Highlight
Enzyme kinetic measurements and positive selection analysis show that C4 species in Suaedoideae have PEPC and Rubisco kinetics similar to other C4 species despite different amino acid convergence.
Key words: Bienertia, C4 photosynthesis, PAML, phosphoenolpyruvate carboxylase, positive selection analysis, Rubisco, Suaedoideae.
Abstract
The two carboxylation reactions performed by phosphoenolpyruvate carboxylase (PEPC) and ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) are vital in the fixation of inorganic carbon for C4 plants. The abundance of PEPC is substantially elevated in C4 leaves, while the location of Rubisco is restricted to one of two chloroplast types. These differences compared with C3 leaves have been shown to result in convergent enzyme optimization in some C4 species. Investigation into the kinetic properties of PEPC and Rubisco from Kranz C4, single cell C4, and C3 species in Chenopodiaceae s. s. subfamily Suaedoideae showed that these major carboxylases in C4 Suaedoideae species lack the same mutations found in other C4 systems which have been examined; but still have similar convergent kinetic properties. Positive selection analysis on the N-terminus of PEPC identified residues 364 and 368 to be under positive selection with a posterior probability >0.99 using Bayes empirical Bayes. Compared with previous analyses on other C4 species, PEPC from C4 Suaedoideae species have different convergent amino acids that result in a higher K m for PEP and malate tolerance compared with C3 species. Kinetic analysis of Rubisco showed that C4 species have a higher catalytic efficiency of Rubisco (k catc in mol CO2 mol–1 Rubisco active sites s–1), despite lacking convergent substitutions in the rbcL gene. The importance of kinetic changes to the two-carboxylation reactions in C4 leaves related to amino acid selection is discussed.
Introduction
When organisms develop the same solution to an abiotic or biotic stress resulting in a similar character state, it is referred to as convergent evolution or phenotypic convergence. One of the most documented convergent phenotypes in plants is the repeated development of C4 photosynthesis, an adaptation that uses four carbon acids to increase photosynthesis under conditions where carbon assimilation can be limited by high photorespiration (Sage et al., 2012). The number of times that C4 independently developed (at least 66) makes it an extremely useful phenotype for analysing the genetics of adaptations (Christin et al., 2010; Sage et al., 2011).
The genetic mechanisms responsible for C4 photosynthesis remain largely unknown, but they are thought to involve co-ordinated changes to genes that affect leaf anatomy, cell ultrastructure, energetics, metabolite transport, and the location, content, and regulation of many metabolic enzymes (Hibberd and Covshoff, 2010). One approach to gain further insight into the underlying genetic regulation of C4 photosynthesis is to analyse how enzymes are optimized for C4 biochemistry.
In C4 plants there is spatial separation between the capture of atmospheric CO2 with synthesis of C4 acids, and the donation of CO2 to Rubisco by decarboxylation of C4 acids, which, in most species, occurs in mesophyll and bundle sheath (BS) cells, respectively. In the mesophyll cells, atmospheric CO2 is initially converted into bicarbonate ( ) by carbonic anhydrase and the chloroplasts generate phosphoenolpyruvate (PEP) from pyruvate by pyruvate, Pi dikinase. Then, in the cytosol, the and PEP are utilized as substrates for phosphoenolpyruvate carboxylase (PEPC, EC 4.1.1.31) for the synthesis of oxaloacetate (Chollet et al., 1996). The oxaloacetate is subsequently reduced in the chloroplast to malate (MA) by NADP-malate dehydrogenase or transaminated to aspartate (Asp) by Asp aminotransferase. The MA and Asp are transported to BS cells where CO2 is donated to Rubisco via C4 acid decarboxylases. How C4 photosynthesis is regulated, by the level of enzymes and their kinetic properties, their state of activation, and their control by allosteric effectors, is important for understanding the mechanism and how they accomplish high rates of photosynthesis under CO2 limiting conditions.
The kinetic properties of PEPC and Rubisco from C4 plants are different from those in C3 plants, which are considered to have optimized their function in the C4 system (Ghannoum et al., 2005; Gowik and Westhoff, 2011; Whitney et al., 2011b). These differences have led to questions about how these changes occurred during the evolution of C4 from C3 by positive selection on certain amino acid residues. PEPC in C4 plants have high enzymatic activities, as much as 20–40-fold higher than C3 plants (per mg of chlorophyll), and K m values for PEP are several fold higher than C3 plants (Ting and Osmond, 1973; Kanai and Edwards, 1999; Engelmann et al., 2003; Lara et al., 2006). The C4 PEPC can have cooperativity with PEP as substrate (reflected in higher Hill coefficients), be less sensitive to the inhibition of catalysis by Asp and MA, and react to the positive allosteric effectors glucose, 6-phosphate (G6P), glyceraldehyde-3P, and glycine (Gowik and Westhoff, 2011). G6P decreases the K m for PEP (Engelmann et al., 2003; Gowik et al., 2006), and lowers the inhibition by MA (Gupta et al., 1994; Chollet et al., 1996; Engelmann et al., 2003). Positive selection analysis to identify amino acid residues under selection, that may account for the observed kinetic properties of the C4 PEPC, have been made in family Asteraceae (in C3, intermediate, and C4 species in the genus Flaveria), Cyperaceae, and Poaceae (Christin et al., 2007; Besnard et al., 2009; Gowik and Westhoff, 2011). This includes the identification of amino acid substitution at residue 780 to a serine in the C4 species which has been considered a key substitution in PEPC for C4 kinetics.
Rubisco in C4 plants functions where the ratio of CO2 to O2 is elevated, resulting in a decrease of the oxygenase reaction with RuBP and photorespiration. The high CO2 concentration provides selective pressure for a faster turnover of the enzyme under saturating CO2, resulting in higher k catc and K m(CO2) values (Yeoh et al., 1981; Seemann et al., 1984; Sage, 2002; Kubien et al., 2003, 2008; Ghannoum et al., 2005). These kinetic changes to Rubisco in C4 plants allow for a reduced investment in the enzyme, as much as half as in C3 leaves, while achieving higher rates of photosynthesis under warm temperatures and current ambient levels of CO2 due to their CO2-concentrating mechanism (Long, 1999; von Caemmerer, 2013). Rubisco is a heterooctomer composed of multiple small and large subunits which are encoded by nuclear RbcS and chloroplast rbcL genes, respectively (Whitney et al., 2011a). Analyses for rbcL amino acid residues under positive selection in C4 lineages have been made mainly in families Poaceae, Cyperaceae, and Amaranthaceae s.l. (Kapralov and Filatov, 2007; Christin et al., 2008, 2009; Kapralov et al., 2011, 2012).
Among eudicot families, Chenopodiaceae s.s. has the largest number of eudicot C4 species and the most diversity in forms of C4, yet there is no information comparing the kinetic properties of the carboxylases and positive selection of amino acid residues in C4 lineages. The focus of the current study was on subfamily Suaedoideae which has diverse forms of C4 along with C3 species (Edwards and Voznesenskaya, 2011; Kadereit et al., 2012). There are four independent origins of C4 in the subfamily, including two distinct Kranz anatomies in Suaeda sections Salsina s.l. and Schoberia, and two independent origins of single-cell C4 anatomy, in Suaeda aralocaspica and in genus Bienertia (Kapralov et al., 2006; Rosnow et al., 2014). A recent positive selection analysis on C4 PEPC in Suaedoideae showed that there was divergence in where positive selection was occurring compared with previous studies in grasses and sedges (Rosnow et al., 2014). In the current study, the kinetic properties of PEPC across C3 and C4 Suaedoideae species, including the affinity for PEP, the kinetic response to allosteric effectors (G6P and MA), and the degree of cooperativity with varying PEP as substrate, were investigated together with additional PEPC sequence information.
With respect to Rubisco, positive selection analysis for rbcL in Amaranthaceae s.l. showed evidence for selection of residues at positions 281 and 309 among C4 species, which has also been observed in C4 monocots (Kapralov et al., 2012). Also a functional analysis with hybrids of Rubiscos utilizing rbcL genes from C3 versus C4 Flaveria species indicated that a substitution in the rbcL gene at position 309 from a methionine to an isoleucine results in a higher Rubisco k catc (Whitney et al., 2011b). However, the three Suaedoideae C4 species which were previously analysed (Suaeda altissima, S. microphylla, and Bienertia cycloptera) lacked substitutions at 281 and 309 (Kapralov et al., 2012). This raises questions about Rubisco kinetics (k catc) and rbcL sequences in Suaedoideae C4 lineages.
In this study, kinetic properties and sequence information for PEPC and Rubisco from the subfamily Suaedoideae were analysed. The results show that the C4 species have divergent amino acid positive selection resulting in convergent C4-type kinetic properties for PEPC and Rubisco.
Materials and methods
Plant material
All plants used in this study were started from seed and grown in controlled environmental chambers (Econair GC-16; Bio Chambers). Seedlings were started under low light [100 photosynthetic photon flux density (PPFD; μmol quanta m–2 s–1)] and temperature conditions with a day/night temperature of 25/22 °C and a photoperiod of 14/10h. The plants were moved to high light and temperature conditions (1,000 PPFD, with a day/night temperature of 35/25 °C and a photoperiod of 14/10h) once well established. A few leaves, for each replication, were sampled from 2–6-month-old plants and used for kinetic analysis.
Enzyme extraction
Chlorophyll content, the quantity of Rubisco binding sites for RuBP, and Rubisco and PEPC activities, were measured on flash-frozen leaves from plants exposed to at least 5h of light in the chambers, using a liquid-nitrogen-chilled mortar and pestle (the extraction included 250mg leaf tissue plus 1ml extraction buffer). For Rubisco assays the extraction buffer consisted of [100mM 4-(2-hydroxyethyl)-1-piperazinepropanesulphonic acid (EPPS, pH 8.0), 1mM EDTA, and 10mM dithiothreitol (DTT)]; preliminary tests showed no difference in activity with or without the protease inhibitor (Sigma Protease Inhibitor Cocktail,P9599). For PEPC assays, the extraction buffer consisted of [100mM 4-(2-hydroxyethyl)piperazine-1-ethanesulphonic acid (HEPES, pH 7.6), 1mM EDTA, 1mM sodium fluoride, and 10mM dithiothreitol (DTT)]. The PEPC extraction included 1mM sodium fluoride to prevent the possible action of phosphatases on the PEP carboxylase protein. The frozen leaf powder was homogenized in the extraction buffer and, prior to centrifugation, a portion of the extract was placed in 80% acetone for chlorophyll determination (Porra et al., 1989). The extract was centrifuged at 10,000 g relative centrifugal force for 1min at room temperature; the supernatant was collected and placed on ice. In the case of extracts for analysis of PEPC, the supernatant was desalted in a cold Sephadex G-50 column pre-equilibrated with the extraction buffer (to remove low-molecular-weight metabolites including the allosteric effectors malate, aspartate, and G6P, as well as cations, which may affect the assay).
PEPC kinetic assays
Assays were performed immediately following desalting, and there was no apparent loss in activity during the assay period. The activity was coupled to the MA dehydrogenase reduction of OAA and measured as a decrease in absorbance at 340nm resulting from the oxidation of NADH. The standard assay mixture contained 100mM HEPES–KOH (pH 7.6), 10mM MgCl2, 10mM NaHCO3, 0.2mM NADH, 12U NADH-MA dehydrogenase (MP Biomedicals), and 10 μl of enzyme extract in a total volume of 1ml. The reaction was started by the addition of PEP (with or without G6P as indicated). In order to determine the K m, V max, and Hill coefficient for PEP, the Hill equation was fitted to the experimental data by non-linear regression analysis with the software package KaleidaGraph 4.5 (Synergy Software):
(where V, velocity; V max, maximum velocity, K, half maximum rate; S, Substrate PEP; and h, Hill coefficient).
For each species, two independent extractions were analysed and each kinetic measurement was repeated.
The IC 50 for MA, the concentration causing 50% inhibition of PEPC activity, was determined using the coupled spectrometric assay as described above. For each species, the MA inhibition was measured at a PEP concentration which was twice the K m (using the value of K m determined in the presence or absence of G6P). Separate assays were performed with a range of MA concentrations from 0mM to 20mM. The MA IC 50 values are from two independent biological replications, with two technical replications on each. The IC 50 for MA of PEPC was calculated from the experimental data by fitting the same normalized three parameter dose–response curve using the following equation for all species using SAS Proc NLIN (SAS, 2011).
(where V, velocity; V min, minimum velocity; IC 50, concentration of MA causing 50% inhibition; S, substrate PEP; n, slope at IC 50). SAS Proc NLIN was coded so that the response equation was simultaneously computed for all 14 combinations of the seven species and two levels of G6P. This allowed for global tests of equality for IC 50 between species and G6P levels.
Rubisco k catc analysis
From measurement of Rubisco catalytic sites and Rubisco activity, k catc values were determined (mol CO2 mol–1 binding site s–1) (Lilley and Walker, 1974; Collatz et al., 1979; Walker et al., 2013). In the leaf extracts, Rubisco catalytic sites were quantified from the stoichiometric binding of radiolabelled 14C-carboxy-arabinitol-bisphosphate (14CABP). For 14CABP binding assays, 20 μl of enzyme extract was incubated in 150mM EPPS, 18mM MgCl2, 17.5mM and 1mM 14CABP. A portion of the sample was then passed through a low-pressure chromatography column (737-4731; Bio-Rad, Hercules, CA, USA) packed with size exclusion beads (Sephadex G-50 Fine; GE Healthcare Biosciences, Pittsburgh, PA, USA). Samples were analysed in a liquid scintillation counter to quantify binding sites. Rubisco activity was determined spectrophotometrically. Rubisco activity was measured in 1ml of assay buffer (100mM EPPS pH 8.0, 20mM MgCl2, 1mM EDTA, 1mM ATP, 5mM creatine phosphate, 20mM NaHCO3, and 0.2mM NADH) containing coupling enzymes, (12.5U creatine phosphokinase, 250U carbonic anhydrase, 23U 3-phosphoglycerate kinase, 20U glyceraldehyde-3-phosphate dehydrogenase, 56U triose-phosphate isomerase, and 20U glycerol-3-phosphate dehydrogenase), and 10 μl of enzyme extract. Rubisco was activated for 10min at 25 °C, before the addition of coupling enzymes and initiation with 0.5mM RuBP. The activity of Rubisco was determined from the rate of conversion of NADH to NAD+, which was monitored by the change in absorbance at 340nm.
DNA sequencing and analysis
PEPC and Rubisco large subunit (L-subunit) genes, ppc-1 and rbcL, were sequenced for 17 Suaeda species and two Bienertia species. DNA was extracted from 250mg of plant material using the CTAB method following the protocol of Doyle and Doyle (1987). Primers were developed based on homology to previously published sequences (see Supplementary Table S1 at JXB online). Initial PCR conditions were 2min at 95 °C, followed by 35 cycles of: 30 s at 95 °C, 30 s at 52 °C annealing step, and a 3min extension at 72 °C. The PCR product was visualized and purified using a PCR clean-up kit according to the manufacturer’s protocol (Qiagen, USA). For ppc-1, purified PCR product was cloned into the pGEM T-easy vector using the manufacturer’s protocol (Promega, USA). Single colonies were grown overnight and plasmid DNA was purified using alkaline lysis with SDS (Sambrook and Russell, 2001). Plasmid inserts were PCR amplified using GOTaq (Promega, USA), Sp6 and T7 primers, and were visualized on a gel. Prior to sequencing, the PCR product was mixed with 2.5U of Antarctic Phosphatase and 4U of Exo-Sap Nuclease in Antarctic Phosphatase buffer (New England BioSciences, USA) to degrade primers and nucleotides, and subsequently diluted 1:10. Sequencing reactions were performed using the Big Dye terminator master mix v3.1 (Applied BioSciences, USA), using sequence specific internal primers along with Sp6 and T7 (see Supplementary Table S1 at JXB online). Sequencing was carried out at Washington State University genomics core. Sequence data was assembled using Sequencher software (USA). Nucleotide sequences were translated, aligned, and visualized using Se-Al and MacVector (USA). All sequences were deposited in GenBank (see Supplementary Table S2 at JXB online). Positive selection analysis on additional N-terminus PEPC residues was performed using the methodologies of Rosnow et al. (2014). The same phylogenetic tree from the previous study was used for selection analysis, but was pruned to exclude S. heterophylla, Salsola genistoides, and Salsola divaricata as these species were not sequenced in this study. Throughout this paper, the numbering of PEPC residues is based on the Zea mays ppc-B2 sequence CAA33317 (Besnard et al., 2003) for easy comparison with previous studies.
δ13C determination
Measurements of carbon isotope fractionation values (δ13C) were made on all Suaedoideae species used for kinetic analysis to verify photosynthetic type (see Supplementary Table S3 at JXB online). Analyses were made at Washington State University on leaf samples taken from plants grown in growth chambers. A standard procedure relative to Pee Dee Belemnite (PDB) limestone as the carbon isotope standard (Bender, 1973). Plant samples were dried at 80 °C for 24h, then 1–2mg was placed in a tin capsule and combusted in a Eurovector elemental analyser. The resulting N2 and CO2 gases were separated by gas chromatography and admitted into the inlet of a Micromass Isoprime isotope ratio mass spectrometer (IRMS) for determination of 13C/12C ratios (R). δ13C values were determined where δ=1 000(R sample/R standard)−1.
Statistical analysis
For PEPC kinetic parameters determined from varying response to PEP, the statistical design was completely randomized with a two-way treatment structure (nine species, with and without G6P). SAS Proc MIXED (SAS, 2011) was used to compute parameter estimates and test statistics. The assumption of equal variances was assessed and determined to have been violated for all three response variables (K m, Hill coefficient, and V max) with a P-value <0.0001. Because of this, the variances were modelled as part of the mixed model analysis that was used to assess the main effects of species and G6P, along with the interaction between species and G6P. Fisher’s LSD was used to assess pairwise comparison between means. In addition, contrasts were also used to assess whether differences existed between linear combinations of the cell means (average value for each parameter) as they related to the different photosynthetic modes and sequence types. In particular, the photosynthetic modes compared with contrasts were the Kranz C4, single-cell C4, and C3 representatives in Suaedoideae, and comparisons made with the monocot Z. mays. In addition, contrasts were also computed to compare species differences at PEPC residues 733 and 780. All comparisons were taken to be significant at the P <0.05 level.
For Rubisco k catc values, one-way analysis of variance (ANOVA) was performed using Sigma-Plot version 11.0 software (Systat Software Inc.). Post-hoc analysis was used to test statistical significance. All comparisons were taken to be significant at the P <0.05 level.
Results
PEPC sequence analysis
To complement previous sequence and phylogenetic information on PEPC in Suaedoideae (Rosnow et al., 2014), N-terminal PEPC sequence was obtained for 19 species (S. heterophylla, Salsola genistoides, and Salsola divaricata were not included) using homologous upstream primers that overlapped with known C-terminal ppc-1 sequence. The region of coverage included part of exon 2 through exon 8, stopping where previous C-terminal sequence analysis had been performed (Rosnow et al., 2014). The sequenced region resulted in an additional 370 N-terminal amino acids of the ppc-1 coding sequence. Based on gene homology to previously sequenced Alternanthera species PEPCs (Gowik et al., 2006), this is approximately 87 N-terminal amino acids short of complete ppc-1 gene coverage.
Positive selection analysis, using phylogenetic relationships, models amino acid change identifying significant non-synonymous amino acid changes; for model descriptions see Rosnow et al. (2014) (Yang, 2007). There were no codons identified as being under positive selection with a posterior probability >0.95 by BEB in the M2A model or M8 model (see Supplementary Table S4 at JXB online) (P value=0.82 and 0.0071, respectively). There were seven codons (99, 171, 324, 333, 364, 365, 368) that were shown to be under positive selection with a posterior probability >0.95 by BEB, when only branches leading to C4 clades were labelled as foreground branches (P value <0.0001). Positions 364 and 368 were the only two residues identified to have a posterior probability >0.99 by BEB in Model A, when only branches leading to C4 clades were labelled as foreground branches (see Supplementary Table S4 at JXB online). Residues 364 and 368 are in the N-terminal region which is shown to be involved in the allosteric regulation of activators like G6P (Blasing et al., 2002; Engelmann et al., 2002; Takahashi-Terada et al., 2005). Residue 364 had four alternative amino acids present in this dataset, Arg present in C3 species, and either Lys, Gln, or Pro in C4 species (in order of prevalence). Residue 368 has Asn present in C3 species and Ser in C4 species. Both residues had a substitution in all C4 species (see Supplementary Fig. S1 at JXB online). There were no codons shown to be under positive selection with a posterior probability >0.95 by BEB, when foreground branches leading to Kranz C4 clades or branches leading to single-cell C4 clades alone were labelled (see Supplementary Table S4 at JXB online) (P values=0.65 and 1, respectively). By labelling all C4 branches as foreground branches, 10 codons were identified as being under positive selection (157, 159, 171, 198, 314, 318, 324, 353, 364, 368) with a posterior probability >0.95 by BEB (see Supplementary Table S4 at JXB online) although the results are not significant (P value=0.22). Four of these residues (171, 324, 364, 368) were identified as being on branches leading to C4 clades.
PEPC kinetics
The K m value for a given substrate in Michaelis–Menten kinetics is the concentration at which the rate of reaction is at half the maximal rate, which generally has an inverse relationship to affinity of enzyme for substrate. Since some forms of PEPC show cooperativity with PEP as substrate, the Hill equation was used to determine the K m, the Hill coefficient (h) for PEP, and V max. The analyses of PEPC kinetics in members of subfamily Suaedoideae representing different photosynthetic types, are shown in Tables 1–4 and Fig. 1. (see Supplementary Table S3 at JXB online for δ13C values of C4 and C3 species in the study)
Table 1.
PEPC K m-PEP values (pH 7.6) in representative species in subfamily Suaedoideae
Values were determined by curve-fitting the Hill equation to the data. Values represent the average of two biological and two technical replicates. The amino acid residues at positions 733 and 780, M (methionine), S (serine), L (leucine), A (alanine), F (phenylalanine), and V (valine) for the species are from Rosnow et al. (2014) and Besnard et al. (2003) for Z. mays.
| Species | Photosynthetic | Amino acid at | Amino acid | K m PEP (mM) | Hill coefficient (h) | Fold increase by G6P | ||
|---|---|---|---|---|---|---|---|---|
| mode | residue 733 | at residue 780 | No G6P | 5mM G6P | No G6P | +G6P | at 0.3mM PEP | |
| S. accuminata | Schoberia Kranz C4 | M | S | 0.83±0.12 | 0.10±0.01 | 2.61±0.12 | 1.13±0.19 | 4.3 |
| S. eltonica | Schoberia Kranz C4 | M | S | 1.04±0.14 | 0.14±0.02 | 2.71±0.24 | 0.90±0.08 | 2.7 |
| S. moquinii | Salsina Kranz C4 | L | A | 0.46±0.12 | 0.14±0.01 | 1.45±0.06 | 0.96±0.07 | 3.2 |
| S. fruticosa | Salsina Kranz C4 | L | A | 0.67±0.11 | 0.14±0.04 | 1.78±0.21 | 0.98±0.06 | 3.8 |
| S. aralocaspica | Single-Cell C4 | L | A | 0.74±0.08 | 0.14±0.02 | 2.13±0.22 | 0.90±0.03 | 2.7 |
| S. linearis | C3 | F | A | 0.21±0.01 | 0.03±0.01 | 1.19±0.15 | 1.29±0.25 | 2.3 |
| S. physophora | C3 | F | A | 0.27±0.02 | 0.05±0.01 | 0.94±0.15 | 0.93±0.11 | 2.1 |
| S. linifolia | C3 | F | A | 0.35±0.03 | 0.04±0.01 | 0.95±0.02 | 0.90±0.07 | 2.6 |
| Zea mays | C4 | V | S | 1.23±0.07 | 0.15±0.08 | 1.18±0.01 | 1.17±0.07 | 5.0 |
Table 4.
Estimates of malate IC 50 values for half-maximum inhibition of PEPC activity at pH 7.6 (PEP concentration, 2× the K m) in representative photosynthetic types in subfamily Suaedoideae
The amino acid residues potentially involved in malate tolerance are presented. For species comparisons, different letters indicate a significant difference within a category of G6P (+ or –) while comparison of G6P levels within a species is indicated by an asterisk (*) for significance at the P <0.05 level.
| Species | Photosynthetic | Residue at | Residue at | Residue at | Residue at | IC 50 (mM) | IC 50 (mM) | Significant |
|---|---|---|---|---|---|---|---|---|
| mode | 780 | 868 | 879 | 890 | No G6P | 5mM G6P | effect of G6P | |
| S. accuminata | Schoberia Kranz C4 | S | R | D | R | 0.6 c | 1.4 c | * |
| S. eltonica | Schoberia Kranz C4 | S | R | D | R | 1.0 b | 1.9 b | * |
| S. moquinii | Salsina Kranz C4 | A | L | N | M | 4.5 a | 5.9 a | – |
| S. fruticosa | Salsina Kranz C4 | A | L | N | M | 4.5 a | 5.2 a | – |
| S. aralocaspica | Single Cell-C4 | A | Q | E | R | 0.9 b | 1.6 bc | * |
| S. physophora | C3 | A | K | D | R | 0.3 d | 0.9 d | * |
| S. linifolia | C3 | A | K | D | R | 0.3 d | 0.8 d | * |
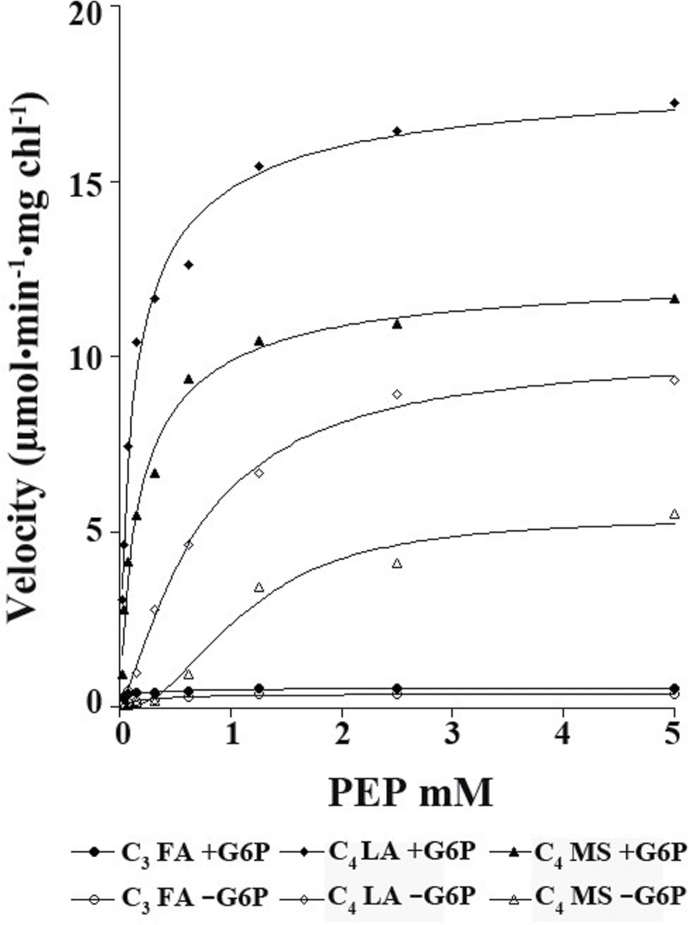
Fig. 1.
Representative PEPC kinetics based on photosynthetic mode (C4 or C3) and the amino acids present at residue 733 (M, L, or F) and residue 780 (A or S). PEPC rates were obtained at pH 7.6 with saturating Mg2+ and bicarbonate, while varying phosphoenolpyruvate (PEP) concentrations, either in the presence or absence of 5mM glucose-6-phosphate (G6P). The mean data points are presented for each PEPC type while species values are presented in Table 3. The solid line is the Hill equation fit to the data, see the Materials and methods for the equation details.
In Table 1, results are shown with PEPC for three forms of C4, Kranz-type Schoberia C4 (two species) Kranz-type Salsina C4 (two species), the single cell C4 S. aralocaspica, along with C3 type Suaeda (three species), and the C4 monocot Z. mays. Substitutions on amino acid residues at positions 733 and 780 are candidates for affecting the K m for PEPC (Rosnow et al., 2014). The four combinations among the Suaeda species for 733 and 780, respectively, are MS Schoberia, LA Salsina, LA single-cell C4, and FA C3 species. The results are shown from kinetic analyses for K m PEP, the Hill coefficient, and the effect of the allosteric effector G6P. In Table 2, statistical analyses are shown for significant differences in K m for PEP, the Hill coefficients, and the V max of PEPC. Contrasts 1–3 in Table 2 show where there are differences in these parameters based on the three combinations of residues in Suaeda species at positions 733 and 780 (MS, LA, and FA). Contrasts 4–9 show where there are differences in these parameters based on the four photosynthetic types of Suaeda.
Table 2.
Contrasts of Suaedoideae PEPC kinetic parameters (K m for PEP, Hill coefficient, and V max) under saturating Mg2+ and
Contrasts were done based on amino acid residues 733 (M, L, or F) and 780 (S or A) or photosynthetic mode; see Table 1. In contrasts 1–3, amino acids are compared, MS (C4) occurs in Schoberia species, LA (C4) occurs in Salsina species and single-cell S. aralocaspica, and FA (C3) represents C3 species. In contrasts 4–9, the four photosynthetic modes are being compared, Schoberia type C4 (S. eltonica and S. accuminata), Salsina type C4 (S. fruticosa and S. moquinii), single-cell C4 (S. aralocaspica), and C3 species (S. linearis, S. linifolia, and S. physophora). *, Significant at the P <0.05 level of significance. (+) Indicates whether the first component of the contrast is larger than the second component and (–) indicates that the first component of the contrast is smaller than the second component. Maize data were excluded from analysis.
| Contrast | K m | Hill coefficient | PEPC V max | |||
|---|---|---|---|---|---|---|
| No G6P | 5mM G6P | No G6P | 5mM G6P | No G6P | 5mM G6P | |
| 1. MS (C4)×LA (C4) | *(+) | (–) | *(+) | (+) | (–) | (–) |
| 2. MS (C4)×FA (C3) | *(+) | *(+) | *(+) | (+) | *(+) | *(+) |
| 3. LA (C4) x FA (C3) | *(+) | *(+) | *(+) | (+) | *(+) | *(+) |
| 4. Schoberia C4 (MS)×Salsina C4 (LA) | *(+) | (–) | *(+) | (+) | *(–) | *(–) |
| 5. Schoberia C4 (MS)×SC C4 (LA) | (–) | (–) | (+) | (+) | *(–) | *(–) |
| 6. Salsina C4 (LA)×SC C4 (LA) | (+) | (+) | (–) | (+) | (+) | (–) |
| 7. Schoberia C4 (MS)×C3 (FA) | *(+) | *(+) | *(+) | (–) | *(+) | *(+) |
| 8. Salsina C4 (LA)×C3 (FA) | *(+) | *(+) | *(+) | (–) | *(–) | *(+) |
| 9. SC C4 (LA)×C3 (FA) | *(+) | *(+) | *(+) | (–) | *(+) | *(+) |
Absence of G6P
In kinetic analyses, in the absence of the allosteric effector G6P, the K m values for PEP were higher in the three types of C4 species than in the C3 species (Table 1; Table 2, contrasts 7–9, P <0.05). Unlike the C4 species, the C3 species have a Phe (F) residue at position 733 (Table 1). Among the C4, the K m PEP was significantly higher in Kranz Schoberia species (with a Met at 733 and a Ser at residue 780) compared with the Kranz Salsina species (with a Leu at 733 and an Ala at 780; Table 1; Table 2, contrast 4). In addition, the K m PEP for MS Schoberia was significantly higher than the K m PEP for the species having LA residues (Salsina C4 and single-cell C4; Table 2, contrast 1). There was no significant difference in the contrast between the single-cell C4 species (SC C4-LA) with either of the Kranz-type C4 species (Table 2, contrasts 5 and 6).
The cooperativity of PEPC for the binding of PEP was investigated by determining the Hill coefficient, from the curve fitting of the Hill equation to the data set. In the absence of G6P, the Hill coefficients were higher in C4 species (from 1.45–2.71) than in the C3 species (0.94–1.19) indicating cooperativity in binding of PEP in C4 species and no cooperativity in the C3 species (Table 1; Fig. 1; Table 2, contrasts 7–9). There was the same pattern of significant differences among species in the Hill coefficients as in the K m for PEP. The C4 species had higher Hill coefficients than C3 species, the Schoberia C4 had higher Hill coefficients than the Salsina C4, while there was no significant difference in the coefficients between the single-cell C4 species S. aralocaspica and either Kranz C4 type (Table 2, contrasts 4–9).
Presence of G6P
In the presence of G6P there was a large decrease in the K m for PEP in both C4 and C3 species (Table 1) which was significantly different in the absence of G6P (see Supplementary Table S5 at JXB online). This difference is highlighted by the fold increase in rate at 0.3mM PEP, where all species had at least a 2-fold increase in rate in the presence of G6P, with the highest increase in rate being found in C4 species (Table 1). The K m values for PEP in the presence of G6P were higher in the C4 than in the C3 species (Table 1; Table 2, contrasts 2, 3, and 7–9). Among the C4 species there were no significant differences in K m for PEP (Table 2, contrasts 1, and 4–6).
In the presence of G6P there was a large decrease in the Hill coefficient in the C4 species, whereas there was no significant difference in the C3 species with and without G6P (Table 1; see Supplementary Table S5 at JXB online). With G6P, in both C3 and C4 species the Hill coefficients were low (0.9–1.29) indicating little or no cooperativity in binding of PEP, and there was no significant difference in the coefficients between the photosynthetic groups (Table 2).
The change in K m values for PEP in Z. mays with or without G6P was similar to that in the C4 Suaeda species. With the addition of G6P, the K m PEP decreased from 1.23 to 0.15mM in Z. mays, with values in the presence of G6P similar to the C4 Suaeda species. Unlike the C4 Suaeda species, the Hill coefficients in Z. mays were low with and without G6P (~1.2) indicating no change in cooperativity with the allosteric effector (Table 1; see Supplementary Table S5 at JXB online).
V max
The maximum velocity (V max on a chlorophyll basis) of the PEPC reaction was determined from the curve-fitting of the Hill equation; as expected, C4 species had much higher PEPC rates than C3 species (Table 3; Table 2, contrasts 7, 8, and 9). There was a significant increase in V max for each C4 species in the presence of G6P, where the mean fold increase was 1.8 (Table 3; see Supplementary Table S5 at JXB online). In the C3 species, G6P had no significant effect on V max in two of the C3 species; in C3 S. linearis, which had very low activity, there was some increase with G6P (Table 3; see Supplementary Table S5 at JXB online). Among the C4 contrasts, the V max in the Salsina C4-LA is higher than Schoberia C4-MS, and SC C4-LA is higher than Schoberia C4-MS, while there is not a significant difference between Salsina C4-LA and SC C4-LA (Table 2, contrasts 4, 5, and 6).
Table 3.
PEPC V max values with saturating Mg2+ and for representative species in subfamily Suaedoideae at pH 7.6
| Species | Photosynthetic mode | V max (μmol min–1 mg–1 chl) | Fold increase in activity in presence of G6P | |
|---|---|---|---|---|
| No G6P | + 5mM G6P | |||
| S. accuminata | Schoberia Kranz C4 | 4.6 | 10.0 | 2.2 |
| S. eltonica | Schoberia Kranz C4 | 6.5 | 12.9 | 2.0 |
| S. moquinii | Salsina Kranz C4 | 9.9 | 15.6 | 1.6 |
| S. fruticosa | Salsina Kranz C4 | 11.5 | 17.6 | 1.5 |
| S. aralocaspica | Single-Cell C4 | 16.5 | 24.1 | 1.5 |
| S. linearis | C3 | 0.1 | 0.4 | 3.5 |
| S. physophora | C3 | 1.0 | 0.8 | 0.8 |
| S. linifolia | C3 | 0.4 | 0.4 | 1.1 |
| Zea mays | C4 | 15.1 | 21.1 | 1.4 |
Figure 1 shows the differences in activity in response to varying PEP for the different types of PEPC according to amino acid residues (LA type in Salina and SC-C4, MS type in Schoberia, and FA type for C3 species), with and without G6P. On a chlorophyll basis at high PEP, the C4 LA type has higher activity than the C4 MS type, while the C3 species have very low activity. Both the LA type and the MS type respond in a similar way. At 5mM PEP, the addition of G6P results in about a 2-fold increase in activity. In both types, at low levels of PEP, there is a large increase in activity with the addition of G6P as a consequence of lowering the K m for PEP. This increase in activity by G6P at low PEP (e.g. at ~0.5mM PEP) is more dramatic in the MS type Schoberia (Fig. 1), which has a higher K m for PEP and a higher Hill coefficient in the absence of G6P than the LA type (Table 1).
Malate inhibition
The concentration of a metabolic inhibitor that reduces the rate of an enzyme by 50% (IC 50) is a useful determination in considering how in vivo metabolites might regulate enzyme activity. Table 4 shows the IC 50 values for MA with species representing different photosynthetic types in Suaedoideae. Amino acid differences are shown for residues 868, 879, and 890, along with residue 780 which are candidates for affecting the IC 50 for MA (Kai et al., 2003; Paulus et al., 2013b). There was a significant increase in the IC 50 values in the presence of G6P in all species except in Salsina. The two Salsina C4 species had IC 50 values that were significantly higher, with and without G6P, than any other species tested; the Salsina species also had different amino acid residues at position 868, 879, and 890. PEPC in the C3 species were the most sensitive to MA, both in the presence and absence of G6P. The two Schoberia C4 species that have Ser at residue 780 and Arg at residue 868, had IC 50 values which were higher than the C3 species, but lower than Salsina C4 species (Table 4). The C3 species, which had the lowest IC 50 values, were different from other species in having Lys at residue 868. The IC 50 values for the single cell C4 S. aralocaspica were similar to the Schoberia type, and was different from other types in having a Gln residue at 868, and a Glu residue at 879. There was no significant difference in PEPC IC 50 values based on the presence of a Ser versus an Ala residue at 780.
Rubisco rbcL sequence information
A full-length rbcL sequence was generated for 20 Suaedoideae species, including at least two species from each Suaedoideae clade. There were 19 polymorphic Rubisco large subunit residues across the Suaedoideae species analysed, but none of the amino acid substitutions was invariantly fixed across C4 species (see Supplementary Table S6 at JXB online). Two C3 (S. linifolia, S. vera) and two C4 (S. accuminata, S. aralocaspica) species, representing four different sections, had identical amino acid sequences (see Supplementary Table S6 at JXB online). The PAML branch-site test for positive selection did not show significant evidence for selection along C4 branches (data not shown).
Rubisco k catc
Measurement of Rubisco k catc using the coupled enzyme assay showed that C4 species had significantly higher values than C3 species (Table 5). The average k catc value for C4 was 2-fold higher than that of C3 species (3.6 versus 1.8mol CO2 mol–1 binding sites s–1). There was no significant difference in Rubisco kcatc between the single-cell C4 and Kranz species.
Table 5.
Rubisco kcatc (mol CO2 mol–1 binding sites s–1) values for representative photosynthetic types in subfamily Suaedoideae
One way analysis of variance a,b=statistically significant difference based on photosynthetic mode (C3 or C4) (P <0.05).
| Species | Photosynthetic mode | Rubisco k catc | SD |
|---|---|---|---|
| S. accuminata | Schoberia Kranz C4 | 2.95 | 0.33 |
| S. eltonica | Schoberia Kranz C4 | 3.34 | 1.18 |
| S. moquinii | Salsina Kranz C4 | 3.76 | 0.30 |
| S. fruticosa | Salsina Kranz C4 | 4.23 | 0.46 |
| S. altisima | Salsina Kranz C4 | 3.19 | 0.17 |
| Mean Kranz C4 | 3.49 b | ||
| S. aralocaspica | Single-Cell C4 | 3.77 | 0.39 |
| Bienertia cycloptera | Single-Cell C4 | 3.90 | 0.27 |
| Bienertia sinuspersici | Single-Cell C4 | 3.46 | 0.34 |
| Mean Single-Cell C4 | 3.71 b | ||
| Zea mays | Kranz C4 | 3.58 | 0.00 |
| S. linearis | C3 | 1.77 | 0.28 |
| S. physophora | C3 | 1.52 | 0.09 |
| S. linifolia | C3 | 2.08 | 0.17 |
| S. vera | C3 | 1.82 | 0.49 |
| Mean C3 | 1.80 a |
Discussion
PEPC kinetic features in Suaedoideae: V max, affinity for PEP, regulation by G6P and MA
The maximum activities of PEPC (V max, μmol mg–1 chlorophyll) from leaves of the C4 Suaeda species were much higher than the C3 species, which is characteristic of C4 plants (Kanai and Edwards, 1999). In addition, compared with the C3 species, all of the C4 species analysed had a significantly higher K m for PEP, both in the absence and presence of G6P (Tables 1, 2), which is the same general trend that has been reported throughout the literature (Svensson et al., 1997; Gowik et al., 2006; Lara et al., 2006; Jacobs et al., 2008). From studies in the genus Flaveria with ppc-2, the location of amino acids responsible for an increase in PEPC K m was shown through reciprocal domain swapping to be in region 2 (amino acids 302–442) and region 5 (amino acids 651–966). In region 5, the single amino acid change to a Ser at residue 780 was suggested to be an important substitution resulting in the increase in K m in C4 PEPC (Blasing et al., 2000; Engelmann et al., 2002). Subsequently, this substitution has been considered to be a key substitution for increasing the K m PEP from analyses of various C4 species (Christin et al., 2007; Besnard et al., 2009; Gowik and Westhoff, 2011). However, the results of the current study, and from analysis of Hydrilla verticillata (a facultative aquatic C4 species) PEPC (Rao et al., 2008), suggest that alternative substitutions can change the affinity for PEP. In Suaedoideae C4 species, a substitution at residue 733 in region 5 is a candidate for raising the K m, and the cooperativity in PEP binding (higher Hill coefficients).
Previous investigations on PEPC in C4 plants showed that the addition of phosphorylated sugars (e.g. G6P and triose-P) reduced the sigmoid nature of Michaelis–Menten kinetics plots, reducing the Hill coefficient to near one, demonstrating that allosteric activators can reduce the cooperative binding of PEP (Coombs and Baldry, 1975; Huber and Edwards, 1975; Nakamoto et al., 1983; Bauwe and Chollet, 1986; Doncaster and Leegood, 1987; Tovar-Mendez et al., 2000; Engelmann et al., 2003; Gowik et al., 2006). In addition, G6P has been shown to crystallize in the active site of the Flaveria trinervia ppc-2 gene, demonstrating that it can also act as a competitive inhibitor (Schlieper et al., 2014). In the present study, inclusion of the allosteric effector G6P in the assay of PEPC (pH 7.6) lowered the K m for PEPC and increased enzyme activity in both the C3 and C4 species of Suaeda. However, in the absence of G6P, the C4 species showed cooperativity with PEP (the mean Hill coefficient for five species is 2.1) while the PEPC in C3 species showed no cooperativity (the mean Hill coefficient for three species is 1.0). This suggests certain substitutions in the C4 PEPC result in both an increase in K m for PEP and an increase in the cooperativity of PEP binding.
Region 2 in the N-terminus was previously identified as the G6P regulatory site in C4 PEPC in Z. mays and it has also been suggested to influence the affinity of the enzyme for PEP (Kai et al., 2003). In a study of representative photosynthetic types in Flaveria, residue 352 in region 2 of ppc-2 (aka ppcA) was the only amino acid that showed differences between the C4 and C4-like Flaveria species which have a Lys residue, while the C3 and C3–C4 intermediate Flaveria have an Arg residue at this position. The C4 PEPC in Z. mays also has a Lys at residue 352 (Engelmann et al., 2003). By contrast, current analysis of the N-terminus in Suaeda species showed position 352 is either a Thr or Ser residue (see Supplementary Fig. S1 at JXB online), and this residue is also an invariant Thr across Alternanthera PEPCs (Gowik et al., 2006). In the Suaeda species, positive selection was found in region 2 at residues 364 (for Gln) and 368 (for Ser; see Supplementary Table S5 and Supplementary Fig. S1 at JXB online). The Alternanthera ppc-1 gene has positive selection for Ser at residue 368, while residue 364 is invariant. Interestingly, the ppc-1 gene in Z. mays and the ppc-2 gene of F. trineriva (C4) has Asn at residue 368 (the same residue observed in all Suaedoideae C3 PEPC), while F. pringeli (C3) has Ser (the same amino acid observed in all Suaedoideae C4 PEPC). These results suggest that paralogous genes (ppc-1 versus ppc-2) have undergone different selection processes. In C4 Suaedoideae and C4 Alternanthera ppc-1, substitution at residue 368 is a candidate for affecting the cooperativity with PEP as substrate, and regulation by binding G6P as an allosteric effector.
From species surveyed in Suaedoideae, the IC 50 values for MA indicate that it is an effective inhibitor of PEPC at mM levels (Table 4). PEPC from C3 Suaeda species was more sensitive to inhibition by MA (assayed either with or without G6P) which is consistent with other studies where C4 PEPCs are generally reported to be more tolerant to MA compared with C3 orthologous genes or paralogous genes (Svensson et al., 1997; Dong et al., 1998; Blasing et al., 2002; Paulus et al., 2013a). Among the C4 Suaeda, the two Salsina Kranz species, had significantly higher IC 50 values indicating higher tolerance to MA (when assayed in the presence or absence of G6P), compared with the Schoberia Kranz species and the single-cell C4 species S. aralocaspica. Also, with the addition of G6P, the IC 50 for MA increased in the C4 Schoberia and C3 species, but not in the C4 Salsina. Studies on Z. mays show C4 PEPC has an allosteric site that binds MA and Asp, which is so close to the catalytic site that these metabolites act competitively with the substrate PEP, resulting in a less active enzyme (Izui et al., 2004). In Suaedoideae, an amino acid substitution at residue 868 (Leu) is observed in all C4 species studied in the subfamily except Bienertia (see Supplementary Fig. S1 at JXB online) which may explain the difference in IC 50 values between C3 and C4 species. The high IC 50 values for MA in the Salsina species may be linked to their PEPC having, in addition to substitution at 868, substitutions at 879 (Asp), and 890 (Met) which is different from the other Suaeda species based on previous C-terminal PEPC sequence information (Rosnow et al., 2014). Amino acid substitution at 868 is also observed in Z. mays and other Amaranthaceae C4 ppc-1 genes, but not Flaveria C4 ppc-2. In other studies on MA inhibition of PEPC, a substitution at residue 884 from an Arg to a Gly in Flaveria was recently shown to increase tolerance to MA (Paulus et al., 2013b). This substitution is also observed in some, but not all, C4 grass species (Paulus et al., 2013a). However, this substitution is not observed in any C4 Suaedoideae species (Rosnow et al., 2014). Using heterologously expressed chimeric ppc-2 enzymes from Flaveria, the replacement of Ala 780 by Ser caused a slight increase in MA tolerance (observed in the presence, but not in the absence of G6P), which was not considered as the main determinant for higher MA tolerance in C4 PEPC (Jacobs et al., 2008). In the present study, the highest tolerance to MA was in the Salsina C4 species, which have an Ala 780 and which also indicates other residues are the main determinants of tolerance.
The lack of strong convergence for a substitution near the MA/Asp allosteric pocket, suggests that there is less selective pressure on increasing MA tolerance than on increasing the K m for PEP (decreased affinity), and G6P activation. Tolerance to MA may increase with alternative substitutions at different residues, without convergent amino acids, together with G6P activation and phosphorylation of PEPC in the light reducing sensitivity to MA. In the light, C4 PEPC is regulated by phosphorylation at a conserved N-terminus Ser residue, which leads to activation of the enzyme by reducing its sensitivity towards the allosteric inhibitors MA and Asp (Jiao and Chollet, 1991; Vidal and Chollet, 1997; Nimmo, 2003).
The current results raise questions about the molecular route for a C4 PEPC to acquire modified kinetic properties; i.e. modification to the allosteric activator site (residue 364/368) before or after increasing PEP K m near the reaction site (733/780), and the impact of the order of mutations on selective pressure. Further analyses are needed to address the influence of amino acid substitutions on PEPC tolerance to MA in C4 Suaedoideae and other C4 species, versus the impact of in vivo phosphorylation of PEPC in the light (in this study extractions were made in the light).
Overall, the results indicate that phylogenetically distant C4 origins can optimize PEPC with divergent amino acid substitutions. The kinetic properties of C4 PEPC are considered to be optimized for function in C4 photosynthesis without interference with other metabolic processes. During the day, the positive allosteric effectors triose-P, G6P, and glycine are produced during photosynthesis in C4 plants (Leegood and von Caemmerer, 1988, 1989; DeVeau and Burris, 1989; Zelitch et al., 2009). These positive allosteric effectors increase the affinity of PEPC for PEP and its effective use in the C4 cycle while the IC 50 values for the C4 acids MA and Asp increases, which minimizes inhibition by products of C4 photosynthesis. Activity of PEPC at night can be controlled by the enzyme having a high K m for PEP, due to relatively low levels of positive allosteric effectors (G6P, triose-P, and glycine) and by the non-phosphorylated form of the enzyme at night having a low IC 50 for C4 acids (Doncaster and Leegood, 1987).
Convergent evolution of Rubisco kinetics in C4 Suaedoideae achieved via non-parallel amino acid substitutions
In the current study of Suaedoideae, the determination of Rubisco k catc showed that the enzyme in C4 species representing four lineages has, on average, approximately 2-fold higher catalytic rates than the C3 species. The mean k catc value for these C4 species (all NAD-ME type), are similar to those of NAD-ME type C4 grasses (Ghannoum et al., 2005). Although the k catc values in Suaedoideae are higher for the C4 than the C3 species, sequence analysis of rbcL did not show any evidence for positive selection across lineages which could account for this adaptation.
The C4 species S. aralocaspica (section Borszczowia) and S. accuminata (section Schoberia) and the C3 species S. linifolia (section Schanginia) and S. vera (section Suaeda) have Rubisco large-subunit sequences that are identical (see Supplementary Table S6 at JXB online). This suggests that, in some C4 species, Rubisco with higher specific activity evolved via amino acid changes in the Rubisco small subunits. Positive selection on the small subunit encoding RbcS gene has previously been demonstrated for C4 Flaveria, which was strongly correlated with higher k catc values and weakly correlated with higher K m(CO2) values (Kapralov et al., 2011). Although Rubisco catalytic sites are located within the large subunits, significant changes in kinetics were shown when small subunits from C3 rice were replaced with those from C4 Sorghum, suggesting a differential role of S-subunits in Rubisco kinetics (Ishikawa et al., 2011). Further work is necessary to determine if there are amino acids encoded by certain RbcS genes that are under positive selection, and candidates for determinant of the higher k catc values in some Suaedoideae C4 lineages.
In C4 species representing sections Salsina and Bienertia, analysis of the Rubisco large-subunit residue polymorphism indicates that there are differences in amino acid residues compared with C3 species which may be associated with increased Rubisco k catc values. The two single-cell C4 species from the genus Bienertia have three amino acid replacements putatively associated with increased Rubisco k catc. These are Ile 225, reported among submerged aquatic macrophytes (Iida et al., 2009) which may be linked to kinetic properties of Rubisco associated with CO2-concentrating mechanisms (Yeoh et al., 1981); Ile 270 which has been shown to be under positive selection along C4 lineages in Poaceae and Cyperaceae (Christin et al., 2008); and Ser 439, found to be under positive selection among terrestrial species representing different phylogenetic lineages (Galmes et al., 2014). Three Kranz C4 species from the section Salsina (see Supplementary Table S6 at JXB online) have Val 145 which is under positive selection along C4 lineages in Poaceae and Cyperaceae (Christin et al., 2008). In addition, S. altissima from this section has Ile 270, suggesting parallel acquisition of this mutation with Bienertia species; while S. moquinii has Ser 281 (see Supplementary Table S6 at JXB online), which is reported to be under positive selection along C4 lineages in Poaceae and Cyperaceae (Christin et al., 2008) and C4 lineages in Amaranthaceae (Kapralov et al., 2012).
Conclusions
The likelihood of a gene being repeatedly and independently recruited and changed to develop a convergent phenotype is most likely linked to the tissue in which it is expressed and its optimization for catalytic regulation. At the molecular level, when gene families are either recurrently recruited or when there are identical amino acid replacements in distant lineages, it suggests that there is limited genetic material suitable for new functions or that there is a restricted number of substitutions which can confer specific enzymatic properties (Christin et al., 2010). For PEPC, the differences in substrate affinity and the reaction towards allosteric effectors, suggest that C4 PEPC’s harbour specific C4 determinants that were acquired during the evolution of C4 photosynthesis (Gowik and Westhoff, 2011). The results presented here suggest that the development of C4 photosynthesis can occur with divergent amino acid substitutions that alter enzyme kinetics to converge on the same function. To our knowledge this is the first report to demonstrate that PEPC from C4 terrestrial plants without Ser at position 780 have C4-like PEPC kinetics (and to identify candidates for positive selection at positions 364, 368, and 733). Similarly, this is the first case which shows that there are C4 species which have C4-type Rubisco kcatc values while lacking amino acid substitutions in the large subunit of Rubisco. This demonstrates that there are multiple molecular routes to the same C4 carboxylase phenotype.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. Name and sequence of primers used.
Supplementary Table S2. Species origin, voucher, and sequence accession numbers.
Supplementary Table S3. Carbon isotope fraction values for leaf biomass.
Supplementary Table S4. ppc-1positive selection results.
Supplementary Table S5. Statistical analysis for PEPC K m for PEP, Hill coefficient, and V max.
Supplementary Table S6. Rubisco large subunit residue polymorphisms.
Supplementary Fig. S1. Phylogeny of Suaedoideae taxa used for ppc-1 positive selection analysis showing key amino acid changes.
Acknowledgements
This material is based upon work supported by the National Science Foundation under funds MCB #1146928 to GEE and funds from the School of Biological Sciences at WSU to EHR. Thanks to Andy McCubbin for help with sequencing and research support, Patrick Ellsworth for discussion on statistical analysis, Ryan Boyd and Anthony Gandin for help with enzyme measurements, and Richard Sharpe for discussion on RbcS genes. This manuscript is in partial fulfillment of a doctoral thesis at Washington State University.
References
- Bauwe H, Chollet R. 1986. Kinetic properties of phosphoenolpyruvate carboxylase from C3, C4, and C3–C4 intermediate species of Flaveria (Asteraceae). Plant Physiology 82, 695–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender MM. 1973. 13C/12C ratio changes in crassulacean acid metabolism plants. Plant Physiology 52, 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Muasya AM, Russier F, Roalson EH, Salamin N, Christin P-A. 2009. Phylogenomics of C4 photosynthesis in Sedges (Cyperaceae): multiple appearances and genetic convergence. Molecular Biology and Evolution 26, 1909–1919. [DOI] [PubMed] [Google Scholar]
- Besnard G, Pincon G, D’Hont A, Hoarau J-Y, Cadet F, Offmann B. 2003. Characterisation of the phosphoenolpyruvate carboxylase gene family in sugarcane (Saccharum spp.). Theoretical and Applied Genetics 107, 470–478. [DOI] [PubMed] [Google Scholar]
- Blasing OE, Ernst K, Streubel M, Westhoff P, Svensson P. 2002. The non-photosynthetic phosphoenolpyruvate carboxylases of the C4 dicot Flaveria trinervia: implications for the evolution of C4 photosynthesis. Planta 215, 448–456. [DOI] [PubMed] [Google Scholar]
- Blasing OE, Westhoff P, Svensson P. 2000. Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria, a conserved serine residue in the carboxyl-terminal part of the enzyme is a major determinant for C4-specific characteristics. Journal of Biological Chemistry 275, 27917–27923. [DOI] [PubMed] [Google Scholar]
- Chollet R, Vidal J, O’Leary MH. 1996. Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annual Review of Plant Physiology and Plant Molecular Biology 47, 273–298. [DOI] [PubMed] [Google Scholar]
- Christin P-A, Salamin N, Kellogg EA, Vicentini A, Besnard G. 2009. Integrating phylogeny into studies of C4 variation in the grasses. Plant Physiology 149, 82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin P-A, Salamin N, Muasya AM, Roalson EH, Russier F, Besnard G. 2008. Evolutionary switch and genetic convergence on rbcL following the evolution of C4 photosynthesis. Molecular Biology and Evolution 25, 2361–2368. [DOI] [PubMed] [Google Scholar]
- Christin P-A, Salamin N, Savolainen V, Duvall MR, Besnard G. 2007. C4 photosynthesis evolved in grasses via parallel adaptive genetic changes. Current Biology 17, 1241–1247. [DOI] [PubMed] [Google Scholar]
- Christin P-A, Weinreich DM, Besnard G. 2010. Causes and evolutionary significance of genetic convergence. Trends in Genetics 26, 400–405. [DOI] [PubMed] [Google Scholar]
- Collatz GJ, Badger MR, Smith C, Berry JA. 1979. A radioimmune assay for RuP2 carboxylase protein. Carnegie Institute Washington Yearbook 78, 171–175. [Google Scholar]
- Coombs J, Baldry CW. 1975. Metabolic regulation in C4 photosynthesis: phosphoenol pyruvate carboxylase and 3C intermediates of the photosynthetic carbon reduction cycle. Planta 124, 153–158. [DOI] [PubMed] [Google Scholar]
- DeVeau EJ, Burris JE. 1989. Photorespiratory rates in wheat and maize as determined by 18O-labeling. Plant Physiology 90, 500–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncaster HD, Leegood RC. 1987. Regulation of phosphoenolpyruvate carboxylase activity in maize leaves. Plant Physiology 84, 82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L-Y, Masuda T, Kawamura T, Hata S, Izui K. 1998. Cloning, expression, and characterization of a root-form phosphoenolpyruvate carboxylase from Zea mays: comparison with the C4-form enzyme. Plant and Cell Physiology 39, 865–873. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19, 11–15. [Google Scholar]
- Edwards GE, Voznesenskaya EV. 2011. C4 photosynthesis: Kranz forms and single-cell C4 in terrestrial plants. In: Raghavendra AS, Sage RF, eds. C4photosynthesis and related CO2concentrating mechanisms , Vol. 32 Dordrecht, The Netherlands: Springer Science, 29–61. [Google Scholar]
- Engelmann S, Blasing OE, Gowik U, Svensson P, Westhoff P. 2003. Molecular evolution of C4 phosphoenolpyruvate carboxylase in the genus Flaveria: a gradual increase from C3 to C4 characteristics. Planta 217, 717–725. [DOI] [PubMed] [Google Scholar]
- Engelmann S, Blasing OE, Westhoff P, Svensson P. 2002. Serine 774 and amino acids 296 to 437 comprise the major C4 determinants of the C4 phosphoenolpyruvate carboxylase of Flaveria trinervia . FEBS Letters 524, 11–14. [DOI] [PubMed] [Google Scholar]
- Galmes J, Kapralov MV, Andralojc PJ, Conesa MA, Keys AJ, Parry MAJ, Flexas J. 2014. Expanding knowledge of the Rubisco kinetics variability in plant species: environmental and evolutionary trends. Plant, Cell and Environment 37, 1989–2001. [DOI] [PubMed] [Google Scholar]
- Ghannoum O, Evans JR, Chow WS, Andrews TJ, Conroy JP, von Caemmerer S. 2005. Faster Rubisco is the key to superior nitrogen-use efficiency in NADP-malic enzyme relative to NAD-malic enzyme C4 grasses. Plant Physiology 137, 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U, Engelmann S, Blasing OE, Raghavendra AS, Westhoff P. 2006. Evolution of C4 phosphoenolpyruvate carboxylase in the genus Alternanthera: gene families and the enzymatic characterisitcs of the C4 isozyme and its orthologues in C3 and C3/C4 Alternantheras . Planta 223, 359–368. [DOI] [PubMed] [Google Scholar]
- Gowik U, Westhoff P. 2011. C4-phosphoenolpyruvate carboxylase. In: Raghavendra AS, Sage RF, eds. C4 photosynthesis and related CO2 concentrating mechanisms , Vol. 32: Springer Science, 257–275. [Google Scholar]
- Gupta SK, Ku MSB, Lin J-H, Zhang D, Edwards GE. 1994. Light/dark modulation of phosphoenolpyruvate carboxylase in C3 and C4 species. Photosynthesis Research 42, 133–143. [DOI] [PubMed] [Google Scholar]
- Hibberd JM, Covshoff S. 2010. The regulation of gene expression required for C4 photosynthesis. Annual Review of Plant Biology 61, 181–207. [DOI] [PubMed] [Google Scholar]
- Huber SC, Edwards GE. 1975. Inhibition of phosphoenolpyruvate carboxylase from C4 plants by malate and aspartate. Canadian Journal of Botany 53, 1925–1933. [Google Scholar]
- Iida S, Miyagi A, Aoki S, Ito M, Kadono Y, Kosuge K. 2009. Molecular adaptation of rbcL in the heterophyllous aquatic plant Potamogeton . PLos ONE 4, e4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa C, Hatanaka T, Misoo S, Miyake C, Fukayama H. 2011. Functional incorporation of sorghum small subunit increases the catalytic turnover rate of rubisco in transgenic rice. Plant Physiology 156, 1603–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izui K, Matsumura H, Furumoto T, Kai Y. 2004. Phosphoenolpyruvate carboxylase: a new era of structural biology. Annual Review of Plant Biology 55, 69–80. [DOI] [PubMed] [Google Scholar]
- Jacobs B, Engelmann S, Westhoff P, Gowik U. 2008. Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria: determinants for high tolerance towards the inhibitorl-malate. Plant, Cell and Environment 31, 793–803. [DOI] [PubMed] [Google Scholar]
- Jiao J-a, Chollet R. 1991. Posttranslational regulation of phosphoenolpyruvate carboxylase in C4 and Crassulacean acid metabolism plants. Plant Physiology 95, 981–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadereit G, Ackerly D, Pirie MD. 2012. A broader model for C4 photosynthesis evolution in plants inferred from the goosefoot family (Chenopodiaceae s.s.). Proceedings of the Royal Society of Biological Sciences 279, 3304–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai Y, Matsumura H, Izui K. 2003. Phosphoenolpyruvate carboxylase: three-dimensional structure and molecular mechanisms. Archives of Biochemistry and Biophysics 414, 170–179. [DOI] [PubMed] [Google Scholar]
- Kanai R, Edwards GE. 1999. The biochemistry of C4 photosynthesis. In: Sage RF, Monson RK, eds. C4plant biology . San Diego: Academic Press, 49–87. [Google Scholar]
- Kapralov MV, Akhani H, Voznesenskaya EV, Edwards G, Franceschi V, Roalson EH. 2006. Phylogenetic relationships in the Salicornioideae/Suaedoideae/Salsoloideae s.l. (Chenopodiaceae) clade and a clarification of the phylogenetic position of Bienertia and Alexandra using multiple DNA sequence datasets. Systematic Botany 31, 571–585. [Google Scholar]
- Kapralov MV, Filatov DA. 2007. Widespread positive selection in the photosynthetic Rubisco enzyme. BMC Evolutionary Biology 7, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapralov MV, Kubien DS, Andersson I, Filatov DA. 2011. Changes in rubisco kinetics during the evolution of C4 photosynthesis in Flaveria (Asteraceae) are associated with positive selection on genes encoding the enzyme. Molecular Biology and Evolution 28, 1491–1503. [DOI] [PubMed] [Google Scholar]
- Kapralov MV, Smith AC, Filatov DA. 2012. Rubisco evolution in C4 eudicots: an analysis of Amaranthaceae sensu lato . PLos ONE 7, e52974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubien DS, von Caemmerer S, Furbank RT, Sage RF. 2003. C4 photosynthesis at low temperature. A study using transgenic plants with reduced amounts of Rubisco. Plant Physiology 132, 1577–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubien DS, Whitney SM, Moore PV, Jesson LK. 2008. The biochemistry of Rubisco in Flaveria . Journal of Experimental Botany 59, 1767–1777. [DOI] [PubMed] [Google Scholar]
- Lara MV, Chuong SDX, Akhani H, Andreo CS, Edwards GE. 2006. Species having C4 single-cell-type photosynthesis in the chenopodiaceae family evolved a photosynthetic phosphoenolpyruvate carboxylase like that of kranz-type C4 species. Plant Physiology 142, 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegood RC, von Caemmerer S. 1988. The relationship between contents of photosynthetic metabolites and the rate of photosynthetic carbon assimilation. Planta 174, 253–262. [DOI] [PubMed] [Google Scholar]
- Leegood RC, von Caemmerer S. 1989. Some relationships between contents of photosynthetic intermediates and the rate of photosynthetic carbon assimilation in leaves of Zea mays L. Planta 178, 258–266. [DOI] [PubMed] [Google Scholar]
- Lilley RM, Walker DA. 1974. An improved spectrophotometric assay for ribulose bisphosphate carboxylase. Biochimica et Biophysica Acta 358, 226–229. [DOI] [PubMed] [Google Scholar]
- Long S. 1999. Environmental responses. In: Sage RF, Monson RK, eds. C4 plant biology . San Diego: CA: Academic Press, 215–249. [Google Scholar]
- Nakamoto H, Ku MSB, Edwards GE. 1983. Photosynthetic characteristics of C3–C4 intermediate Flaveria species. II. Kinetic properties of phosphoenolpyruvate carboxylase from C3, C4, and C3–C4 intermediate species. Plant and Cell Physiology 24, 1387–1393. [Google Scholar]
- Nimmo HG. 2003. Control of the phosphorylation of phosphoenolpyruvate carboxylase in higher plants. Archives of Biochemistry and Biophysics 414, 189–196. [DOI] [PubMed] [Google Scholar]
- Paulus JK, Niehus C, Groth G. 2013. a Evolution of C4 phosphoenolpyruvate carboxylase: enhanced feedback inhibitor tolerance is determined by a single residue. Molecular Plant 6, 1996–1999. [DOI] [PubMed] [Google Scholar]
- Paulus JK, Schlieper D, Groth G. 2013. b Greater efficiency of photosynthetic carbon fixation due to single amino-acid substitution. Nature Communications 4, 1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. 1989. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta 975, 384–394. [Google Scholar]
- Rao SK, Reiskind JB, Bowes G. 2008. Kinetic analyses of recombinant isoforms of phosphoenolpyruvate carboxylase from Hydrilla verticillata leaves and the impact of substituting a C4-signature serine. Plant Science 174, 475–483. [Google Scholar]
- Rosnow JJ, Edwards GE, Roalson EH. 2014. Positive selection of Kranz and non-Kranz C4 phosphoenolpyruvate carboxylase amino acids in Suaedoideae (Chenopodiaceae). Journal of Experimental Botany doi: 10.1093/jxb/eru053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF. 2002. Variation in the k cat of Rubisco in C3 and C4 plants and some implications for photosynthetic performance at high and low temperature. Journal of Experimental Botany 53, 609–620. [DOI] [PubMed] [Google Scholar]
- Sage RF, Christin P-A, Edwards EJ. 2011. The C4 plant lineages of planet Earth. Journal of Experimental Botany 62, 3155–3169. [DOI] [PubMed] [Google Scholar]
- Sage RF, Sage TL, Kocacinar F. 2012. Photorespiration and the evolution of C4 photosynthesis. Annual Review of Plant Biology 63, 19–47. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. 2001. Molecular cloning . New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- SAS II. 2011. SAS/STAT . Cary, NC, USA. [Google Scholar]
- Schlieper D, Forster K, Paulus JK, Groth G. 2014. Resolving the activation site of positive regulators in plant phosphoenolpyruvate carboxylase. Molecular Plant 7, 437–440. [DOI] [PubMed] [Google Scholar]
- Seemann JR, Badger MR, Berry JA. 1984. Variations in the specific activity of Ribulose-1,5-bisphosphate carboxylase between species utilizing differing photosynthetic pathways. Plant Physiology 74, 791–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson P, Blasing OE, Westhoff P. 1997. Evolution of the enzymatic characteristics of C4 phosphoenolpyruvate carboxylase: a comparison of the orthologous PPCA phosphoenolpyruvate carboxylases of Flaveria trinervia (C4) and Flaveria pringlei (C3). European Journal of Biochemistry 246, 452–460. [DOI] [PubMed] [Google Scholar]
- Takahashi-Terada A, Kotera M, Ohshima K, Furumoto T, Matsumura H, Kai Y, Izui K. 2005. Maize phosphoenolpyruvate carboxylase: mutations at the putative binding site for glucose 6-phosphate caused desensitization and abolished responsiveness to regulatory phosphorylation. Journal of Biological Chemistry 280, 11798–11806. [DOI] [PubMed] [Google Scholar]
- Ting IP, Osmond CB. 1973. Photosynthetic phosphoenolpyruvate carboxylases. Plant Physiology 51, 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar-Mendez A, Mujica-Jimenez C, Munoz-Clares R. 2000. Physiological implications of the kinetics of maize leaf phosphoenolpyruvate carboxylase. Plant Physiology 123, 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal J, Chollet R. 1997. Regulatory phosphorylation of C4 PEP carboxylase. Trends in Plant Science 2, 230–237. [Google Scholar]
- von Caemmerer S. 2013. Steady-state models of photosynthesis. Plant, Cell and Environment 36, 1617–1630. [DOI] [PubMed] [Google Scholar]
- Walker B, Ariza LS, Kaines S, Badger MR, Cousins AB. 2013. Temperature response of in vivo Rubisco kinetics and mesophyll conductance in Arabidopsis thaliana: comparisons to Nicotiana tabacum . Plant, Cell and Environment 36, 2108–2119. [DOI] [PubMed] [Google Scholar]
- Whitney SM, Houtz RL, Alonso H. 2011. a Advancing our understanding and capacity to engineer nature’s CO2-sequestering enzyme, Rubisco. Plant Physiology 155, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Sharwood RE, Orr D, White SJ, Alonso H, Galmes J. 2011. b Isoleucine 309 acts as a C4 catalytic switch that increases ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) carboxylation rate in Flaveria . Proceedings of the National Academy of Sciences, USA 108, 14688–14693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: Phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24, 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yeoh H-H, Badger MR, Watson L. 1981. Variations in kinetic properties of ribulose-1,5-bisphosphate carboxylases among plants. Plant Physiology 67, 1151–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelitch I, Schultes NP, Peterson RB, Brown P, Brutnell TP. 2009. High glycolate oxidase activity is required for survival of maize in normal air. Plant Physiology 149, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.