads were counted when their position was within the length of the mature miRNA sequence. The miRNA read that was mapped to both of the A and C subgenomes was counted twice. At least a twofold change was taken to define the differentially expressed miRNAs between offspring and the mean values of parental lines in regard to the read number with corresponding mature pre-miRNAs.
Sequences alignment and visualization
MAFFT (http://mafft.cbrc.jp/alignment/server/, version 7) was employed for multiple sequences alignment. The WebLogo (http://weblogo.threeplusone.com/create.cgi) was used to create the logo for the alignment results and CLC Sequence Viewer 6 (CLC Bio, Aarhus, Denmark) was used for the display of the conservation among whole sequences. The distribution of miRNAs and their partners was plotted in a 1Mb window size by Circos (Krzywinski et al., 2009). The UEA sRNA workbench (Stocks et al., 2012) was employed to visualize the hairpin structure of miRNAs with the corresponding mature sequence and parameters generated by Mireap.
Annotation of transposable elements
As no annotation data for transposable elements in the newly released B. napus genome were available, transposable elements were identified in the genome using a combination of de novo and homology-based approaches as follows. First, the RepeatModeler pipeline was used to search for repeats in the genome and RepeatMask was then used to identify repeats in Repbase (Tempel, 2012). Long terminal repeat (LTR) retrotransposons were found using LTR_FINDER with default parameters (Xu and Wang, 2007).
Search for miRNAs previously identified in the B. napus genome
BlastN was employed to map the miRNA precursor sequence onto the B. napus genome with an E-value of <1e–5 and a matched length longer than 90% of the query sequence.
RNA ligase-mediated (RLM)-5′RACE analysis
To validate the predicted targets of miRNAs, 5′RACE was carried out with 2 µg of total RNA using a GeneRacerTM Kit (Invitrogen) as described by Shen et al. (2014). The reverse transcription of mRNAs was performed using the random primers. For amplification of cDNA ends, a pair of gene-specific primers was designed (Supplementary Table S2). A touchdown PCR was performed. One microlitre of this initial touchdown PCR was used as template for the following nested PCR. The reaction products were separated on a 1.7% agarose gel. The bands with the expected size were excised, purified, and cloned into the pGEM-T vector (Promega) for sequencing.
Results
Identification of miRNAs
For a genome-wide analysis of B. napus miRNAs, 136 263 599 raw reads of 18–44 nt were generated from 12 small RNA libraries (two replicates) of young leaves of two parental lines (Tapidor and Ningyou7) and their four DH lines (Supplementary Table S1). From these, 105 219 098 clean reads were obtained. The majority of the clean reads were 21–24 nt small RNAs, consistent with a previous report (Shen et al., 2014; Supplementary Fig. S1, available at JXB online). Of the clean reads, 64.9% could be mapped to the B. napus genome. After removing the non-coding RNAs, the remaining reads, referred to as endogenous small RNAs, were used for miRNA prediction.
We identified 645 miRNAs, including 280 conserved miRNAs from 34 families and 365 novel miRNAs from 225 families (Table 1 and Supplementary Table S3, available at JXB online). Out of 645 miRNAs, 275 and 364 originated from the An and Cn subgenomes, respectively (six miRNAs remained unassigned). Besides the known B. napus miRNAs deposited in the current international miRNA database miRBase (release 21) and reported in recent studies (Huang et al., 2013; Lukasik et al., 2013; Wang et al., 2013; Shen et al., 2014), 351 miRNAs including 47 conserved from 19 families and 304 novel miRNAs from 187 families were newly identified by this study. To confirm the expression of the predicted miRNAs, eight conserved and 14 novel miRNAs were chosen for stem–loop reverse transcription PCR analysis. The results confirmed the expected expression of all eight conserved and 13 of the 14 novel miRNAs, except for a non-specific amplification of miRX132 in B. napus seedlings (Supplementary Fig. S2, available at JXB online).
Table 1.
miRNAs and their targets in B. napus identified in this study
| Type | miRNAs | miRNAs target to: | ||
|---|---|---|---|---|
| Total | An/Cn | Glucosinolatesa | Lipida | |
| Conserved | 280 | 143/136 | 1 (1) | 55 (27) |
| Novel | 365 | 132/228 | 10 (6) | 103 (95) |
| Total | 645 | 639b | 11 (7) | 158 (122) |
a Numbers in parentheses indicate the number of glucosinolates and lipid biosynthesis-related genes that were predicted to be targeted by miRNAs.
b Six miRNAs located in unplaced scaffolds.
Prediction and validation of miRNA targets
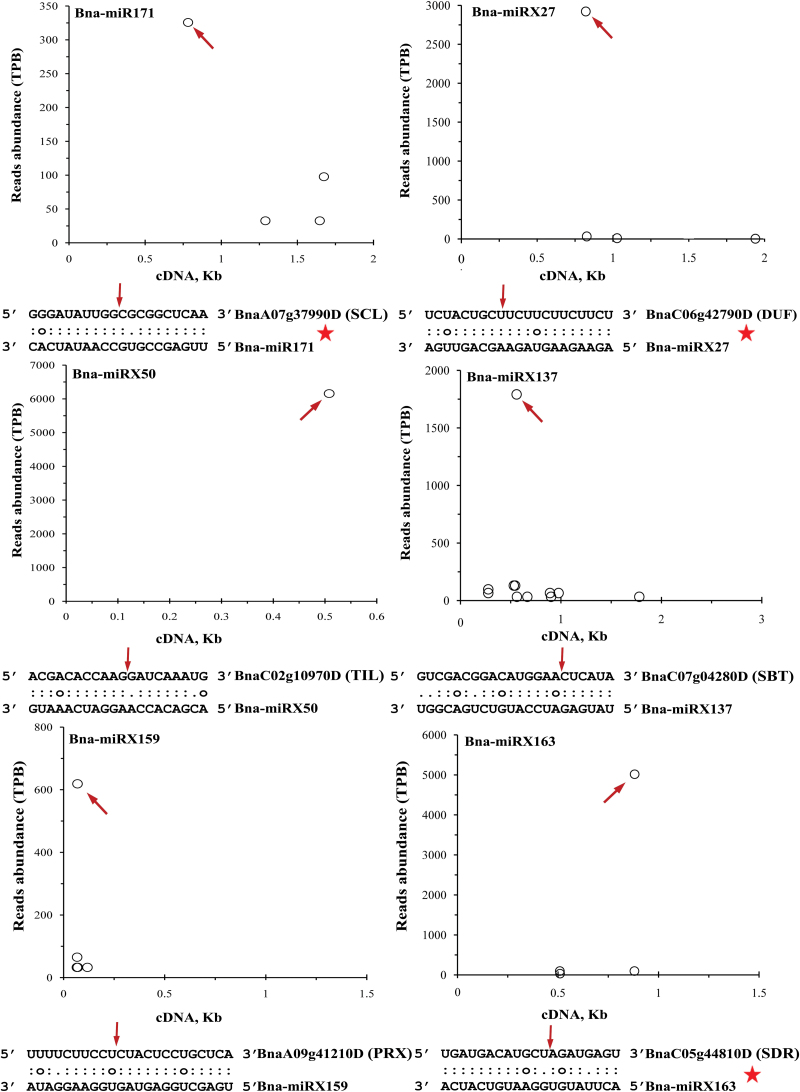
The targets of the newly identified miRNAs were predicted (Supplementary Table S4, available at JXB online). Consistent with the results reported previously, the majority of potential targets of the conserved miRNAs were transcription factors and shared high homology with Arabidopsis orthologues. For example, both miR160 and miR167 target auxin response factor (ARF) transcription factors (Mallory et al., 2005; Wu et al., 2006), miR156/miR157 and miR162 target squamosa promoter-binding-like protein (SPL) and Dicer-liker (DCL) protein in Arabidopsis and B. napus. Of the 365 novel miRNAs, 363 (99.4%) had putative targets in the annotated gene sets of B. napus. To validate the miRNA–target interaction, degradome sequencing was performed with RNA samples from Tapidor roots and young leaves, confirming 849 targets predicted for 263 conserved and 291 novel miRNAs (31 and 168 families), respectively (Fig. 1, Supplementary Table S4). Among the targets of 291 novel miRNAs, a number of genes were stress or environmental adaptation related. The Arabidopsis orthologue of the miRX50 target TIL is a heat stress-related gene (Chi et al., 2009). The orthologue of the miRX137 target SBT is associated with the density and distribution of stomates (Berger and Altmann, 2000). miRX159 and miRX163 target the genes PRX and SDR encoding dehydrogenase/reductase and peroxidase, respectively (Passardi et al., 2004; Kavanagh et al., 2008).
Fig. 1.
Identification of candidate targets of miRNAs by degradome experiments. Six targets of one conserved and five novel miRNAs are shown in the panels as an example. The x-axis indicates the position of target genes while the y-axis represents the abundance of sequenced reads. Each circle is a degradome fragment that can be mapped to the corresponding target gene and the arrows indicate the expected miRNA positions. The putative miRNA-binding sites are depicted within the corresponding target transcripts within the sequence alignments. The cleavage sites deduced from the degradome data are indicated by the arrows. The stars indicate the cleavages validated by the 5’RLM-RACE assays. (This figure is available in colour at JXB online.)
Ten miRNAs (nine novel miRNAs) were predicted to target genes involved in biosynthesis or breakdown of glucosinolates (GSLs) (Chalhoub et al., 2014) (Table 1 and Supplementary Table S5, available at JXB online). The miRNA-mediated regulation was confirmed for six of the 10 miRNAs either by degradomic data or by RLM-5’RACE (Supplementary Table S4, Fig. 3, Supplementary Fig. S4, available at JXB online). Three of these miRNAs play a role in GSL metabolism (Supplementary Table S5). 1) miRX95 was predicted to target BnaC05g12520D, an orthologue of the GSL biosynthesis-related gene CYP79F1 (AT1G16410) in Arabidopsis (Chalhoub et al., 2014; Liu et al., 2014). The biosynthesis of GSL is mediated by CYP79F1 (Reintanz et al., 2001). There were two single-nucleotide polymorphisms between Tapidor and Ningyou7 in the predicted miRX95 target region, which resulted in additional (G:U) mismatches in the miRX95 binding site (binding prediction score >3.0) in Tapidor, which has a potential impact on the miRX95-mediated regulation of BnaC05g12520D and consequently the GSL biosynthesis in Tapidor. miR6032a was predicted to target BnaA03g38670D. Its Arabidopsis orthologue APK1 (AT2G14750) is involved in the biosynthesis of a major class of sulfated secondary GSL (Mugford et al., 2009). Lastly, miRX113 was predicted to target BnaA07g19610D, an orthologue of Arabidopsis PEN3 (AT1G59870). PEN3 is required for breakdown of GSLs (Wittstock and Burow, 2010).
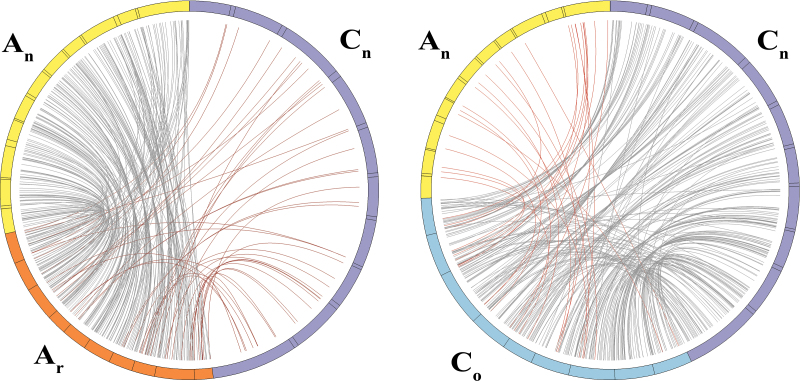
Fig. 3.
Synteny of miRNAs between B. napus (AnAnCnCn) and its two progenitor genomes (ArAr and CoCo). The 851 MIRNAs from the B. rapa and B. oleracea genomes were mapped to the B. napus genome. The lines refer to the best matches of the miRNAs from the two progenitors in the B. napus genome. An and Cn represents the A and C subgenome of B. napus, Ar indicates the genome of B. rapa, and Co stands for the genome of B. oleracea. The black lines indicate the miRNA best match in An-Ar and Cn-Co, and the red lines represent the miRNA best match in An-Co and Cn-Ar.
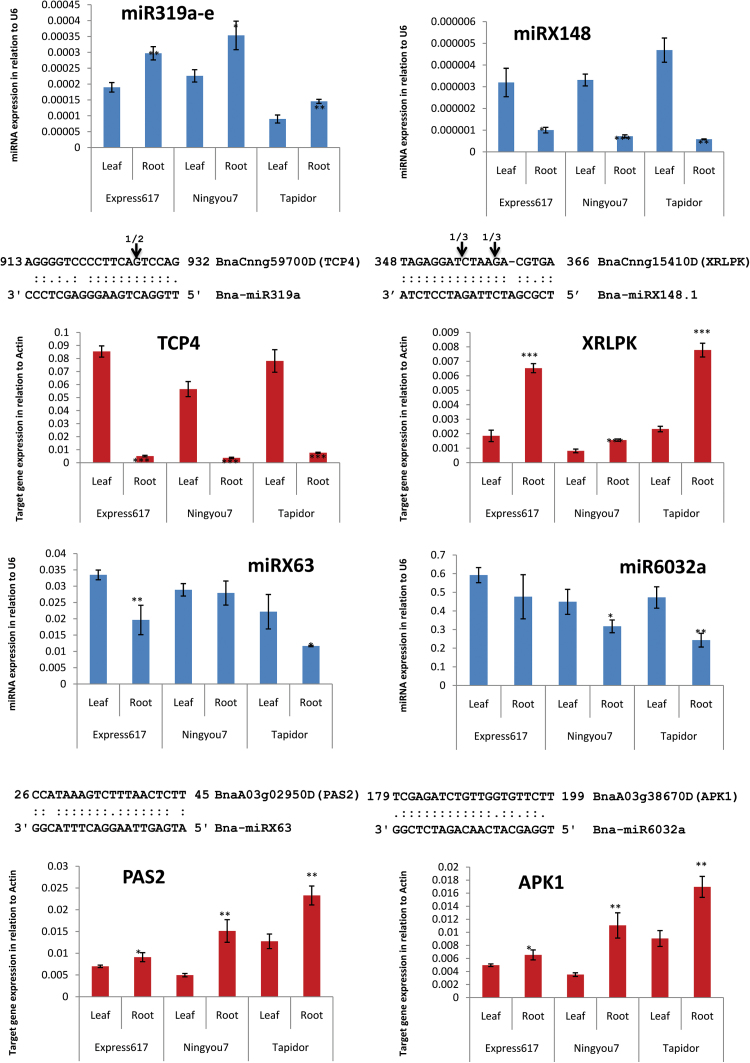
Based on a homologue search using the 738 annotated lipid biosynthesis-related genes in Arabidopsis (http://aralip.plantbiology.msu.edu), 2606 putative lipid biosynthesis-related genes were identified in B. napus. Of these, 122 genes were predicted targets of 158 miRNAs, comprising 55 conserved and 103 novel miRNAs (Table 1 and Supplementary Table S6, available at JXB online), from which 27 targets of 45 miRNAs were identified by the degradomic data (Supplementary Table S4). For example, one novel miRNA (miRX63) was predicted to target BnaA03g02950D, an orthologue of Arabidopsis PAS2 or AT5G10480 that has acyl-CoA dehydratase activity and has been shown to be involved in the biosynthesis of very long chain fatty acids (see http://www.tair.org). An opposite change in the expression of miRX63 and BnaA03g02950D in leaves and roots of two of the three cultivars was obvious (Fig. 2), suggesting that miRX63-mediated regulation of the lipid biosynthesis-related gene BnaA03g02950D might play a role in the development of leaf and root. As GSL and lipid biosynthesis is regulated by complex mechanisms and by developmental stages (Grubb and Abel, 2006; Pollard et al., 2008; Niu et al., 2009), more detailed investigations are needed to clarify the role of these miRNA–target interactions in the biosynthesis of GSL and lipids in the future.
Fig. 2.
Differential expression of miRNAs and their targets in roots and leaves of three oilseed rape cultivars. The sequences depict the miRNA-binding site within the target transcript in the middle of each panel. The arrows represent the miRNA cleavage sites on targets with degradome/RACE evidence. (This figure is available in colour at JXB online.)
To identify miRNAs involved in regulating different physiological processes in roots and leaves, respectively, the small RNA populations from leaves were compared with those from roots of B. napus (Shen et al., 2014) and 59 miRNAs were identified including 35 novel ones, which gave a significant difference in the read numbers (>2-fold) between the roots and leaves (Supplementary Table S7, available at JXB online). Differential expression was confirmed for 10 selected miRNAs (four conserved and six novel miRNAs) by stem–loop qPCR (Supplementary Fig. S3, available at JXB online). Among these, several miRNAs (such as miR159 and miR319) have a pronounced role in regulating plant leaf/root development (Khraiwesh et al., 2012).
The expression profiles of four of these miRNAs and their targets in roots and leaves were further investigated using RT-qPCR. As shown in Fig. 2, in all four cases (miR319, miR6032, miRX63, and miR148), a reciprocal change in the expression levels between the miRNA and its target was evident. Thus, these data suggested a direct miRNA–target expression modulation in regulation of developmental and/or physiological processes in the root and leaf.
Evolution of miRNAs after the allopolyploidization event
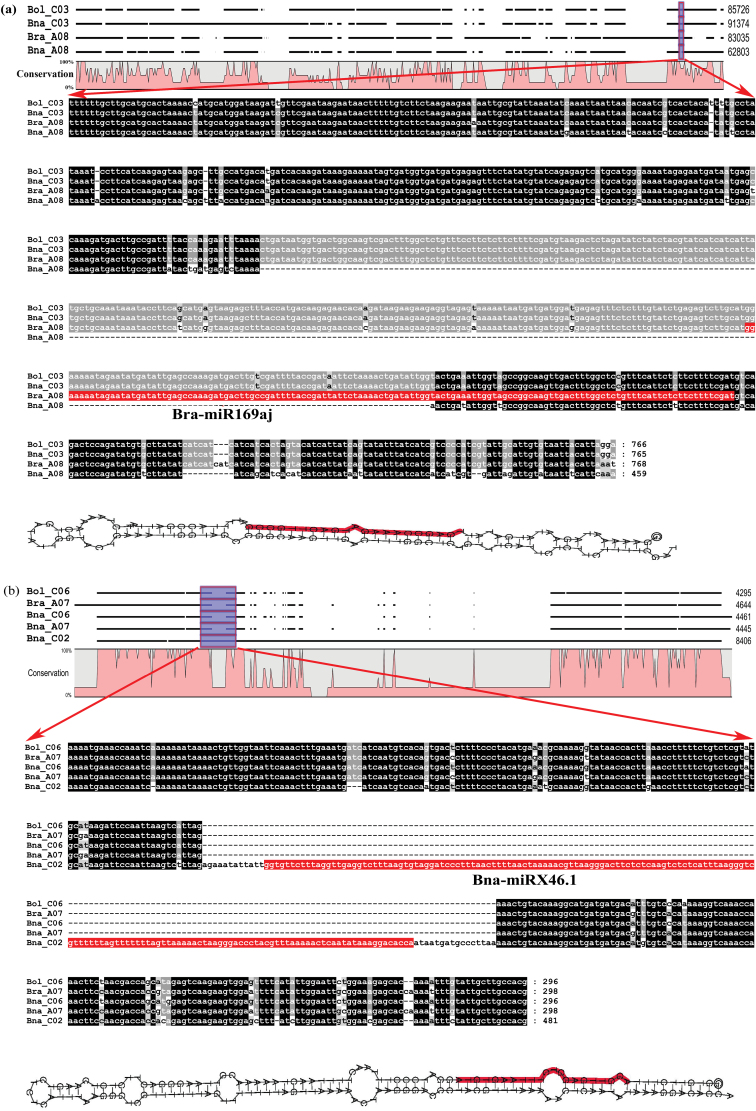
The availability of the genomes of B. napus and their two progenitors provides an opportunity to look at the expansion and loss of miRNAs in B. napus. The synteny of MIRNAs between B. napus (AnAnCnCn) and its two progenitor genomes (ArAr and CoCo) was first investigated by mapping the 851 MIRNAs identified from the two progenitor genomes in a previous study (Shen et al., 2014) to the B. napus genome. To ascertain genomic synteny of the predicted MIRNAs between the genomes of B. napus and its two progenitors, a microsynteny analysis was performed using the MIRNAs and their flanking protein-coding sequences. As expected, the majority (646/851, 75.9%) of the two progenitor MIRNA orthologues existed in the corresponding subgenomes of B. napus (Supplementary Table S8, available at JXB online), although a small number (72) of these MIRNAs showed a match to the opposite subgenome of B. napus (Fig. 3). These results support the observation of a high conservation between the subgenomes of B. napus and its two progenitor genomes (Chalhoub et al., 2014). In addition, 115 MIRNAs, including 104 novel MIRNAs, failed to show any homologous sequence hit in the B. napus genome (Supplementary Table S9, available at JXB online). Of these, five conserved MIRNAs (e.g. members of the miR169 and miR172 families) were obviously lost due to sequence deletions or mutations as revealed by comparative genome analysis between B. napus and it two progenitors (Table 2; an example of miR169 member is shown in Fig. 4a).
Table 2.
Changes of copy numbers of miRNA families between B. napus and its two progenitors (B. rapa and B. oleracea)
Only part of the miRNA families are shown here and a full list is provided in Supplementary Table S10, available at JXB online.
| Family | B. napus | B. rapa a | B. oleraceaa | (AnAnCnCn)-(ArAr+CoCo) | ||
|---|---|---|---|---|---|---|
| AnAn | CnCn | ArAr | CoCo | Based on small RNAs | Based on genomic synteny | |
| miR156 | 17 | 19 | 17 | 15 | 4 | 5 |
| miR171 | 11 | 7 | 7 | 3 | 8 | 3 |
| miR166 | 13 | 11 | 6 | 4 | 14 | 2 |
| miR168 | 9 | 9 | 5 | 7 | 6 | 1 |
| miR169 | 16 | 15 | 26 | 19 | –14 | –2 |
| miR172 | 8 | 6 | 11 | 9 | –6 | –2 |
| miR390 | 6 | 2 | 6 | 6 | –4 | –1 |
| miR395 | 5 | 5 | 9 | 6 | –5 | 0 |
| miR159 | 1 | 2 | 4 | 4 | –5 | 0 |
| miR398 | 1 | 1 | 4 | 3 | –5 | 0 |
| miR160 | 9 | 9 | 7 | 7 | 4 | 0 |
| miR165 | 2 | 0 | 3 | 3 | –4 | 0 |
a Based on Shen et al. (2014).
Fig. 4.
Gain and loss of miRNAs in B. napus. (a) An example of miR169 loss in B. napus. The upper part was the synteny region among the three species. The deletion of seveval bases on chromosome A08 of B. napus, which caused the loss of one member of miR169 family in B. napus. (b) The novel-miRX46.1 as example for the gain of miRNAs in B. napus. The upper part indicates the synteny region found by MCScan, in which an insertion sequence was detected on the chromosome C02 of B. napus. The sequence can form a perfect hairpin structure as indicated in the lower part.
In the next step, the 645 miRNAs of B. napus were compared with Arabidopsis miRNAs (miRBase, release 21). A significant expansion of several miRNA families, including miR156, miR160, and miR169 in B. napus, was observed (Supplementary Table S10). Since such expanded miRNA families could also be found in the two progenitor genomes, it was concluded that these expansion events must have occurred before the AA-CC speciation hybridization (Supplementary Table S10).
However, 18 B. napus MIRNAs (six novel and 12 conserved MIRNAs) were not present in the syntenic regions of the two progenitor genomes, suggesting that they might be newly generated after the allopolyploidization event in B. napus (Supplementary Table S11, available at JXB online). The 12 newly generated conserved MIRNAs, including six, two, and three from the miR156, miR171, and miR166 families, respectively (Table 2 and Supplementary Table S11), obviously originated from the existing MIRNAs via the gene or genomic segmental duplication events. The expression of three of the six novel miRNAs (bna-miRX46 as an example shown in Fig. 4b) could be confirmed in B. napus seedlings by the stem–loop reverse transcription PCR analysis (Supplementary Fig. S2). The predicted targets of the six novel miRNAs could be confirmed by the degradome sequencing (Supplementary Table S4). These newly evolved B. napus miRNAs might be associated with adaptation to changed environmental conditions under natural and artificial selection pressure. In support of this, the targets of miRX132.1 (BnaA05g30540D for the putative defensin-like protein 73 and BnaA03g50380D for the putative F-box/LRR-repeat protein 19) were related to plant defence responses. Also, the target of miR46.1 (BnaC03g35590D for phosphatidylinositol 4-phosphate 5-kinase 6) interacts with the lipid pathway (Supplementary Table S8).
Partitioning and heritability of miRNA expression
To investigate the partitioned expression of miRNAs between the An and Cn subgenomes, the expression levels and patterns of miRNAs in the An or Cn subgenome were compared based on small RNA reads. Although 275 and 364 MIRNAs were identified from the An and Cn subgenomes, respectively, a slightly higher genomic density of MIRNAs was observed in the An subgenome (1.03 and 0.73 MIRNAs per Mb for the An and Cn subgenomes), which was similar to those observed in the two progenitor genomes (Shen et al., 2014). To measure miRNA expression (see Materials and methods), the number of small RNA reads of corresponding mature miRNAs were compared, and a significantly higher abundance of reads (per unique mature miRNA sequence per million reads) was observed in the An subgenome than in the Cn subgenome (P<0.01) (Table 3, Fig. 5). To confirm this, small RNA populations were generated from Tapidor and Ningyou7 seedlings grown under independent environmental conditions and sequenced. A similar bias of miRNA expression was obvious between the two subgenomes (Table 3). The comparison of miRNA expression patterns of four Tapidor/Ningyou7-derived DH lines with those of their parental lines also gave similar biased expression patterns of miRNAs (P<0.01 and P<0.02 in two environments, respectively; Table 3). When the individual miRNA families (for 32 with members > 3) were compared, only 13 of the 32 families were found, which were the main contributors to the different miRNA read abundance or miRNA expression levels between the An and Cn subgenomes, while the remaining 19 miRNA families showed a similar expression pattern between the An and Cn subgenomes. These data strongly suggested a partitioned expression of miRNA between the two subgenomes after the hybridization of speciation in B. napus.
Table 3.
Comparison of miRNA expression in the two subgenomes in B. napus
The expression levels were measured by Illumina sequencing reads per unique mature sequence of miRNA or per Mb per million reads in two DH parental lines (Tapidor and Ningyou7 or T/N) and their four DH lines.
| Expression level (read density)a | An | Cn | Cn/An | P value | Based on: |
|---|---|---|---|---|---|
| Reads (E1) per mature miRNA | 4293.26 | 2360.90 | 0.55 | <0.01 | T/N |
| Reads (E2) per mature miRNA | 3119.39 | 2261.51 | 0.72 | <0.01 | T/N |
| Reads (E1) per Mb | 3928 | 1493 | 0.52 | <0.01 | T/N |
| Reads (E2) per Mb | 2854 | 4231.09 | 0.68 | <0.01 | T/N |
| Reads (E1) per mature miRNA | 6226.15 | 4231.09 | 0.68 | <0.01 | 4 DH lines |
| Reads (E2) per mature miRNA | 2006.64 | 1449.13 | 0.72 | 0.02 | 4 DH lines |
| Reads (E1) per Mb | 5696.26 | 2792.52 | 0.49 | <0.01 | 4 DH lines |
| Reads (E2) per Mb | 1835.86 | 956.43 | 0.52 | <0.01 | 4 DH lines |
a Small RNA populations in two parental lines (T/N) were collected and sequenced in two different environments (E1 and E2), respectively.
Fig. 5.
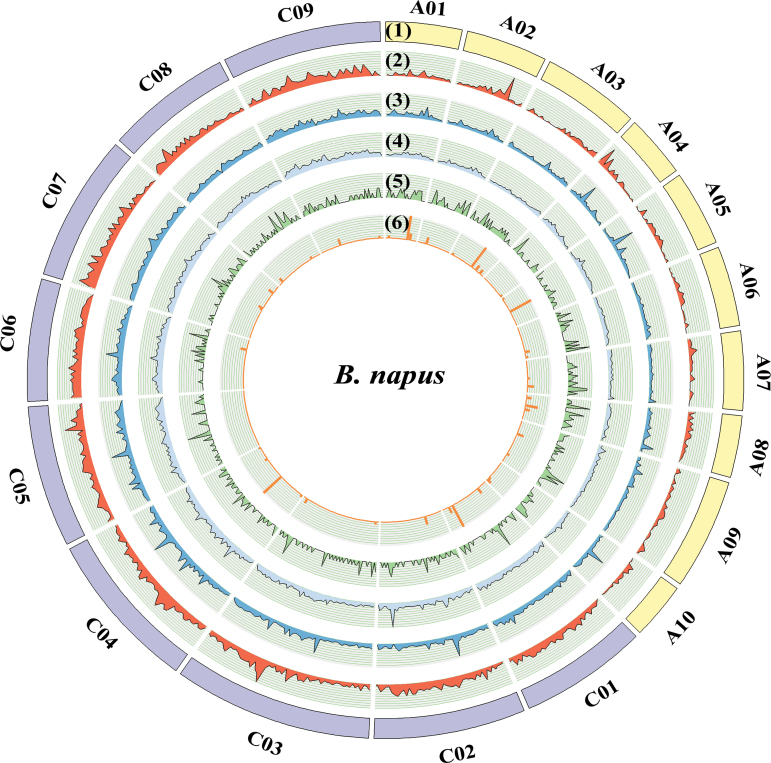
Genomic distribution of miRNAs in B. napus. From outer to inter circles: (1) chromosomes of B. napus; (2) LTR retrotransposon; (3) small RNA reads; (4) 24 nt small RNA reads; (5) miRNAs identified by this study; (6) small RNA reads that can be mapped to the mature sequences of miRNAs. The peak value represents the density of each element with a window of 1Mb in the B. napus genome.
To investigate the heredity of miRNA expression, small RNA populations from seedlings of the same developmental stage (six leaves) of the two parental lines (Tapidor and Ningyou7) and their four DH lines were analysed. After normalization of small RNA reads, the expression levels of miRNAs in Tapidor and Ningyou7 were compared with those in the four DH lines. Using a 2-fold change of the average expression level of a miRNA in the four DH lines relative to their parental lines as the threshold value (Stupar et al., 2007), the additive genetic effect of miRNAs in B. napus was estimated. Significantly, a higher number of miRNAs with a non-additive rather than with an additive genetic effect (2- to 3-fold) were observed in the four DH lines (Supplementary Table S12, available at JXB online), suggesting the potential dominance of non-additive genetics of miRNAs in B. napus.
Finally, the expression levels of other small RNAs in the B. napus genome were investigated and a genomic bias was also observed for siRNA expression (Fig. 5). However, this was totally in contrast to the An-biased miRNA expression, i.e. a higher abundance of small RNA reads was observed in the Cn but not in the An subgenome (2.1-fold difference between the two subgenomes), suggesting that more siRNAs are generated or expressed from the Cn subgenome. Consistent with this, the content of LTR retrotransposons was found to be 10.3-fold higher in the Cn subgenome than in the An subgenome (Fig. 5), a phenomenon also observed previously (Chalhoub et al., 2014; Liu et al., 2014), implying the possible involvement of these repetitive transposable elements in the biogenesis of siRNAs.
Discussion
This study carried out the first genome-wide investigation of miRNAs in B. napus using the newly sequenced B. napus genome. In addition to known B. napus miRNAs, identified mainly based on the two progenitor genomes of B. napus (Zhou et al., 2012; Korbes et al., 2012; Xu et al., 2012; Zhao et al., 2012, Shen et al., 2014), a new set of B. napus miRNAs was identified, including both conserved and novel ones. As expected, the number of B. napus miRNAs identified so far is significantly higher than that in the model dicot species Arabidopsis thaliana and monocot species rice (Oryza sativa). To the best of our knowledge, this represents the largest miRNA population ever found in a single plant species up to now, thus offering a unique dataset for the study of the origin, genomic structure, and evolution of miRNAs in allopolyploid plant genomes.
The results of this study support the observation of a high genomic conservation between the subgenomes of B. napus and its two progenitor genomes (Chalhoub et al., 2014). In many cases, it was not possible to distinguish the miRNA reads between the A and C subgenomes as they share the same mature sequence. The genomic synteny regions applied in this study allowed a precise localization of miRNA loci in the orthologous regions of both subgenomes. Only miRNAs with unique mature sequences in the A or C subgenome were defined as A or C subgenome-specific sequences, respectively. The majority of the two progenitor MIRNA orthologues were found to exist in the corresponding subgenomes of B. napus, and only a small group of MIRNAs showed an opposite match to the subgenomes. These mismatches might be a result of the homeologous recombination or gene-level duplication events during/after the AA-CC hybridization. Also, 115 MIRNAs were found only in the genomes of the two progenitors and not in the B. napus genome, strongly suggesting a possible loss of these MIRNAs due to sequence deletion or mutation (Table 2, Fig. 4a), although these MIRNAs could be undetected due to limited assembly of the two progenitor genomes. Because of a high genetic redundancy after the AA-CC hybridization, it is conceivable that the miRNA loss occurred in the B. napus genome. As B. napus has a very recent origin (~7500 years ago; Chalhoub et al., 2014), it would be expected that the numbers of conserved miRNAs in B. napus will be decreased to a relatively stable level in future, as observed in the tetraploid cotton genome (Xie and Zhang, 2015). However, the allopolyploidization event resulted in an increase in the number of MIRNAs in B. napus by generating new MIRNAs through gene or genomic segmental duplication events. As revealed by the genomic microsynteny analysis, at least 12 of the 18 newly generated MIRNAs in B. napus obviously originated from the existing conserved MIRNAs (miR156, miR171, and miR166 families) in the two progenitors via such genomic reorganization. For instance, miR156 affects the flowering time and responses to biotic and abiotic stress (Sunkar et al., 2012; Stief et al., 2014). Similarly, miR171 also participates in the biotic and abiotic stress responses (Zhou et al., 2007; Liu et al., 2008), while miR166 can alter the development and polarity of the leaf via an interaction with HD-ZIP genes (Emery et al., 2003; Williams et al., 2005). Following this, it is reasonable to speculate that the 12 new copies of these conserved MIRNAs might greatly contribute to the environmental adaptation of B. napus. Their origins might be attributed to natural selection. In addition, a significant expansion of a few conserved miRNA families, such as miR156, miR160, and miR169, have obviously enlarged the number of miRNAs in B. napus (Supplementary Table S10), but these expansions have mostly occurred before the AA-CC hybridization, as such expansion events could also be observed in the two progenitor genomes (Supplementary Table S10). This is consistent with the very recent origin of the AACC genome in B. napus. Unequal genomic/segmental duplications as well as tandem duplications of MIRNAs obviously might also have greatly contributed to the MIRNA expansion in the B. napus genome (Shen et al., 2014).
An interesting finding in this study was the partitioned expression of miRNAs between the two subgenomes of B. napus. A significantly higher abundance of miRNAs in the An subgenome than in the Cn subgenome was observed in at least 13 miRNA families in two different genetic backgrounds (Tapidor and Ningyou7) and under different environmental conditions. Furthermore, similar results observed in the four Tapidor/Ningyou7-derived DH lines provide further genetic evidence for partitioning of miRNA expression between the two subgenomes in B. napus. It is of great interest and importance to explore the regulation and functional relevance of the partitioning of miRNA expression in oilseed rape in the future. In this study, a potential dominance of non-additive genetics for miRNAs in B. napus was suggested for the first time by heredity analyses of the expression patterns of each miRNA using small RNA populations from the parental lines (Tapidor and Ningyou7) and four Tapidor/Ningyou7-derived DH lines. Nevertheless, further investigations using more DH lines or offspring lines from different parental lines and in particular their degradome data are needed to confirm this observation.
miRNAs are believed to have played an important role in genome polyploidy (Ng et al., 2012), as well as in regulating network in speciation and subsequent evolution of polyploidy crops (Yao et al., 2007; Kenan-Eichler et al., 2011; Tang et al., 2012; Gong et al., 2013; Xie and Zhang, 2015). In upland cotton (AADD, Gossypium hirsutum L.), miRNA loss and gain after the hybridization of AA and DD species were detected at the genomic level. However, significant expansions of several AADD-specific miRNA families were identified in cotton but not in B. napus. This might be attributed to the post-Neolithic origin of B. napus (Chalhoub et al., 2014). The allotetraploid cotton arose from the union of two types of diploid cotton genomes about 1–2 million years ago (Li et al., 2015; Zhang et al., 2015). Similar to the observations here in B. napus, the biased expressions of miRNAs in the two subgenomes of a tetraploid were also observed in cotton (Gong et al., 2013; Xie and Zhang, 2015). Significantly higher expression of ~20 conserved miRNAs in the AA subgenome than in the DD subgenome in cotton were reported (Gong et al., 2013). These results suggest that miRNAs play an important role in the regulation network in speciation and subsequent evolution of polyploidy crops.
Increasing data suggest that miRNAs are involved in regulating plant responses to diverse developmental and physiological processes, as well as plant responses to diverse biotic and abiotic stress (Khraiwesh et al., 2012). By comparison with previously sequenced small RNA populations from roots (Shen et al., 2014), a number of miRNAs were identified here that are differentially expressed between the root and leaf of B. napus. Among these are miR159, miR164, miR166, miR168, miR172, miR319, and miR390, which have been demonstrated previously to have an effect on leaf development in Arabidopsis (Emery et al., 2003; Palatnik et al., 2003; Achard et al., 2004; Allen et al., 2005; Laufs et al., 2004; Vaucheret et al., 2004; Guo et al., 2005; Schwab et al., 2005; Williams et al., 2005), as well as miR160 and miR393, which have been shown to influence the development of roots via inhibition of their target genes (ARF and TIR, respectively) in Arabidopsis (Navarro et al., 2006; Liu et al., 2007). The opposite expression patterns between miRNAs and their targets occurred in the root and leaf of B. napus, suggesting their conserved role in the regulation of developmental or physiological processes in the root and leaf of B. napus. Of great importance, a subset of novel miRNAs were identified that could have a specific function in regulating the expression of specific traits, such as biosynthesis of lipids and GSLs in oilseed rape. Further experimental investigation and analysis are needed to gain insights into the underlying mechanisms.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1. Small RNAs generated from B. napus by this study.
Supplementary Table S2. Primers used in this study.
Supplementary Table S3. A list of miRNAs identified in B. napus by this study.
Supplementary Table S4. Predicted miRNA targets and targets with degradomic evidence.
Supplementary Table S5. Details about miRNAs predicted to target glucosinolate-related genes in Darmor (reference genome), Tapidor, and Ningyou7.
Supplementary Table S6. Lipid-related miRNAs identified in B. napus in this study.
Supplementary Table S7. Differential expressed miRNAs in root and leaf at the seedling stage.
Supplementary Table S8. The 646 miRNAs (Shen et al., 2014) with high similarity (>95%) between B. napus and its two progenitors.
Supplementary Table S9. The 851 miRNAs previously identified in the two progenitor genomes (Shen et al., 2014) mapped to the B. napus genome.
Supplementary Table S10. Changes of conserved miRNA family sizes in B. napus and its two progenitors compared with A. thaliana.
Supplementary Table S11. The loss and gain of miRNAs in B. napus after the hybridization of its two progenitors.
Supplementary Table S12. The additive genetic effects of miRNA expression between DH lines and their parents.
Supplementary Fig. S1. Frequency distribution of small RNA reads with different length by this study.
Supplementary Fig. S2. Expression analysis of nine conserved and 13 novel miRNAs by stem–loop RT-PCR.
Supplementary Fig. S3. Eleven differentially expressed miRNAs in leaf and roots in three B. napus cultivars.
Supplementary Fig. S4. RACE reactions with three selected miRNAs as examples.
Acknowledgements
This work was supported by the National Basic Research Program of China (2011CB109306/2015CB150200), the Ministry of Science and Technology of China (2013IM030700), the National Science Foundation of China (31100876), and the FNR (Fachagentur Nachwachsende Rohstoffe), Germany (22410312). The authors thank DAAD and the China Scholarship Council for travel grants and the Jiangsu Collaborative Innovation Center for Modern Crop Production (JCIC-MCP).
References
- Achard P, Herr A, Baulcombe DC, Harberd NP. 2004. Modulation of floral development by a gibberellin-regulated microRNA. Development 131, 3357–3365. [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. [DOI] [PubMed] [Google Scholar]
- Bancroft I, Morgan C, Fraser F, et al. 2011. Dissecting the genome of the polyploid crop oilseed rape by transcriptome sequencing. Nature Biotechnology 29, 762–766. [DOI] [PubMed] [Google Scholar]
- Berger D, Altmann T. 2000. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes and Development 14, 1119–1131. [PMC free article] [PubMed] [Google Scholar]
- Chalhoub B, Denoeud F, Liu S, et al. 2014. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. [DOI] [PubMed] [Google Scholar]
- Chen X. 2009. Small RNAs and their roles in plant development. Annual Review of Cell and Developmental Biology 25, 21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi WT, Fung RW, Liu HC, Hsu CC, Charng YY. 2009. Temperature-induced lipocalin is required for basal and acquired thermotolerance in Arabidopsis . Plant, Cell and Environment 32, 917–927. [DOI] [PubMed] [Google Scholar]
- Dai X, Zhao PX. 2011. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Research 39, W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. 2003. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Current Biology 13, 1768–1774. [DOI] [PubMed] [Google Scholar]
- Gong L, Kakrana A, Arikit S, Meyers BC, Wendel JF. 2013. Composition and expression of conserved microRNA genes in diploid cotton (Gossypium) species. Genome Biology and Evolution 5, 2449–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb CD, Abel S. 2006. Glucosinolate metabolism and its control. Trends in Plant Science 11, 89–100. [DOI] [PubMed] [Google Scholar]
- Guo HS, Xie Q, Fei JF, Chua NH. 2005. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. The Plant Cell 17, 1376–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XF, Fang YY, Feng L, Guo HS. 2008. Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica . FEBS Letters 582, 2445–2452. [DOI] [PubMed] [Google Scholar]
- Huang D, Koh C, Feurtado JA, Tsang EW, Cutler AJ. 2013. MicroRNAs and their putative targets in Brassica napus seed maturation. BMC Genomics 14, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. 2004. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular Cell 14, 787–799. [DOI] [PubMed] [Google Scholar]
- Kavanagh KL, Jornvall H, Persson B, Oppermann U. 2008. Medium- and short-chain dehydrogenase/reductase gene and protein families: the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cellular and Molecular Life Sciences 65, 3895–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenan-Eichler M, Leshkowitz D, Tal L, Noor E, Melamed-Bessudo C, Feldman M, Levy AA. 2011. Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics 188, 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B, Zhu JK, Zhu J. 2012. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochimica et Biophysica Acta 1819, 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbes AP, Machado RD, Guzman F, et al. 2012. Identifying conserved and novel microRNAs in developing seeds of Brassica napus using deep sequencing. PLoS One 7, e50663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Research 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs P, Peaucelle A, Morin H, Traas J. 2004. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131, 4311–4322. [DOI] [PubMed] [Google Scholar]
- Li FG, Fan GY, Lu CR, et al. 2015. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nature Biotechnology 33, 524–U242. [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, et al. 2013. Acyl-lipid metabolism. The Arabidopsis Book 11, e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. 2008. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana . RNA 14, 836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC. 2007. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. The Plant Journal 52, 133–146. [DOI] [PubMed] [Google Scholar]
- Liu S, Liu Y, Yang X, et al. 2014. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nature Communications 5, 3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Meyers BC, Green PJ. 2007. Construction of small RNA cDNA libraries for deep sequencing. Methods 43, 110–117. [DOI] [PubMed] [Google Scholar]
- Lukasik A, Pietrykowska H, Paczek L, Szweykowska-Kulinska Z, Zielenkiewicz P. 2013. High-throughput sequencing identification of novel and conserved miRNAs in the Brassica oleracea leaves. BMC Genomics 14, 801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma ZR, Coruh C, Axtell MJ. 2010. Arabidopsis lyrata small RNAs: transient MIRNA and small interfering RNA loci within the Arabidopsis genus. The Plant Cell 22, 1090–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B. 2005. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. The Plant Cell 17, 1360–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, et al. 2008. Criteria for annotation of plant MicroRNAs. The Plant Cell 20, 3186–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford SG, Yoshimoto N, Reichelt M, et al. 2009. Disruption of adenosine-5’-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. The Plant Cell 21, 910–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. 2006. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439. [DOI] [PubMed] [Google Scholar]
- Ng DW, Lu J, Chen ZJ. 2012. Big roles for small RNAs in polyploidy, hybrid vigor, and hybrid incompatibility. Current Opinion in Plant Biology 15, 154–161. [DOI] [PubMed] [Google Scholar]
- Niu Y, Wu GZ, Ye R, Lin WH, Shi QM, Xue LJ, Xu XD, Li Y, Du YG, Xue HW. 2009. Global analysis of gene expression profiles in Brassica napus developing seeds reveals a conserved lipid metabolism regulation with Arabidopsis thaliana . Molecular Plant 2, 1107–1122. [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. 2003. Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. [DOI] [PubMed] [Google Scholar]
- Passardi F, Longet D, Penel C, Dunand C. 2004. The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry 65, 1879–1893. [DOI] [PubMed] [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB. 2008. Building lipid barriers: biosynthesis of cutin and suberin. Trends in Plant Science 13, 236–246. [DOI] [PubMed] [Google Scholar]
- Qiu D, Morgan C, Shi J, et al. 2006. A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theoretical and Applied Genetics 114, 67–80. [DOI] [PubMed] [Google Scholar]
- Reintanz B, Lehnen M, Reichelt M, Gershenzon J, Kowalczyk M, Sandberg G, Godde M, Uhl R, Palme K. 2001. bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. The Plant Cell 13, 351–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K, Chen X. 2013. Biogenesis, turnover, and mode of action of plant microRNAs. The Plant Cell 25, 2383–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. 2005. Specific effects of microRNAs on the plant transcriptome. Developmental Cell 8, 517–527. [DOI] [PubMed] [Google Scholar]
- Shen D, Suhrkamp I, Wang Y, Liu S, Menkhaus J, Verreet JA, Fan L, Cai D. 2014. Identification and characterization of microRNAs in oilseed rape (Brassica napus) responsive to infection with the pathogenic fungus Verticillium longisporum using Brassica AA (Brassica rapa) and CC (Brassica oleracea) as reference genomes. New Phytologist 204, 577–594. [DOI] [PubMed] [Google Scholar]
- Song C, Fang J, Wang C, Guo L, Nicholas KK, Ma Z. 2010. a MiR-RACE, a new efficient approach to determine the precise sequences of computationally identified trifoliate orange (Poncirus trifoliata) microRNAs. PloS One 5, e10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Jia Q, Fang J, Li F, Wang C, Zhang Z. 2010. b Computational identification of citrus microRNAs and target analysis in citrus expressed sequence tags. Plant Biology 12, 927–934. [DOI] [PubMed] [Google Scholar]
- Stief A, Altmann S, Hoffmann K, Pant BD, Scheible WR, Baurle I. 2014. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. The Plant Cell 26, 1792–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocks MB, Moxon S, Mapleson D, Woolfenden HC, Mohorianu I, Folkes L, Schwach F, Dalmay T, Moulton V. 2012. The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics 28, 2059–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar RM, Hermanson PJ, Springer NM. 2007. Nonadditive expression and parent-of-origin effects identified by microarray and allele-specific expression profiling of maize endosperm. Plant Physiology 145, 411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Li YF, Jagadeeswaran G. 2012. Functions of microRNAs in plant stress responses. Trends in Plant Science 17, 196–203. [DOI] [PubMed] [Google Scholar]
- Tang S, Wang Y, Li Z, Gui Y, Xiao B, Xie J, Zhu QH, Fan L. 2012. Identification of wounding and topping responsive small RNAs in tobacco (Nicotiana tabacum). BMC Plant Biology 12, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempel S. 2012. Using and understanding RepeatMasker. Methods in Molecular Biology 859, 29–51. [DOI] [PubMed] [Google Scholar]
- Nagaharu N. 1935. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Japanese Journal of Botany 7, 389–452. [Google Scholar]
- Vaucheret H, Vazquez F, Crete P, Bartel DP. 2004. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes and Development 18, 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Legrand S, Windels D. 2010. The biosynthetic pathways and biological scopes of plant small RNAs. Trends in Plant Science 15, 337–345. [DOI] [PubMed] [Google Scholar]
- Voinnet O. 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687. [DOI] [PubMed] [Google Scholar]
- Wang F, Li H, Zhang Y, Li J, Li L, Liu L, Wang L, Wang C, Gao J. 2013. MicroRNA expression analysis of rosette and folding leaves in Chinese cabbage using high-throughput Solexa sequencing. Gene 532, 222–229. [DOI] [PubMed] [Google Scholar]
- Wang L, Yu S, Tong C, et al. 2014. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biology 15, R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, et al. 2011. The genome of the mesopolyploid crop species Brassica rapa . Nature Genetics 43, 1035–1039. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tang H, Debarry JD, et al. 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research 40, e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Grigg SP, Xie M, Christensen S, Fletcher JC. 2005. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132, 3657–3668. [DOI] [PubMed] [Google Scholar]
- Wittstock U, Burow M. 2010. Glucosinolate breakdown in Arabidopsis: mechanism, regulation and biological significance. The Arabidopsis Book 8, e0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Tian Q, Reed JW. 2006. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133, 4211–4218. [DOI] [PubMed] [Google Scholar]
- Xie F, Zhang B. 2015. microRNA evolution and expression analysis in polyploidized cotton genome. Plant Biotechnology Journal 13, 421–434. [DOI] [PubMed] [Google Scholar]
- Xu MY, Dong Y, Zhang QX, Zhang L, Luo YZ, Sun J, Fan YL, Wang L. 2012. Identification of miRNAs and their targets from Brassica napus by high-throughput sequencing and degradome analysis. BMC genomics 13, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wang H. 2007. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Research 35, W265–W268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Guo G, Ni Z, Sunkar R, Du J, Zhu JK, Sun Q. 2007. Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biology 8, R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Wang H, Lu Y, de Ruiter M, Cariaso M, Prins M, van Tunen A, He Y. 2012. Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa . Journal of Experimental Botany 63, 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. 2015. MicroRNA: a new target for improving plant tolerance to abiotic stress. Journal of Experimental Botany 66, 1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TZ, Hu Y, Jiang WK, et al. 2015. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nature Biotechnology 33, 531–537. [DOI] [PubMed] [Google Scholar]
- Zhao YT, Wang M, Fu SX, Yang WC, Qi CK, Wang XJ. 2012. Small RNA profiling in two Brassica napus cultivars identifies microRNAs with oil production- and development-correlated expression and new small RNA classes. Plant Physiology 158, 813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wang G, Zhang W. 2007. UV-B responsive microRNA genes in Arabidopsis thaliana . Molecular Systems Biology 3, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZS, Song JB, Yang ZM. 2012. Genome-wide identification of Brassica napus microRNAs and their targets in response to cadmium. Journal of Experimental Botany 63, 4597–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.