Highlight
StWRKY1 transcription factor mediates secondary cell wall thickening in potato by regulating expression of tyramine-related hydroxycinnamic acid amide biosynthetic genes, thus contributing to resistance to late blight disease.
Key words: Hydroxycinnamic acid amides, Phytophthora infestans, quantitative resistance, secondary cell wall thickening, transcription factors
Abstract
Quantitative resistance is polygenically controlled and durable, but the underlying molecular and biochemical mechanisms are poorly understood. Secondary cell wall thickening is a critical process in quantitative resistance, regulated by transcriptional networks. This paper provides compelling evidence on the functionality of StWRKY1 transcription factor, in a compatible interaction of potato–Phytophthora infestans, to extend our knowledge on the regulation of the metabolic pathway genes leading to strengthening the secondary cell wall. A metabolomics approach was used to identify resistance-related metabolites belonging to the phenylpropanoid pathway and their biosynthetic genes regulated by StWRKY1. The StWRKY1 gene in resistant potato was silenced to decipher its role in the regulation of phenylpropanoid pathway genes to strengthen the secondary cell wall. Sequencing of the promoter region of StWRKY1 in susceptible genotypes revealed the absence of heat shock elements (HSEs). Simultaneous induction of both the heat shock protein (sHSP17.8) and StWRKY1 following pathogen invasion enables functioning of the latter to interact with the HSE present in the resistant StWRKY1 promoter region. EMSA and luciferase transient expression assays further revealed direct binding of StWRKY1 to promoters of hydroxycinnamic acid amide (HCAA) biosynthetic genes encoding 4-coumarate:CoA ligase and tyramine hydroxycinnamoyl transferase. Silencing of the StWRKY1 gene was associated with signs of reduced late blight resistance by significantly increasing the pathogen biomass and decreasing the abundance of HCAAs. This study provides convincing evidence on the role of StWRKY1 in the regulation of downstream genes to biosynthesize HCAAs, which are deposited to reinforce secondary cell walls.
Introduction
Late blight caused by Phytophthora infestans (Mont.) de Bary is the most destructive disease of potato. It reduces plant biomass or even may kill the plant, thus decreasing crop quantity and quality (Fry, 2008). The resistance in potato can be improved by selecting resistance genes that are induced following pathogen invasion, either by molecular breeding or cis/trans genetic engineering. Plant responses to stress are complex and involve numerous physiological, molecular, and cellular adaptations (Rejeb et al., 2014). The plant cell wall is one of the barriers that pathogens need to overcome to successfully colonize plant tissues. Cell wall thickening reduces the spread of pathogens in plants beyond the site of infection, reducing lesion expansion (Dangl et al., 2013). The biosynthesis of the secondary cell wall is a developmental process that involves the co-ordinated expression of secondary cell wall biosynthetic genes regulated by a cascade of transcription factors (Cosgrove, 2005). Understanding such processes is vital for the manipulation of secondary cell wall thickening to resist pathogen development in plants (War et al., 2012).
Plants have evolved intricate strategies to recognize pathogen infection and devise effective defence responses (Jones and Dangl, 2006). The first line of defence following infection involves the recognition of pathogen-associated molecular patterns (PAMPs), which trigger basal levels of plant defence responses referred to as PAMP-triggered immunity (PTI). To combat PTI, pathogens produce haustoria inside the plant cell to release effector proteins. This invokes a second line of defence in plants to produce effector-specific resistance (R) proteins encoded by R genes. The resulting hypersensitive reaction or effector-triggered immunity (ETI) constitutes qualitative resistance (Kushalappa and Gunnaiah, 2013). PAMP also triggers phytohormones, such as jasmonic acid, salicylic acid, ethylene, abscisic acid, and gibberellins, to provide broad biochemical resistance and reduce pathogen growth (Robert-Seilaniantz et al., 2011). These phytohormones fine-tune the pathways involved in the production of secondary metabolites that impart quantitative resistance. Following recognition of PAMPs, the receptors trigger a phosphorylation cascade through the mitogen-activated protein kinase (MAPK) signalling pathway (Chisholm et al., 2006), leading to the induction or repression of target genes (Czernic et al., 1999). Among the induced target genes are a variety of transcription factors that regulate plant defence responses involving up- or down-regulation of resistance-related (RR) genes by binding to specific DNA sequences in their promoter regions (Alves et al., 2014).
WRKY domain-containing proteins are one of the largest families of transcriptional regulators in plants. They have a DNA-binding domain called a W-box (TTGACC/T or WRKYGQK) and are involved in plant immune responses (Rushton et al., 2010). Overexpression of WRKY transcription factor increased tolerance to pathogen infection through induction of phytohormone-mediated signalling pathways in grapes and tobacco (Dang et al., 2014; Marchive et al., 2013). In contrast, the overexpression of WRKY reduced resistance mechanisms in rice (Chujo et al., 2013; Yokotani et al., 2013) and Capsicum plants (Wang Y et al., 2013). Although WRKY proteins might act as crucial nodes in the cross-talk between PTI and ETI, little is known regarding the molecular events occurring during the establishment of resistance or susceptibility to pathogens in plants. Also, details of molecular mechanisms of individual proteins of the WRKY superfamily are still very limited, especially in non-model plants.
The present study was designed to elucidate the mechanisms by which StWRKY1 regulates the downstream genes leading to quantitative resistance to late blight in potato based on a metabolomics approach. We identified high-fold-change RR metabolites in potato stems based on non-targeted metabolomics of two genotypes: a resistant (F06025) and a susceptible (Shepody) genotype. The two genotypes with contrasting levels of resistance to Phytophthora infestans induced different metabolic pathways. As reported by others, a high fold change in abundance of phenylpropanoid metabolites was observed in stems that appears to play a major role in late blight resistance by enhancing cell wall thickening through deposition of polyaromatic domains of suberin (Franke et al., 2012). Phenylpropanoids were also observed in the current authors’ previous studies (Pushpa et al., 2014; Yogendra et al., 2014), but the molecular events in their biosynthesis and regulation were not revealed. A hierarchy of genes involved in the induction of the phenylpropanoid pathway has been identified, although this is limited to model plants. Therefore, candidate metabolites with high fold change in abundances were mapped onto metabolic pathways to identify their biosynthetic candidate genes based on genomic databases. WRKYs are implicated in the regulation of plant defence against pathogens, but only a few members of the WRKY superfamily in potato have been isolated or functionally characterized. StWRKY1 was reported in potato following infection with P. infestans (Dellagi et al., 2000; Machinandiarena et al., 2012), but no details on metabolic pathway regulation were revealed. The current study uses a forward genetics approach to discover StWRKY1 from the high fold change in abundance of hydroxycinnamic acid amides (HCAAs) detected based on metabolic profiling, and reverse genetics to validate in depth the transcriptional activation of StWRKY1, possible interactions between StWRKY1 and HCAA biosynthetic genes, and their link to secondary cell wall strengthening to contain pathogen spread following P. infestans invasion.
Materials and methods
Plant production
A resistant genotype (F06025) and a susceptible genotype (Shepody) were obtained from Mrs Agnes Murphy (Potato Research Centre, Agriculture and Agri-Food Canada, New Brunswick, Canada). F06025 was derived from AWN86514-2×N06993-13 and Shepody was derived from Bake King×F58050. Sprouted tubers, either one piece with at least two eyes or one whole small tuber, were planted in a pot containing promix BX® and perlite. Temperature (20±3 °C with 16h photoperiod) and relative humidity (70±10%) were maintained in the greenhouse throughout the growing period.
Pathogen production, inoculation, and incubation
P. infestans culture [clonal lineage US-8, A2 mating type, isolate (1661), obtained from Dr. H. Platt (AAFC, Charlottetown, Prince Edward Island, Canada)] was maintained on potato dextrose agar media. For inoculation, fresh sporangia were produced by inoculating thin potato tuber slices and incubating in Petri dishes lined with moist filter paper at 18 °C. A spore suspension was prepared by extracting sporangia in sterile water and the spore concentration was adjusted to 1×105 spores ml–1.
Young stems of 5-week-old plants were inoculated with 20 µl sporangial suspension or mock-solution at two spots, corresponding to opposite sides of the stem, using a Hamilton syringe fitted with an autodispenser (Gastight 1750 DAD W/S; Hamilton, Reno, NV, USA). The inoculated drops were covered with Whatman filter paper discs (4–5mm diameter) to prevent spread of the spore suspension. Immediately after inoculation, the plants were covered with transparent plastic bags sprayed with sterile water on the inside to maintain high humidity to facilitate infection. The covers were removed 72h post-inoculation (hpi).
Disease severity assessment and pathogen biomass quantification
The experiment was conducted as a randomized complete block design with two genotypes (resistant and susceptible) inoculated with P. infestans and three replicates over time. Each experimental unit consisted of five pots, each with two plants and five stems inoculated on opposite sides. Following inoculation, the plants were covered with plastic bags for 72h. Disease severity was assessed by measuring lesion length using a digital caliper at 3 d intervals until 9 d post-inoculation (dpi). Lesion length (mm) was used to calculate area under the disease progress curve (AUDPC). Relative biomass of P. infestans in the infected samples was quantified based on quantitative PCR (qPCR) (Asai et al., 2008). Genomic DNA was isolated from P. infestans-infected stems (6 dpi) using a DNeasy Plant Mini Kit (Qiagen, Canada). qPCR was performed using IQ SYBR Green Supermix (Bio-Rad, Canada) in a CFX384TM Real-Time System (Bio-Rad, Canada) according to the manufacturer’s instructions, using specific primers to amplify and detect P. infestans DNA and potato DNA (Supplementary Table S1 at JXB online).
Sample collection, metabolite extraction, and LC-high-resolution MS (LC-HRMS) analysis
The experiment was conducted as a completely randomized block design with two genotypes (F06025 and Shepody) and two inoculations (pathogen and mock) with five replicates over time. The experimental units consisted of five plants with a total of 10 stems inoculated for each treatment. At 72 hpi, 1cm stem segments, containing the inoculated spot, were cut using a sterile scalpel. Samples were flash frozen in liquid nitrogen and stored at –80 °C. Metabolites were extracted using 60% aqueous methanol with 0.1% formic acid (De Vos et al., 2007). These were analysed in negative ionization mode using a LC-HRMS system (LC-ESI-LTQ Orbitrap; Thermo Fisher, Waltham, MA, USA), with a 5cm kinetex column, as described previously (Yogendra et al., 2014).
LC-HRMS output processing
The LC-HRMS output Xcalibur RAW files were converted into mzXML format. Data was processed using the interactive LC-MS data processing software MZmine-2 with the high sensitivity peak detection algorithm XCMS centWave (Pluskal et al., 2010). The observed masses and their abundance (relative intensity) were imported to MS Excel; peaks that were not consistent among replicates and those annotated as isotopes and adducts were excluded from further analyses (Yogendra et al., 2014).
Identification of RR metabolites
Monoisotopic mass peak intensity (m/z) data was subjected to pairwise Student’s t-test analysis in RM versus SM, RP versus RM and SP versus SM (RM, mock-inoculated resistant genotype; SM, mock-inoculated susceptible cultivar; RP, pathogen-inoculated resistant genotype; SP, pathogen-inoculated susceptible cultivar) as described previously (Kushalappa and Gunnaiah, 2013). Metabolites with significantly higher abundance in the resistant compared with the susceptible genotype were considered RR metabolites. RR metabolites were further classified into RR constitutive (RRC) and RR induced (RRI) metabolites. The RR metabolites were putatively identified based on two criteria: (i) accurate mass match (accurate mass error <5 ppm) with metabolites reported in different databases [METLIN, KNApSAcK, Plant Metabolic Network (PMN), LIPIDMAPS, and KEGG] and (ii) fragmentation pattern match with those in databases or in silico verification (Yogendra et al., 2014).
RNA isolation and quantitative real-time PCR (qRT-PCR)
For qRT-PCR, total RNA was isolated from stems inoculated with pathogen or mock-solution at 48 hpi in three replicates (five stems were collected from five plants for each replicate) using a RNeasy Plant Mini Kit (Qiagen, Canada). Purified RNA (3 µg from each sample) was reverse transcribed using an Affinity Script qRT-PCR cDNA Synthesis Kit (Agilent Technologies, USA). For qRT-PCR, Primer-BLAST software was used to design primer sets for StWRKY1, 4-coumarate:CoA ligase (St4-CL), tyramine hydroxycinnamoyl transferase (StTHT), hydroxycinnamoyl transferase (SpHCT), and tyrosine decarboxylase (SpTyDC) (Supplementary Table S1), and elongation factor-1α (StEF1α) and β-tubulin (Stβ-tubulin) were used as reference genes (Nicot et al., 2005). A sample of 25ng cDNA was used in a qRT-PCR reaction using IQ SYBR Green Supermix (Bio-Rad, Canada) in a CFX384TM Real-Time System (Bio-Rad) in a 10 µl reaction volume according to the manufacturer’s instructions. The relative gene expression level was calculated using the 2–∆∆CT method (Livak and Schmittgen, 2001).
Histochemical analysis of potato stems
Fifteen young potato internodes were collected from pathogen- and mock-inoculated plants at 72 hpi for histochemical study. Internodes were cut 0.5cm above and 0.5cm below the inoculation site using a scalpel and immediately frozen at –20 °C. For cryosectioning, tissues were embedded in Shandon Cryomatrix (Richard-Allan Scientific, USA). The cross-sectioning was carried using a cryotome (Leica, Canada) with 35 µm thickness and collected on glass slides. Sections were washed with distilled water for 2min, stained with Neu’s reagent [1% 2-amino ethyl diphenyl borinate (Sigma-Aldrich, Canada) in absolute methanol] for 5min, washed with water again for 30 s, and mounted in 15% glycerol (Alemanno et al., 2003). The cross-sections of five internodes for each treatment with at least five sections from each internode were observed under a fluorescent microscope (Nikon, USA) for chemifluorescence with blue laser diode excitation at 405nm using a HQ442/45 emission filter.
Virus-induced gene silencing (VIGS) vector construction
Tobacco rattle virus [pTRV1 (YL192) and pTRV2 (pYL154)] vectors (Liu et al., 2002) were obtained from the Arabidopsis Biological Resource Center, The Ohio State University, Columbus, OH, USA. To generate pTRV2-StWRKY1, a 519bp segment of potato WRKY1 was PCR amplified from resistant potato cDNA using primers WRKY1-F (5′-TACGTCGAATTCATGGAGAATTATGCAACAATA-3′) and WRKY1-R (5′-TAGCTGCTCGAGTTAAAAGGAAGTA TAGATTTGCAT-3′) with EcoRI and XhoI restriction sites (underlined) at the 5′ and 3′ ends of the amplified fragment, respectively. The PCR product was digested with EcoRI/XhoI and inserted into the same site of the pTRV2 vector, resulting in pTRV2-StWRKY1. For silencing phytoene desaturase (PDS), a 396bp segment of the potato PDS gene was amplified from the resistant genotype and cloned into pTRV2 vector using primers (PDS-F: 5′-TACGTCGAATTCCAGTGGATA TTTTCAAGCTGCTT-3′; PDS-R: 5′-TAGCTGCTCGAGAAAC AGACCTTGGAGTTTTGACA-3′) with EcoRI and XhoI restriction sites (underlined) at the 5′ and 3′ ends of the amplified fragment, respectively. The positive clones in the pTRV2 plasmid were confirmed by sequencing. All TRV-VIGS clones were transformed into Agrobacterium tumefaciens strain GV3101 using a standard transformation protocol.
Agrobacterium-mediated virus infiltration (agroinfiltration)
Cultures of A. tumefaciens strain GV3101 containing each of the constructs derived from pTRV2, with an empty vector as a control, and pTRV1 were grown at 28 °C for 24h in Luria–Bertani (LB) medium supplemented with 10mM MES, 200 μM acetosyringone, 50mg l–1 gentamycin, and 50mg l–1 kanamycin. Agrobacterium cells were harvested and suspended in the infiltration buffer (10mM MgCl2, 200 μM acetosyringone, and 10mM MES, pH 5.6). Agrobacterium cultures were pelleted, resuspended in infiltration buffer, and adjusted to an OD600 of 1.0. Mixtures of Agrobacterium cultures containing pTRV1 and pTRV2 and its derivatives, in a ratio of 1:1 (v/v), were placed in the dark at room temperature for 4h before infiltration. For infiltration, 0.1mm holes were made in the leaflets of 2-week-old potato plants using the corner of a new razor blade and infiltrated with Agrobacterium cultures (agroinfiltration) using a 1ml needleless syringe. Plants were maintained at 18 °C in a growth chamber.
Induction of protein expression and purification of recombinant protein
Full-length cDNA of the StWRKY1 gene was amplified by RT-PCR using Solanum tuberosum cDNA as template and StWRKY1-F (5′-GGATCCTGAGAATTATGCAACAATATTTC-3′) and StWRKY1-R (5′-AGATCTTTAAAAGGAAGTATAG ATTTGC-3′) primers containing BamHI and BglII restriction sites (underlined). PCR product was digested with BamHI and BglII, and the fragment was ligated to pTrcHis B (Invitrogen, USA) vector and transformed into Escherichia coli BL21 cells. E. coli cells harbouring StWRKY1 were grown in 200ml LB containing 50 µg ml–1 ampicillin incubated at 37 °C and 250rpm. Induction of protein expression and purification was achieved using a Ni-NTA column (Qiagen, Germany) as described previously (Kumar et al., 2012; Kumar et al., 2014). The purified protein fractions of recombinant StWRKY1 were detected by 12% SDS-PAGE by staining with Coomassie Brilliant Blue.
EMSA
The promoter region of THT, having the putative StWRKY1-binding site, was used to design synthetic single-stranded forward (5′-TCGTCAGAGTTGACCTCCACCAACCA-3′) and reverse (5′-TGGTTGGTGGAGGTCAACTCTGACGA-3′) 26bp olig- onucleotide primers. The underlined region of the primers represents the WRKY-binding domain. Single-stranded primers were annealed at room temperature to generate a double-stranded probe, which was labelled using a Pierce Biotin 3′ End DNA Labeling Kit (Thermo Scientific, USA). EMSA was performed using a LightShift® Chemiluminescent EMSA Kit (Thermo Scientific, USA) as per the manufacturer’s instructions. Briefly, each reaction contained 2 μl 10× binding buffer, 1.0 μl each 1 µg µl–1 poly(dI–dC), 50% glycerol, 1% NP-40, 1M KCl, 100mM MgCl2, 200mM EDTA, 3 µl (3 μg) purified protein, and 2 µl (20fmol) labelled probe or unlabelled DNA as the competitor made up to a final volume of 20 µl. The binding reactions were incubated at 25 °C for 20min. A 6% polyacrylamide minigel (containing 3% glycerol) was pre-run for 30min using 0.5× TBE as a running buffer. Then 5 μl 5× loading buffer was added to the reaction and a 25 μl aliquot of the reaction mixture was subjected to electrophoresis. The gel was transferred to a nitrocellulose membrane (Amersham Biosciences, USA) using a Mini Trans-Blot® Electrophoretic Transfer Cell (Bio-Rad, USA), followed by cross-linking of the membrane below a UV lamp. The biotin-labelled DNA was detected using a Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific, USA) as per the manufacturer’s instructions. The change in band position representing different molecular weights was used to confirm the shift.
Bimolecular fluorescence complementation (BiFC) assay
For the BiFC assay, sequences encoding StWRKY1 and StMEK1 were amplified and fused with the N- and C-terminal parts of Yellow Fluorescent Protein (YFPn and YFPc, respectively). BiFC plasmids containing YFPn and YFPc fusions were co-transformed into potato protoplasts as described previously (Walter et al., 2004). The protoplasts were incubated under weak light for 12–16h before observation. YFP fluorescence signals were checked under a fluorescent microscope (Nikon, USA). The presence of yellow fluorescence was used to discriminate treatments.
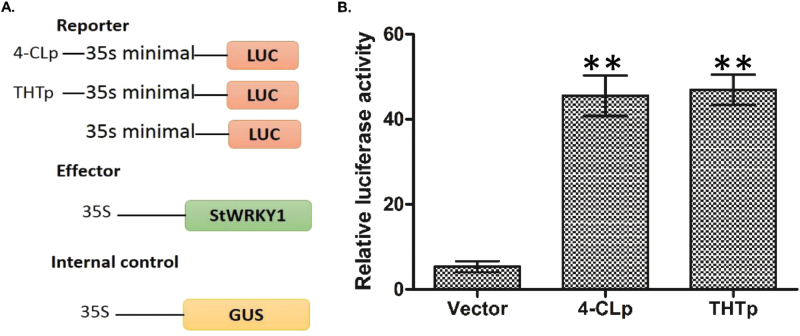
Luciferase (LUC) transient expression assay
For LUC transient expression assays, reporter plasmids (4-CLp-LUC or THTp-LUC), effector constructs containing StWRKY1, and 35S::β-glucuronidase (GUS) internal control were co-transformed into potato protoplasts. The protoplasts were pelleted and resuspended in 1× cell culture lysis reagent (Promega, USA). GUS fluorescence was measured using a Modulus luminometer/fluorometer with a UV fluorescence optical kit (Fluorescence Microplate Reader; BioTek, USA). The experiment was carried out in three replicates; each replicate contained 20 µl protoplast lysate and 100 µl LUC mix. LUC activity was detected with a luminescence kit using LUC assay substrate (Fluorescence Microplate Reader). The relative reporter gene expression levels were expressed as LUC/GUS ratios, which were used to discriminate treatments.
Results
Late blight severity and P. infestans biomass
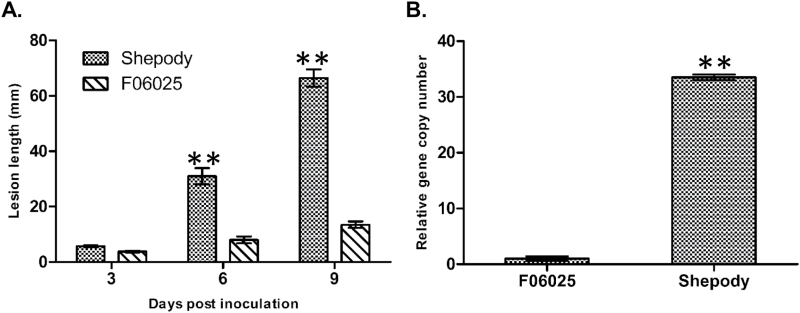
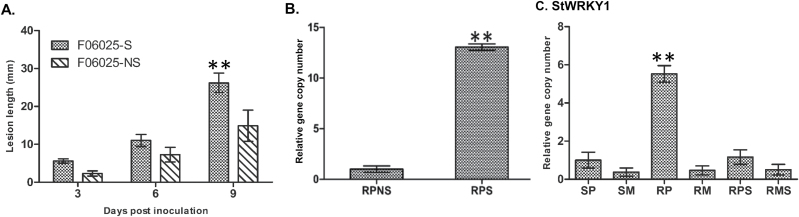
Late blight severity in the potato genotypes F06025 and Shepody was assessed by measuring lesion length over time following inoculation with P. infestans clonal lineage US-8. The initial symptoms were observed as brown-coloured lesions on the inoculated stems at 3 dpi in both F06025 and Shepody. By 9 dpi, the stem lesion length was 66.44mm (AUDPC=201.09) in Shepody, differing (P<0.001) from the resistant genotype F06025 with a lesion length of 13.44mm (AUDPC=49.71) (Fig. 1A and Supplementary Fig. S1 at JXB online).
Fig. 1.
Disease progress curve and pathogen biomass in infected stems of resistant (F06025) and susceptible (Shepody) genotypes inoculated with P. infestans: (A) Late blight disease severity progress curve based on lesion length (mm) and (B) biomass of pathogen quantified as relative P. infestans-specific (O-8) gene expression at 6 dpi. Significant differences between susceptible and resistant plants using Student’s t-test: *P<0.05; **P<0.01.
Pathogen biomass in the infected stems of genotypes F06025 and Shepody were quantified based on qPCR. At 6 dpi, P. infestans-specific gene copy numbers (DNA, O-8) were 33.52-fold higher (P<0.001) in the susceptible Shepody than in the resistant F06025 genotype (Fig. 1B).
Differential accumulation of metabolites in stems of potato genotypes
Metabolites were profiled in the stems of resistant and susceptible genotypes at 3 dpi following inoculation with P. infestans or with water and analysed based on LC-HRMS. A total of 8408 consistent peaks of monoisotopic masses were detected in all the replicate and treatment combinations. Among the mock-inoculated plants, a total of 679 metabolites had higher abundance in the resistant than in the susceptible genotype, and these were designated as RRC (RM>SM) metabolites (Table 1, and Supplementary Fig. S2A and Supplementary Table S2 at JXB online). These metabolites belonged to different chemical groups (Supplementary Fig. S2B) and some important metabolites had high fold changes in the resistant genotype [phenylpropanoids: 1,2-di-O-sinapoyl-β-d-glucose (14.55), 1-O-sinapoyl-β-d-glucose (6.31) and scopolin (3.15); glycerophospholipids: 1-(9Z-nonadecenoyl)-glycero-3-phosphoserine (3.61), 1-hexadecyl-2-propionyl-sn-glycero-3-phosphocholine (3.50) and 1-octadecyl-2-ethyl-sn-glycero-3-phosphocholine (3.14); sphingolipids: (3′-sulfo) Galβ-Cer (d18:0/20:0(2OH)) (3.77), N-palmitoylsphingosine (2.90) and N-(eicosanoyl)-sphinganine (2.37)].
Table 1.
Fold change in abundance of RR metabolites detected in potato stem following P. infestans or mock-solution inoculation
| Observed mass (Da) | Exact mass (Da) | Name | Fold change |
|---|---|---|---|
| Phenylpropanoids | |||
| 148.0528 | 148.0524 | trans-Cinnamic acid | 1.24* RRI |
| 162.032 | 162.0317 | 4-Hydroxycoumarin | 1.36* RRC |
| 165.0793 | 165.0790 | l-Phenylalanine | 1.18* RRI |
| 166.0633 | 166.0630 | Dihydro-3-coumaric acid | 2.13** RRI |
| 174.0531 | 174.0528 | Shikimate | 1.31* RRI |
| 181.0742 | 181.0739 | α-Amino oxy-β-phenyl propionate | 8.22* RRI; 1.74* RRC |
| 192.0273 | 192.0270 | Citrate | 2.27* RRI |
| 192.0637 | 192.0634 | l-Quinic acid | 2.46* RRC; 1.18* RRI |
| 194.0586 | 194.0579 | Ferulic acid | 12.44** RRC; 3.04** RRI |
| 208.0552 | 208.0558 | Methyl 5-(1-propynyl)-2-thiophenepropanoate | 2.95** RRC |
| 264.1472 | 264.1474 | Feruloylputrescine | 5.03** RRI |
| 300.0844 | 300.0845 | Salicylate β-d-glucose ester | 1.59** RRI |
| 306.1690 | 306.1692 | Feruloylagmatine | 5.01** RRI; 3.11** RRC |
| 313.1313 | 313.1314 | N-feruloyltyramine | 12.85** RRI; 1.99** RRC |
| 329.1262 | 329.1263 | N-feruloyloctopamine | 8.90** RRI |
| 342.0946 | 342.0951 | Caffeic acid 3-glucoside | 2.72* RRC |
| 354.0950 | 354.0951 | Scopolin | 3.15* RRC; 1.11* RRI |
| 368.1103 | 368.1107 | 5-O-feruloylquinic acid | 2.63* RRC |
| 386.1210 | 386.1213 | 1-O-sinapoyl-β-d-glucose | 6.31** RRC; 2.92* RRI |
| 537.1823 | 537.1846 | 4-Demethylsimmondsin 2′-(E)-ferulate | 2.98* RRC |
| 582.1941 | 582.1949 | 10-Acetoxyligustroside | 2.74* RRC |
| 592.1790 | 592.1792 | 1,2-Di-O-sinapoyl-β-d-glucose | 14.55** RRC; 1.13* RRI |
| Sphingolipids | |||
| 479.4357 | 479.4338 | Cer(d14:2(4E,6E)/16:0) | 1.30* RRI |
| 537.5138 | 537.5121 | Cer(d18:1/16:0) | 2.90* RRC; 1.44* RRI |
| 595.5880 | 595.5903 | Cer(d18:0/20:0) | 2.37* RRC; 1.15* RRI |
| 683.5331 | 683.5336 | GlcCer(d15:2(4E,6E)/18:0) | 1.30* RRC |
| 711.5643 | 711.5649 | GlcCer(d15:2(4E,6E)/20:0) | 1.24* RRI |
| 851.5776 | 851.5793 | (3′-Sulfo)Galβ-Cer(d18:0/20:0(2OH)) | 3.77** RRC |
| Glycerophospholipids | |||
| 537.309 | 537.3067 | 1-(9Z-Nonadecenoyl)-glycero-3-phosphoserine | 3.61* RRI |
| 537.379 | 537.3794 | 1-Hexadecyl-2-propionyl-sn-glycero-3-phosphocholine | 3.50*RRI |
| 537.4151 | 537.4158 | 1-Octadecyl-2-ethyl-sn-glycero-3-phosphocholine | 3.14* RRI |
Detailed compound identification is presented in Supplementary Table S2. Fold change calculation was based on relative intensity of metabolites: RRC=RM/SM and RRI=(RP/RM)/(SP/SM) (see main text for details). Significance (t-test): *P<0.05, **P<0.01.
Among the pathogen-inoculated plants, 167 metabolites were induced with greater abundances in the resistant than in the susceptible genotype, and these were designated as RRI [(RP>RM)>(SP>SM)] metabolites (Supplementary Fig. S2A and Supplementary Table S2). Out of 167 RRI metabolites, 97 were putatively identified (Table 1 and Supplementary Table S2); the identity of the most significant metabolites was confirmed based on fragmentation patterns using LC-HRMS (Supplementary Fig. S3 at JXB online). Thirty-four of the identified metabolites were phenylpropanoids, including the HCAAs N-feruloyltyramine, N-feruloyloctopamine, feruloylputrescine, and feruloylagmatine. In addition to phenylpropanoids, 24 fatty acids and four alkaloids were induced in higher abundance in the resistant than in the susceptible genotype following pathogen inoculation.
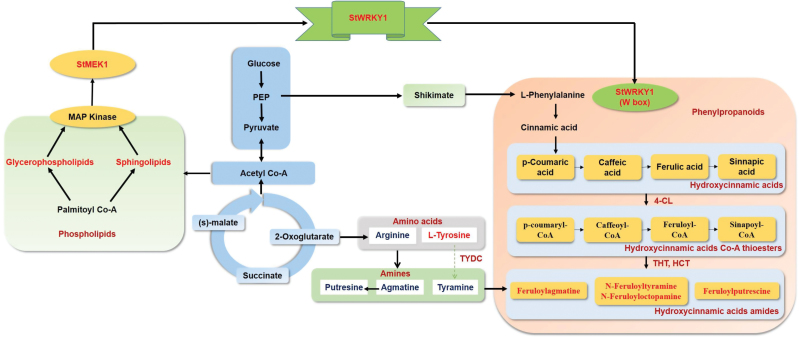
Among the RRI metabolites identified, most of the high-fold-change metabolites belonged to the phenylpropanoid pathway, including the HCAAs N-feruloyltyramine (12.85), N-feruloyloctopamine (8.90), feruloylputrescine (5.03), and feruloylagmatine (5.01), which are known to be involved in cell wall thickening. To confirm the deposition of HCAAs in cell walls, a histochemical staining technique was used to visualize the location of deposition of these metabolites. Deposition of HCAAs (blue fluorescence intensity) was greater in pathogen-treated resistant than in mock-treated resistant and pathogen- or mock-treated susceptible genotypes (Supplementary Fig. S4 at JXB online). The candidate RRI metabolites with the highest fold changes in abundance following pathogen inoculation were mapped onto their metabolic pathways (Fig. 2) to identify the catalytic enzymes, which were then searched in the Spud database (http://solanaceae.plantbiology.msu.edu/; accessed on 25 February 2015) to identify candidate RR genes. The RR genes that encoded the enzymes of these RRI metabolites were 4-coumarate:CoA ligase (4-CL) (GenBank accession number AF150686.1; chromosome 3), hydroxycinnamoyl transferase (HCT) (Gene ID: PGSC0003DMT400036695; chromosome 3), tyramine hydroxycinnamoyl transferase (THT) (GenBank accession number JX896425.1; chromosome 10), and tyrosine decarboxylase (TyDC) (Gene ID: PGSC0003DMT400038501; chromosome 3) (Fig. 2). The expression of RR genes is transcriptionally regulated by the co-ordinated action of transcription factors which bind to cis-acting elements and regulate the expression of RR genes. Previous studies reported that StWRKY1 (GenBank accession number AJ278507.1; chromosome 5) is induced and co-regulated with class I endochitinase during the compatible interaction of potato and P. infestans (Dellagi et al., 2000). The promoter regions of RR genes were sequenced and the presence of the W1-box of the WRKY1 transcription factor was confirmed (Supplementary Table S3 at JXB online).
Fig. 2.
Satellite metabolic pathway of potato–Phytophthora interaction showing the RR metabolites and their catalysing enzymes involved in their biosynthesis detected in resistant potato inoculated with P. infestans or mock-solution. Metabolites and genes detected in this study are marked in red. Detailed interactions are presented in Fig. 7. (This figure is available in colour at JXB online.)
Metabolic regulation of phenylpropanoid pathway genes by StWRKY1
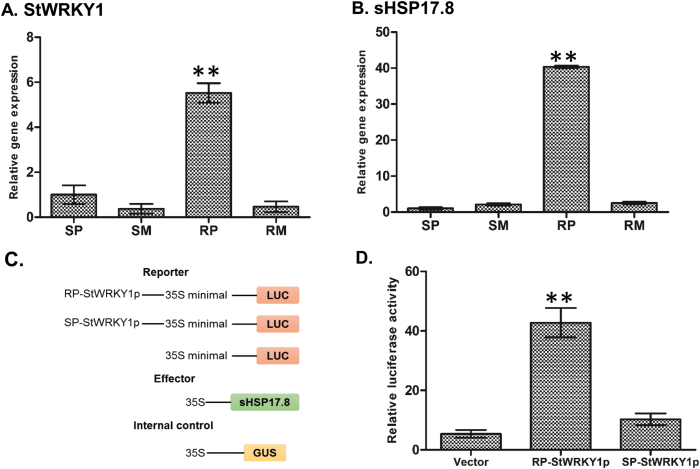
To investigate whether StWRKY1 is involved in defence responses, StWRKY1 transcripts were quantified based on qRT-PCR. The relative transcript abundance of StWRKY1 was higher (P<0.001) in resistant (5.52) than in susceptible genotypes (1.00) (Fig. 3A). Furthermore, the coding and promoter regions of StWRKY1 were amplified from both resistant and susceptible genotypes, and cloned into the pGEM®-T Easy vector and verified by Sanger sequencing. No sequence variation was observed for the coding region of StWRKY1 in both resistant and susceptible genotypes. However, the heat shock element (HSE) domain was detected in the promoter region of the resistant genotype and was completely absent in the susceptible genotype (Supplementary Fig. S5 and Supplementary Table S4 at JXB online). HSEs act as heat shock protein (HSP)-binding sites and help to induce hypersensitive responses by generating oxidative bursts.
Fig. 3.
Relative transcript expression and regulation of StWRKY1 by sHSP17.8. The relative transcript expression for (A) StWRKY1 and (B) sHSP17.8 was confirmed in the resistant genotype relative to the susceptible genotype following P. infestans and mock-solution inoculation at 48 hpi using qRT-PCR in comparison with reference genes for elongation factor-1α and β-tubulin. See main text for further details. Regulation of StWRKY1 by sHSP17.8 was confirmed by LUC transient expression assay: (C) constructs used and (D) relative LUC reporter activity by HSP17.8. Values are averages of three replicates. Significant differences in relative expression levels using Student’s t-test: *P<0.05; **P<0.01.
Transcriptional regulation of StWRKY1 by small HSPs (sHSPs)
RNA sequencing was carried out between the resistant genotype F06025 and another susceptible genotype, Russet Burbank, using 48 hpi potato leaf samples. Sequencing was performed with an Illumina HiSeq 2000 sequencer with 100bp paired-end reads (Genome Québec-McGill University Innovation Center, Montreal, Canada). The reads were aligned to the potato reference genome (Potato Genome Sequencing Consortium, 2011). We observed a high fold change in expression for sHSP17.8 (77.78) and sHSP26.5 (9.00) in the resistant genotype (F06025), compared with the susceptible genotype (Russet Burbank) upon P. infestans infection, based on RNA sequencing analysis (unpublished data and Supplementary Table S5). The high fold change in expression of sHSP17.8 was further validated using qRT-PCR. Interestingly, qRT-PCR revealed a higher fold change of 40.35 transcripts in the resistant (F06025) than in the susceptible (Shepody) genotype (Fig. 3B). As HSPs are transcriptionally activated by heat shock factors (HSF) to interact with HSEs present on the promoter region of a gene, the possible regulation between sHSP17.8 and HSE was tested by in vivo LUC assay in potato protoplasts (Fig. 3C, D). The StWRKY1 promoter from the resistant genotype had a relative LUC value of 42.77 as compared (P<0.001) with the susceptible genotype (10.25) and control vector (5.38). Taken together, these results suggest that sHSP17.8 interacts with the HSE conserved domain present in the promoter region of StWRKY1 in the resistant genotype and is likely involved in mediating the high transcriptional activity of StWRKY1 proteins.
Activation of transcription factor StWRKY1 by MAPK
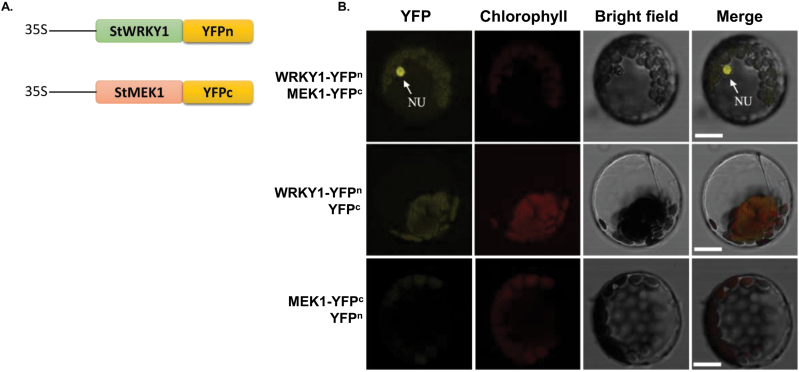
We detected a high fold change in abundance of induced phospholipid metabolites in the resistant genotype following pathogen infection [sphingolipids: (3′-sulfo)Galβ-Cer(d18:0/20:0(2OH)) (3.77), N-palmitoylsphingosine (2.90), and N-(eicosanoyl)-sphinganine (2.37); glycerophospholipids: 1-(9Z-nonadecenoyl)-glycero-3-phosphoserine (3.61), 1-hexadecyl-2-propionyl-sn-glycero-3-phosphocholine (3.50), and 1-octadecyl-2-ethyl-sn-glycero-3-phosphocholine (3.14)]. These phospholipids are structural components of membranes and act as novel secondary messengers in plant defence signalling pathways. These activate the MAPK cascade by triggering oxidative bursts (Laxalt and Munnik, 2002). MAPK (StMEK1) phosphorylates StWRKY1, which in turn activates defence gene expression (Katou et al., 2003). To substantiate the interaction between StWRKY1 and StMEK1 in potato cells, a BiFC assay was performed in which YFPn fused to StWRKY1 (YFPn-StWRKY1) and YFPc fused to StMEK1 (StMEK1-YFPc) were transiently co-expressed in potato protoplasts (Fig. 4A). Co-expression of YFPn-StWRKY1 and StMEK1-YFPc reconstituted a functional YFP in the nucleus, whereas co-expression with either control vector failed to generate YFP fluorescence, indicating StMEK1 is required for activation of StWRKY1 (Fig. 4B).
Fig. 4.
BiFC assay for StWRKY1 and StMEK1 interaction: (A) constructs used in the BiFC expression assay, and (B) BiFC assay showing the interaction between StWRKY1-YFPc and YFPn-StMEK1. The nucleus (NU) is shown in yellow and chlorophyll autofluorescence is shown in red. Bars, 5mm. (This figure is available in colour at JXB online.)
StWRKY1 physically interacts with promoters of HCAA biosynthetic genes and exhibits transcriptional activity
To investigate whether the StWRKY1 proteins are involved in transcriptional regulation of HCAA biosynthesis, the coding region of the StWRKY1 gene and promoters of 4-CL and THT from the resistant genotype were amplified and cloned into pGEM®-T Easy vector (Promega, USA), as confirmed by sequencing. This was followed by subcloning into the FU63 (CD3-1841) (Wang X et al., 2013) vector with a LUC gene driven by the 35S minimal promoter (Fig. 5A). The constructs were transformed into potato protoplasts. We found that StWRKY1 bound to the promoter region of RR genes, and drastically activated LUC reporter expression in 4-CL (45.55) and THT (47.00) promoters compared with vector alone (4.16) (Fig. 5B), suggesting the transcriptional regulation of HCAA biosynthesis.
Fig. 5.
Transcriptional regulation of HCAA biosynthetic genes by StWRKY1: (A) constructs used in the transient expression assay and (B) relative LUC reporter activity by StWRKY1. Values are averages of three replicates. Significant differences in expression levels in promoters compared with vector using Student’s t-test: *P<0.05; **P<0.01.
The HCAAs, including N-feruloyltyramine, N-feruloyloctopamine, feruloylagmatine, and feruloylputrescine, play a critical role in secondary cell wall thickening (Gunnaiah et al., 2012; Pushpa et al., 2014; Yogendra et al., 2015; Yogendra et al., 2014). Accordingly, the physical interaction of StWRKY1 with HCAA biosynthetic gene THT was tested by in vitro EMSA as additional evidence (Supplementary Fig. S6 at JXB online). For EMSA, the purified recombinant StWRKY1 protein fractions had a molecular weight of 25kDa (Supplementary Fig. S6A) as analysed on 12% SDS-PAGE. This was in agreement with the theoretical molecular weight of 23kDa (19.86kDa for the native protein plus 3kDa for the His-tag). The purified recombinant protein was used to study the interaction with the W1-box of the THT promoter. EMSA results showed a shift in signal of biotinylated THT promoter that was inhibited in the presence of excess unlabelled competitor (Supplementary Fig. S6B). These results indicate that StWRKY1 can specifically bind to the W1-box of the THT promoter and can regulate its gene expression.
Functional validation of StWRKY1
The loss-of-function experiment was performed with StWRKY1-silenced potato seedlings to elucidate the biological function of StWRKY1 in late blight resistance mechanisms. Nucleotide sequences of 519bp of StWRKY1 and 396bp of StPDS (GenBank accession number AY484445.1) were amplified from the resistant potato genotype F06025 and cloned into the pTRV2 vector to develop the TRV-VIGS construct (Supplementary Fig. S7A at JXB online). A mixture of Agrobacterium cultures containing pTRV1 and pTRV2-StWRKY1 or pTRV2-StPDS was infiltrated onto 2-week-old resistant potato seedlings. Agroinfiltrated plants were healthy and no plant died from this treatment, although stunted growth was observed. Plants expressing pTRV2-StPDS showed photobleaching after 30 d of agroinfiltration and used as positive controls to identify plants that were successfully silenced (Supplementary Fig. S7B). The PDS gene was used to demonstrate the effectiveness of VIGS, as this gene participates in the carotenoid metabolic pathway. StWRKY1-silenced leaves exhibited higher levels of P. infestans susceptibility compared with non-silenced plants. At this phase, P. infestans spores (~1000 sporangia) were inoculated to stems of StWRKY1-silenced plants, and transcript expression levels, metabolites, and pathogen biomasses were used to validate the resistance function.
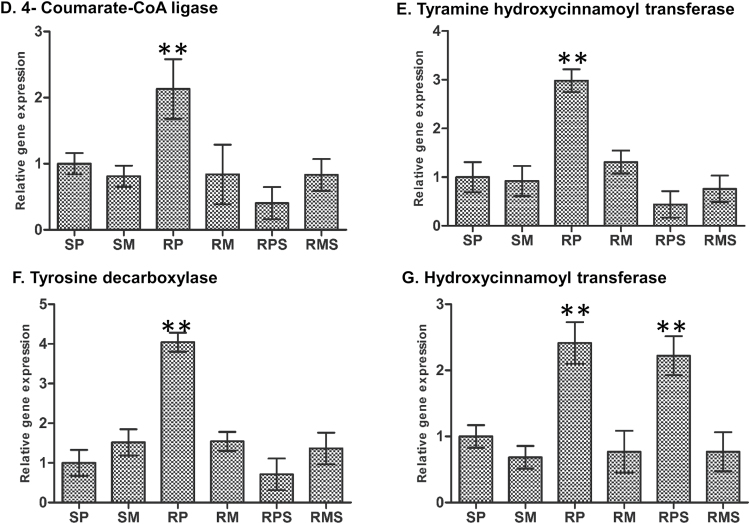
To assess the effect of StWRKY1 in late blight resistance, the disease severity and biomass were quantified in silenced and non-silenced resistant plants. The lesion length was higher (P<0.001) in silenced plants (26.19mm; AUDPC=80.68), significantly differing from the non-silenced plants (14.90mm; AUDPC=47.60). Likewise, pathogen biomass was higher (P<0.001) in silenced plants (fold change=13.06) compared with non-silenced plants at 6 dpi (Fig. 6A, B). Furthermore, the relative abundances of the transcripts of phenylpropanoid genes (4-CL, THT, TyDC, and HCT) were determined in silenced and non-silenced resistant plants with reference to the susceptible genotype (Fig. 6C–F). StWRKY1 and the regulated RR genes (4-CL, THT and TyDC) were reduced (P<0.001) in silenced plants (fold change=1.16, 0.40, 0.44, and 0.71, respectively) as compared with resistant plants (5.52, 2.13, 2.98, and 4.04). However, no significant reduction was observed in HCT transcripts in silenced plants (fold change=2.22) compared with resistant plants (2.41) and this was probably due to the absence of the W1-box in the HCT promoter (Fig. 6G and Supplementary Table S3). We then tested the effect of StWRKY1 silencing on the production of HCAAs. The RRI metabolites N-feruloyltyramine (fold change=10.13), N-feruloyloctopamine (6.65), feruloylputrescine (3.80), and feruloylagmatine (3.44) were reduced (P<0.05) after silencing (Table 2). Taken together, these results provide compelling evidence that StWRKY1 is a positive regulator of late blight resistance through the reinforcement of secondary cell walls.
Fig. 6.
Effect of StWRKY1 silencing on late blight resistance mechanisms. (A) Late blight disease severity progress. Disease severity was quantified as lesion length (mm) at 3 d intervals. (B) Biomass of P. infestans on silenced (S) and non-silenced (NS) resistant potato genotypes. Biomass was quantified as relative P. infestans-specific (O-8) gene expression at 6 dpi. Relative transcript expression of (C) StWRKY1, and phenylpropanoid genes for (D) 4-coumarate:CoA ligase, (E) tyramine hydroxycinnamoyl transferase, (F) tyrosine decarboxylase, and (G) hydroxycinnamoyl transferase. Relative transcript expression was measured at 48 hpi in comparison with reference genes for elongation factor-1α and β-tubulin. RPS indicates resistant silenced and inoculated with P. infestans; RMS indicates resistant silenced and inoculated with mock-solution; see main text for further details. Significant differences in expression levels in RP compared with SP using Student’s t-test: *P<0.05, **P<0.01.
Table 2.
Effect of StWRKY1 silencing on RR phenylpropanoid metabolites upon P. infestans or mock-solution inoculation
| Observed mass (Da) | Exact mass (Da) | Name | Fold change before silencing | Fold change after silencing |
|---|---|---|---|---|
| 264.1472 | 264.1474 | Feruloylputrescine | 5.03** RRI | 3.80* RRI |
| 306.1690 | 306.1692 | Feruloylagmatine | 5.01** RRI; 3.11** RRC | 3.44* RRI |
| 313.1313 | 313.1314 | N-feruloyltyramine | 12.85** RRI; 1.99** RRC | 10.13** RRI; 1.70** RRC |
| 329.1262 | 329.1263 | N-feruloyloctopamine | 8.90** RRI | 6.65** RRI; 2.67* RRC |
Fold change calculation was based on relative intensity of metabolites: RRC=RM/SM and RRI=(RP/RM)/(SP/SM) (see main text for details). Significance (t-test): *P<0.05, **P<0.01.
Discussion
In the present study, an integrated metabolomics, gene expression, and gene sequencing information approach was used to discover and explore the role of transcription factor StWRKY1, and also to explain the late blight resistance mechanisms in potato genotypes F06025 and Shepody. These genotypes varied in their resistance levels and also in accumulation of metabolites. Most of the high-fold-change metabolites identified in the resistant genotype belonged to the phenylpropanoid pathway, especially HCAAs. These metabolites are well known for their role in cell wall thickening, forming antimicrobial barriers (Dangl et al., 2013). Plants have developed a system to maintain cell wall integrity by activation of signalling molecules, transcriptional reprogramming and biosynthesis of defence metabolites that limit pathogen infection. Cell wall thickening reduces the spread of pathogens in plants beyond the site of infection, reducing lesion expansion. These complex molecules are not generally broken down by plant pathogens. Here, a forward genetics approach was followed to explore the upstream genes involved in the transcriptional regulation of the HCAAs biosynthetic pathway genes in potato stem, mainly through functional elucidation of StWRKY1.
Induction of StWRKY1 in potato against P. infestans infection
During potato–P. infestans interaction, significantly higher fold expression of the StWRKY1 gene was identified in the resistant genotype (fold change=5.52) as compared with the susceptible genotype (1.00). Despite the existence of numerous links between WRKY transcription factors and plant defence genes, direct evidence for WRKY proteins binding with RR genes and further accumulation of metabolites remains limited. Several studies have focused on model plants such as Arabidopsis and tobacco, but only a few focused on potato WRKYs. In potato, StWRKY1 is strongly induced following a compatible interaction with P. infestans (Dellagi et al., 2000) and treatment with potassium phosphite during infection inhibited pathogen spread (Machinandiarena et al., 2012). Overexpression of VvWRKY1 in grapevines provided higher resistance to downy mildew Plasmopara viticola through higher expression of genes involved in the jasmonic acid pathway (Marchive et al., 2013). Likewise, the overexpression of CaWRKY27 in tobacco plants was associated with resistance to Ralstonia solanacearum infection through induction of jasmonic acid-, salicylic acid-, and ethylene-mediated signalling pathways (Dang et al., 2014). Resistance to poplar canker was associated with overexpression of PtoWRKY60, which regulated high expression of defence-associated genes, such as PR5.1, PR5.2, PR5.4, PR5.5, and CPR5, in poplar (Ye et al., 2014). Two CaWRKY1 genes, CaWRKY1a and CaWRKY1b (which bind to the pW1a promoter region), were identified in Coffea arabica and conferred resistance to the rust fungus Hemileia vastatrix (Petitot et al., 2013). These findings depict the involvement of the WRKY transcription factors in disease resistance, as described here in potato against P. infestans.
We also looked into the signalling pathways which involved induction of the WRKY transcription factor and detected high fold changes in several induced phospholipid metabolites in the resistant genotype. These phospholipids act as novel secondary messengers in plant defence signalling pathways and activate the MAPK cascade by triggering oxidative bursts (Laxalt and Munnik, 2002). MAPK converts the signals generated from receptors to cellular responses. Upon activation, MAPK translocates into the nucleus and phosphorylates the WRKY transcription factor, which in turn activates downstream gene expression (Kim and Zhang, 2004). In potato, StMEK1 was functionally characterised during defence reactions (Katou et al., 2003), which are also associated with StWRKY1, as was confirmed here by BiFC assay. StMEK1 and StWRKY1 function together to induce the expression of downstream RR genes involved in plant defence.
Transcriptional activation of StWRKY1 by sHSPs only in the resistant potato genotype
We detected significantly high fold changes of sHSP17.8 (fold change=40.35) in a resistant genotype of potato against late blight infection. The presence of HSEs on the promoter region of the StWRKY1 gene in the resistant genotype, but not in the susceptible genotype (Supplementary Table S4), helped the sHSPs to interact and induce the transcription factor StWRKY1. Small HSPs interact and provide stability to the proteins by acting as molecular chaperones (Haslbeck and Vierling, 2015). LUC assay confirmed the transcriptional regulation of StWRKY1 by sHSP17.8. The HSEs are highly conserved and consist of inverted repeats of the pentameric sequence nGAAn (Amin et al., 1988). The HSE present in the promoter region of the Arabidopsis APX1 gene acts as a functional component that helps to protect plants from oxidative stress (Storozhenko et al., 1998). In tobacco, the sHSP Ntshsp17 acts as a molecular chaperone and helps in the maintenance of cellular conditions suitable for inducing defence responses against bacterial wilt pathogen Ralstonia solanacearum by stabilizing signalling-related proteins (Maimbo et al., 2007). In tomato, the sHSP RSI2 is a HSP20 member that specifically interacts and provides stability to the Fusarium oxysporum R protein I-2 (Van Ooijen et al., 2010). Likewise, the induction of mitochondrial HSP22 occurred in tomato during oxidative stress and helped to provide adoptive responses (Banzet et al., 1998). Conflicting with these results, wheat sHSP, Mds1 present on chromosome 3AS increases susceptibility to wheat gall midge and powdery mildew fungus Blumeria graminis f. sp. tritici (Liu et al., 2013).
Transcriptional regulation of phenylpropanoid pathway RRI genes by StWRKY1
To combat pathogen infection, plants have developed specific metabolic pathways to biosynthesize metabolites that reinforce secondary cell walls as mechanical barriers. Following pathogen inoculation, genes involved in the production of tyramine-derived HCAAs were 4-CL, THT, and TyDC, which were upregulated 2.13-, 2.98-, and 4.04-fold, respectively, in the resistant genotype. Concurrently, metabolites produced from these candidate RRI genes, i.e. N-feruloyltyramine and N-feruloyloctopamine, were induced with significantly high fold changes in the resistant genotype. The authors previous studies reported high-fold-change expression of THT and TyDC genes, along with high-fold-change accumulation of RRI metabolites N-feruloyltyramine, N-caffeoyltyramine, and N-feruloyloctopamine in leaves of resistant genotypes (Pushpa et al., 2014; Yogendra et al., 2014; Yogendra et al., 2015). However, here, for the first time, StWRKY1 was associated with the promoters of downstream HCAA biosynthetic genes 4-CL and THT in vivo and in vitro. The results of the EMSA and LUC assays confirmed that StWRKY1 physically interacts with and induces high-fold-change expression of downstream genes involved in secondary cell wall thickening. Likewise, the overexpression of VvWRKY1 induced a cinnamyl alcohol dehydrogenase and a caffeic acid O-methyl transferase gene in grape vines, providing higher resistance to downy mildew (Marchive et al., 2013). Taken together, these findings suggest that StWRKY1 takes part in a regulatory network, directly controlling secondary cell wall thickening by acting on promoters of HCAA biosynthetic genes.
Silencing of StWRKY1 results in profound effects on secondary cell wall thickening
To further assess the effect of StWRKY1 on the resistance mechanisms, StWRKY1 was silenced in a resistant genotype, and silenced and non-silenced resistant plants were compared based on relative gene expression of HCAA biosynthetic genes, accumulation of their downstream biosynthesized RRI metabolites, amount of fungal biomass, and disease severity. The transcript abundance of StWRKY1 and its regulating RRI genes, 4-CL, THT, and TyDC, was reduced (P<0.05) in silenced plants (fold change=1.16, 0.40, 0.44, and 0.71, respectively) compared with resistant plants (5.52, 2.13, 2.98, and 4.04, respectively). However, no significant reduction was observed for HCT transcripts, probably due to the absence of the W1-box. The abundance of the HCAA metabolites N-feruloyltyramine and N-feruloyloctopamine was reduced 2-fold after silencing, resulting in a higher (P<0.001) biomass in silenced plants (fold change=13.06). This may be due to the metabolic profiling protocol, where metabolite extraction, detection, and identification were not comprehensive, and thus it is possible that other RR genes and associated metabolites are also involved in imparting resistance to late blight. Correspondingly, silencing the hydroxycinnamoyl-CoA:hydroxycinnamoyl transferase affected cell wall strength by decreasing the dimethoxylated syringyl units of the lignin polymer in Nicotiana benthamiana stems (Hoffmann et al., 2004). Silencing of cellulose synthase genes affected cell wall formation in flax (Chantreau et al., 2015). Likewise, silencing of ethylene response factor Pti5, which is involved in ethylene signalling, increased the susceptibility to aphids of a near-isogenic genotype that carried the Mi-1.2 R gene in tomato (Wu et al., 2015). These findings, together with the data reported here provide insights into the effect of silencing of StWRKY1 on downstream HCAA biosynthetic genes crucial for secondary cell wall thickening in potato.
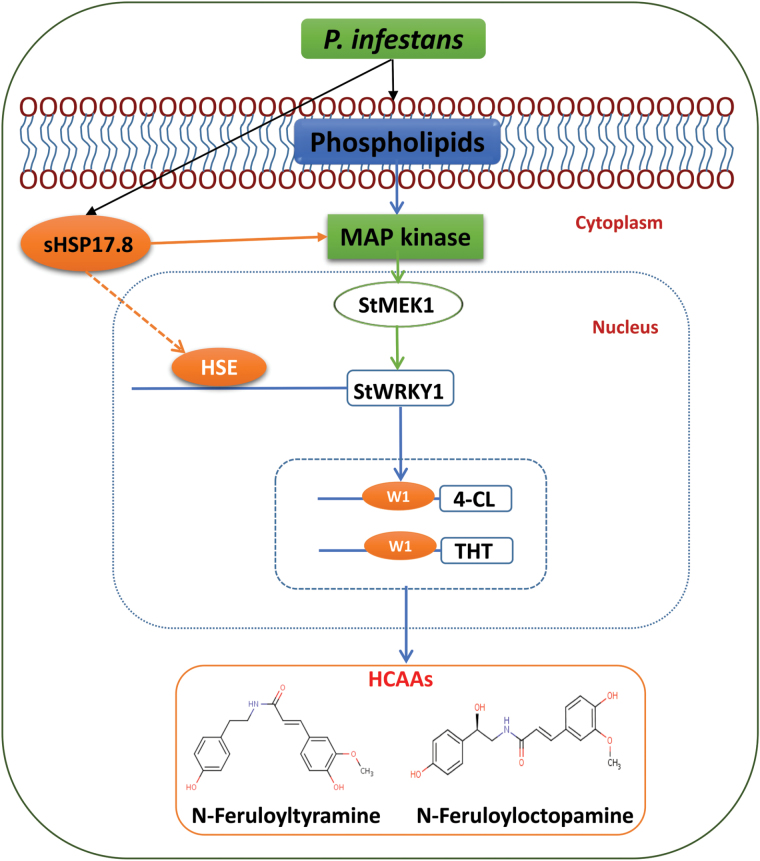
Proposed model for the role of StWRKY1 in potato secondary cell wall thickening
Based on integrated metabolomics and genomics data, the authors propose a model to explain the role of StWRKY1, together with HCAA biosynthetic genes, in mediating potato secondary cell wall thickening (Fig. 7). Following P. infestans invasion, a hierarchy of genes is induced in the phenylpropanoid pathway starting from the induction of phospholipid metabolites (glycerophospholipids and sphingolipids), possibly by enzymes produced by the pathogen, activating MAPK (StMEK1), which in turn phosphorylates the transcription factor (StWRKY1). Concurrently, sHSP17.8 is also induced in the resistant genotype. sHSP17.8 interacts with HSEs present in the StWRKY1 promoter region to enable functioning of StWRKY1. Consequently, StWRKY1 protein interacts with HCAA biosynthetic genes 4-CL, TyDC, THT, and possibly others, inducing high-fold-change production of the HCAA metabolites N-feruloyltyramine, N-feruloyloctopamine, and others. Taken together, these results suggest that StWRKY1 is required for the induction of HCAAs and defence by secondary cell wall thickening in potato against P. infestans. These candidate genes can be used to replace non-functional genes in susceptible commercial cultivars, e.g. Shepody and Russet Burbank, based on genome-editing technologies to enhance quantitative resistance (Shan et al., 2014).
Fig. 7.
Proposed model of StWRKY1 in regulating secondary cell wall thickening. MEK1, MAPK1; see main text for further details.
Supplementary Data
Supplementary data are available at JXB online.
Figure S1. P. infestans-infected potato stems with susceptible (Shepody) and resistant (F06025) genotypes at 9 dpi.
Figure S2. (A) The number of RR metabolites identified in stems of the resistant potato genotype inoculated with water or P. infestans. (B) RR metabolites identified in the resistant potato genotype inoculated with a mock-solution or P. infestans and their classification according to chemical groups.
Figure S3. In silico fragmentation of RR metabolites.
Figure S4. Laser scanning confocal micrographs of stem sections exhibiting secondary cell wall thickening due to HCAAs (HCAA blue fluorescence).
Figure S5. Comparison of sequence variation between resistant and susceptible StWRKY1 promoter regions.
Figure S6. (A) Analysis of purified recombinant StWRKY1 protein on 12% SDS-PAGE. (B) EMSA showing DNA–protein interaction of the THT promoter with StWRKY1.
Figure S7. (A) Schematic representation of TRV-VIGS vectors. (B) Silencing of the PDS gene.
Table S1. Primer sequences used for quantifying RR gene expression in potato genotypes.
Table S2. RR metabolites detected in the resistant potato genotype following P. infestans or mock-solution inoculation.
Table S3. Promoter analysis of RR genes in the resistant potato genotype.
Table S4. Promoter analysis of StWRKY1 gene in resistant and susceptible potato genotypes.
Table S5. The differentially expressed HSPs detected in potato following P. infestans inoculation using RNA sequencing analysis.
Acknowledgments
This work was carried out with the aid of a grant from the International Development Research Center (IDRC), Canada and with the financial support of the Department of Foreign Affairs, Trade and Development Canada (DFATD) and Natural Sciences and Engineering Council of Canada (NSERC).
References
- Amin J, Ananthan J, Voellmy R. 1988. Key features of heat shock regulatory elements. Molecular and Cellular Biology 8, 3761–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemanno L, Ramos T, Gargadenec A, Andary C, Ferriere N. 2003. Localization and identification of phenolic compounds in Theobroma cacao L. somatic embryogenesis. Annals of Botany 92, 613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves MS, Dadalto SP, Gonçalves AB, de Souza GB, Barros VA, Fietto LG. 2014. Transcription factor functional protein-protein interactions in plant defense responses. Proteomes 2, 85–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai S, Ohta K, Yoshioka H. 2008. MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana . The Plant Cell Online 20, 1390–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banzet N, Richaud C, Deveaux Y, Kazmaier M, Gagnon J, Triantaphylidès C. 1998. Accumulation of small heat shock proteins, including mitochondrial HSP22, induced by oxidative stress and adaptive response in tomato cells. The Plant Journal 13, 519–527 [DOI] [PubMed] [Google Scholar]
- Chantreau M, Chabbert B, Billiard S, Hawkins S, Neutelings G. 2015. Functional analyses of cellulose synthase genes in flax (Linum usitatissimum) by virus‐induced gene silencing. Plant Biotechnology Journal , 1–13. [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. 2006. Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Chujo T, Miyamoto K, Shimogawa T, Shimizu T, Otake Y, Yokotani N, Nishizawa Y, Shibuya N, Nojiri H, Yamane H. 2013. OsWRKY28, a PAMP-responsive transrepressor, negatively regulates innate immune responses in rice against rice blast fungus. Plant Molecular Biology 82, 23–37. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. 2005. Growth of the plant cell wall. Nature Reviews Molecular Cell Biology 6, 850–861. [DOI] [PubMed] [Google Scholar]
- Czernic P, Visser B, Sun W, Savouré A, Deslandes L, Marco Y, Van Montagu M, Verbruggen N. 1999. Characterization of an Arabidopsis thaliana receptor‐like protein kinase gene activated by oxidative stress and pathogen attack. The Plant Journal 18, 321–327. [DOI] [PubMed] [Google Scholar]
- Dang F, Wang Y, She J, Lei Y, Liu Z, Eulgem T, Lai Y, Lin J, Yu L, Lei D. 2014. Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiologia Plantarum 150, 397–411. [DOI] [PubMed] [Google Scholar]
- Dangl JL, Horvath DM, Staskawicz BJ. 2013. Pivoting the plant immune system from dissection to deployment. Science 341, 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos RC, Moco S, Lommen A, Keurentjes JJ, Bino RJ, Hall RD. 2007. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nature Protocols 2, 778–791. [DOI] [PubMed] [Google Scholar]
- Dellagi A, Heilbronn J, Avrova AO, Montesano M, Palva ET, Stewart HE, Toth IK, Cooke DE, Lyon GD, Birch PR. 2000. A potato gene encoding a WRKY-like transcription factor is induced in interactions with Erwinia carotovora subsp. atroseptica and Phytophthora infestans and is coregulated with class I endochitinase expression. Molecular Plant–Microbe Interactions 13, 1092–1101. [DOI] [PubMed] [Google Scholar]
- Franke RB, Dombrink I, Schreiber L. 2012. Suberin goes genomics: use of a short living plant to investigate a long lasting polymer. Frontiers in Plant Science 3, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry W. 2008. Phytophthora infestans: the plant (and R gene) destroyer. Molecular Plant Pathology 9, 385–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnaiah R, Kushalappa AC, Duggavathi R, Fox S, Somers DJ. 2012. Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat QTL (Fhb1) contributes to resistance against Fusarium graminearum . PloS One 7, e40695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Vierling E. 2015. A first line of stress defence: small heat shock proteins and their function in protein homeostasis. Journal of Molecular Biology 427, 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D, Lapierre C, Pollet B, Legrand M. 2004. Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. The Plant Cell Online 16, 1446–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Katou S, Yamamoto A, Yoshioka H, Kawakita K, Doke N. 2003. Functional analysis of potato mitogen-activated protein kinase kinase, StMEK1. Journal of General Plant Pathology 69, 161–168. [Google Scholar]
- Kim CY, Zhang S. 2004. Activation of a mitogen‐activated protein kinase cascade induces WRKY family of transcription factors and defense genes in tobacco. The Plant Journal 38, 142–151. [DOI] [PubMed] [Google Scholar]
- Kumar A, Dutt S, Bagler G, Ahuja PS, Kumar S. 2012. Engineering a thermo-stable superoxide dismutase functional at sub-zero to >50°C, which also tolerates autoclaving. Scientific Reports 2, 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Kaachra A, Bhardwaj S, Kumar S. 2014. Copper, zinc superoxide dismutase of Curcuma aromatica is a kinetically stable protein. Process Biochemistry 49, 1288–1296. [Google Scholar]
- Kushalappa AC, Gunnaiah R. 2013. Metabolo-proteomics to discover plant biotic stress resistance genes. Trends in Plant Science 18, 522–531. [DOI] [PubMed] [Google Scholar]
- Laxalt AM, Munnik T. 2002. Phospholipid signalling in plant defence. Current Opinion in Plant Biology 5, 332–338. [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh‐Kumar S. 2002. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N‐mediated resistance to tobacco mosaic virus. The Plant Journal 30, 415–429. [DOI] [PubMed] [Google Scholar]
- Liu X, Khajuria C, Li J, Trick HN, Huang L, Gill BS, Reeck GR, Antony G, White FF, Chen MS. 2013. Wheat Mds-1 encodes a heat-shock protein and governs susceptibility towards the Hessian fly gall midge. Nature Communications 4, 2070. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Machinandiarena MF, Lobato MC, Feldman ML, Daleo GR, Andreu AB. 2012. Potassium phosphite primes defense responses in potato against Phytophthora infestans . Journal of Plant Physiology 169, 1417–1424. [DOI] [PubMed] [Google Scholar]
- Maimbo M, Ohnishi K, Hikichi Y, Yoshioka H, Kiba A. 2007. Induction of a small heat shock protein and its functional roles in Nicotiana plants in the defense response against Ralstonia solanacearum . Plant Physiology 145, 1588–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchive C, Léon C, Kappel C, Coutos-Thévenot P, Corio-Costet M-F, Delrot S, Lauvergeat V. 2013. Over-Expression of VvWRKY1 in grapevines induces expression of jasmonic acid pathway-related genes and confers higher tolerance to the downy mildew. PloS One 8, e54185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot N, Hausman J-F, Hoffmann L, Evers D. 2005. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. Journal of Experimental Botany 56, 2907–2914. [DOI] [PubMed] [Google Scholar]
- Petitot A-S, Barsalobres-Cavallari C, Ramiro D, Freire EA, Etienne H, Fernandez D. 2013. Promoter analysis of the WRKY transcription factors CaWRKY1a and CaWRKY1b homoeologous genes in coffee (Coffea arabica). Plant Cell Reports 32, 1263–1276. [DOI] [PubMed] [Google Scholar]
- Pluskal T, Castillo S, Villar-Briones A, Orešič M. 2010. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potato Genome Sequencing Consortium. 2011. Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. [DOI] [PubMed] [Google Scholar]
- Pushpa D, Yogendra KN, Gunnaiah R, Kushalappa AC, Murphy A. 2014. Identification of late blight resistance-related metabolites and genes in potato through nontargeted metabolomics. Plant Molecular Biology Reporter 32, 584–595. [Google Scholar]
- Rejeb IB, Pastor V, Mauch-Mani B. 2014. Plant responses to simultaneous biotic and abiotic stress: molecular mechanisms. Plants 3, 458–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. 2011. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annual Review of Phytopathology 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. 2010. WRKY transcription factors. Trends in Plant Science 15, 247–258. [DOI] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, Gao C. 2014. Genome editing in rice and wheat using the CRISPR/Cas system. Nature Protocols 9, 2395–2410. [DOI] [PubMed] [Google Scholar]
- Storozhenko S, De Pauw P, Van Montagu M, Inzé D, Kushnir S. 1998. The heat-shock element is a functional component of the Arabidopsis APX1 gene promoter. Plant Physiology 118, 1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen G, Lukasik E, Van Den Burg HA, Vossen JH, Cornelissen BJ, Takken FL. 2010. The small heat shock protein 20 RSI2 interacts with and is required for stability and function of tomato resistance protein I‐2. The Plant Journal 63, 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C. 2004. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. The Plant Journal 40, 428–438. [DOI] [PubMed] [Google Scholar]
- Wang X, Fan C, Zhang X, Zhu J, Fu Y-F. 2013. BioVector, a flexible system for gene specific-expression in plants. BMC Plant Biology 13, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dang F, Liu Z, Wang X, Eulgem T, Lai Y, Yu L, She J, Shi Y, Lin J. 2013. CaWRKY58, encoding a group I WRKY transcription factor of Capsicum annuum, negatively regulates resistance to Ralstonia solanacearum infection. Molecular Plant Pathology 14, 131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- War AR, Paulraj MG, Ahmad T, Buhroo AA, Hussain B, Ignacimuthu S, Sharma HC. 2012. Mechanisms of plant defense against insect herbivores. Plant Signaling & Behavior 7, 1306–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Avila CA, Goggin FL. 2015. The ethylene response factor Pti5 contributes to potato aphid resistance in tomato independent of ethylene signalling. Journal of Experimental Botany 66, 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Jiang Y, Duan Y, Karim A, Fan D, Yang L, Zhao X, Yin J, Luo K. 2014. Constitutive expression of the poplar WRKY transcription factor PtoWRKY60 enhances resistance to Dothiorella gregaria Sacc. in transgenic plants. Tree Physiology 34, 1118–1129. [DOI] [PubMed] [Google Scholar]
- Yogendra KN, Kushalappa AC, Sarmiento F, Rodriguez E, Mosquera T. 2015. Metabolomics deciphers quantitative resistance mechanisms in diploid potato clones against late blight. Functional Plant Biology 42, 284–298. [DOI] [PubMed] [Google Scholar]
- Yogendra KN, Pushpa D, Mosa KA, Kushalappa AC, Murphy A, Mosquera T. 2014. Quantitative resistance in potato leaves to late blight associated with induced hydroxycinnamic acid amides. Functional & Integrative Genomics 14, 285–298. [DOI] [PubMed] [Google Scholar]
- Yokotani N, Sato Y, Tanabe S, Chujo T, Shimizu T, Okada K, Yamane H, Shimono M, Sugano S, Takatsuji H. 2013. WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. Journal of Experimental Botany 64, 5085–5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.