Abstract
Cancer cells meet their needs for energy and biomass production by consuming high levels of nutrients and rewiring metabolism to support macromolecular biosynthesis. Mitochondrial enzymes play central roles in anabolic growth, and acetylation may provide a key layer of regulation over mitochondrial metabolic pathways. As a major mitochondrial deacetylase, SIRT3 regulates the activity of enzymes to coordinate global shifts in cellular metabolism. SIRT3 promotes the tricarboxylic acid (TCA) cycle, electron transport chain function, and reduces oxidative stress. Loss of SIRT3 triggers oxidative damage, reactive oxygen species (ROS)-mediated signaling, and metabolic reprogramming to support proliferation and tumorigenesis. Thus, SIRT3 is an intriguing example of how nutrient-sensitive, post-translational regulation may provide integrated regulation of metabolic pathways to promote metabolic homeostasis in response to diverse nutrient signals.
Tumor metabolism, SIRT3, and post-translational modification
A hallmark of tumor cells is altered metabolism to support rapid cell proliferation [1]. Many metabolic intermediates that support cell growth are provided by mitochondria; consequently, there is great interest in elucidating how mitochondrial metabolic pathways are regulated. In this review, we discuss the role of the conserved sirtuin family of deacetylases in nutrient-sensitive regulation of metabolic pathways, focusing on the mitochondrial sirtuin, SIRT3. By deacetylating proteins involved in multiple mitochondrial processes, SIRT3 can coordinate global shifts in mitochondrial activity, with important implications for tumor growth.
Expanding roles for sirtuins in post-translational regulation
Sirtuins are a conserved family of NAD-dependent enzymes that regulate metabolism, stress responses, and longevity in model organisms such as yeast, worms, and flies [2]. Initially classified as class III histone deacetylases, sirtuins are now known to possess diverse enzymatic activities that include ADP-ribosylation, desuccinylation, demalonylation, depropionylation, and debutyrylation, in addition to deacetylating a wide variety of protein substrates [2–6]. These deacylase and ADP-ribosyltransferase activities share a common catalytic mechanism that requires cleavage of NAD, generating either nicotinamide (NAM), 2′-O-acyl-ADP-ribose, and a de-acylated substrate or NAM and an ADP-ribosylated substrate [2, 3, 5]. In parallel, several recent studies have shed light on the diversity and prevalence of post-translational acyl modifications in organisms ranging from bacteria to humans [3, 7–9]. Unsurprisingly, the list of sirtuin targets is lengthening rapidly, and sirtuins are emerging as important regulators of diverse cellular processes.
Lysine acetylation is the best studied post-translational modification regulated by sirtuins. Recent proteomic surveys of mammalian cells have identified more than 2200 acetylated proteins [9]. A common theme to all of these studies is that enzymes involved in central metabolism are highly acetylated [9]. This observation is especially intriguing in light of the fact that acetyltransferases use acetyl-CoA as an acetyl donor and sirtuins use NAD for deacetylation; these metabolites are both important products and regulators of multiple metabolic pathways. Acetyl-CoA is formed from the breakdown of proteins, lipids, and amino acids and is a primary entry point for nutrient catabolism by the tricarboxylic acid (TCA) cycle, and NAD is a major electron acceptor for multiple metabolic reactions. Consequently, changes in nutrient availability or metabolic pathway activation can influence levels of both acetyl-CoA and NAD. Both glucose and fatty-acid-derived acetyl-CoA can provide an important source of acetyl-CoA for protein acetylation in cultured cells, and shifts in glucose or fatty acid availability correlate with changes in protein acetylation [10, 11]. Similarly, both fatty acid oxidation and glucose catabolism through glycolysis will increase NADH at the expense of NAD, and diet and intracellular signaling can further influence NAD levels [12, 13]. Together, these observations suggest that acetylation of metabolic enzymes may provide critical feedback regulation for cellular metabolism (Figure 1). Metabolites have long been recognized to play important roles in elegant feedback loops by allosterically regulating metabolic enzyme activity. The discovery of nutrient-sensitive post-translational acyl modifications adds another layer of control over metabolic pathways. Sirtuins—in contrast to class I and II histone deacetylases that catalyze deacetylation through simple hydrolysis—may play a special role in maintaining metabolic homeostasis by regulating acetylation. Furthermore, deacetylation via sirtuins generates a novel metabolite, 2′-O-acetyl-ADP-ribose, that may have a metabolic function on its own [14].
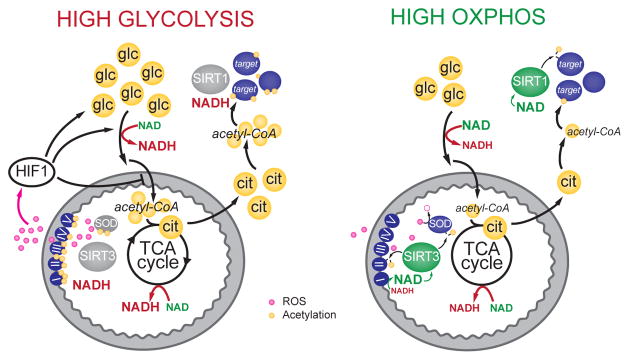
Figure 1. Speculative model for nutrient-sensitive regulation of protein acetylation by sirtuins.
Glucose is catabolized through glycolysis, producing NADH and pyruvate. Once transported to the mitochondria, pyruvate is converted to acetyl-CoA (large yellow circles), which is used to form citrate. Citrate is shuttled back to the cytoplasm, where it provides the major source of acetyl-CoA for protein acetylation [10]. Abundant glucose increases cellular acetyl-CoA pools, providing ample substrate for protein acetylation. High glycolytic activity will increase the nucleocytoplasmic NADH/NAD ratio, potentially reducing sirtuin activity (SIRT1 is shown as an example), and blocking deacetylation. Within the mitochondria, high acetyl-CoA levels may also drive protein acetylation, reducing the activity of multiple oxidative pathways. Increased acetylation of OXPHOS complexes may reduce flux through the ETC, preventing oxidation of NADH to NAD and increasing the likelihood of ROS production. At the same time, acetylation of SOD hampers ROS scavenging. Elevated ROS serve as a retrograde signal to stabilize HIF1α and promote the transcription of genes that will further activate glycolysis while inhibiting pyruvate conversion into acetyl-CoA, potentially providing a feedback reprieve that will block further protein acetylation and ROS production. Conversely, during conditions low glucose availability, acetyl-CoA pools will be reduced and the NAD/NADH ratio will increase, favoring protein deacetylation. Within the mitochondria, deacetylation may promote efficient electron transport through the ETC, promoting ATP production and reducing ROS generation. As NADH is efficiently converted to NAD at the ETC, SIRT3 will be active and maintain enzymes in a deacetylated state. Metabolites shown in yellow, ROS in pink, proteins in blue, sirtuins in grey (inactive) and green (active). Glc, glucose; cit, citrate.
Acetylated proteins are found in the nucleus, cytoplasm, and mitochondria, and the seven mammalian sirtuins (SIRT1-7) are similarly distributed throughout the cell. In particular, mitochondrial proteins appear to be acetylated at a high frequency, with current estimates indicating that approximately 20% of mitochondrial proteins are acetylated, often at multiple lysine residues [15]. The data suggest that acetylation serves as an important regulatory mechanism within mitochondria, possibly by altering protein-protein interactions, affecting complex stability, dictating intramitochondrial localization, or modulating enzyme activity. However, little is known about the enzymes regulating mitochondrial acetylation. While a recent report identified GCN5-like 1 (GCN5L1) as a positive regulator of mitochondrial acetylation [16], no mitochondrial protein with acetyltransferase activity has been identified. Although three members of the sirtuin family (SIRT3-5) and some histone deacetylases (HDAC7) localize to mitochondria, only SIRT3 has been shown to robustly deacetylate mitochondrial proteins [17, 18]. SIRT4 and SIRT5 may have stringent substrate specificity, or they may possess different deacylase activities. Indeed, SIRT5 was recently identified as a desuccinylase and demalonylase [3]. Consistent with the high deacetylase activity of SIRT3, mitochondrial proteins are hyperacetylated in SIRT3 knockout (KO) mice, suggesting that SIRT3 is a major mitochondrial deacetylase [19].
Regulation of mitochondrial metabolism by acetylation
Mitochondria are dynamic organelles that integrate multiple physiological stimuli to regulate energy production, apoptosis, and intracellular signaling. Not surprisingly, mitochondrial dysfunction is implicated in diseases as varied as diabetes, cardiovascular disease, neurodegeneration, and cancer [20]. Mitochondria serve many metabolic functions beyond ATP production: fatty acid oxidation, amino acid catabolism, metabolism of acetyl-CoA by the TCA cycle, and biosynthesis of various molecules all occur in the mitochondrial matrix. Many enzymes in these pathways are acetylated and, in several cases, their deacetylation is regulated by SIRT3. Consequently, SIRT3 is emerging as a major regulator of mitochondrial metabolic pathways, which may play an important role in cellular and organismal metabolic homeostasis.
Highlighting the global role of SIRT3, recent studies have demonstrated that SIRT3 activity can rewire cellular metabolism. SIRT3-driven metabolic reprogramming may be particularly important in the context of cancer, as cancer cell growth is associated with profound metabolic alterations, many of which support cancer cell proliferation and may provide an important therapeutic window for blocking tumor growth. In this review, we will discuss our current knowledge of how metabolism is regulated by SIRT3 and how these metabolic pathways may contribute to tumorigenesis.
Cancer cells have altered metabolic requirements
Although Otto Warburg first noted in the 1920s that cancer cells have altered metabolism relative to their normal counterparts, we are only recently beginning to appreciate the scope of these metabolic changes and their potential oncogenic advantages. Warburg argued that cancer was the result of irreversible damage to cellular respiration followed by a compensatory increase in fermentation [21]. The result—conversion of glucose to lactate through glycolysis, even in the presence of ample oxygen—is known as the Warburg effect. Today, we know that disrupted respiration is not required for increased glycolysis in cancer cells because most cancer cells have normal respiration [22]. Rather, the increased glycolysis characteristic of tumor cells is the result of active metabolic reprogramming designed to support cell growth and proliferation. Importantly, we also recognize that cancer cells take up many nutrients, not just glucose, that are used to fuel macromolecular biosynthesis. These observations indicate that cancer cells have fundamentally different metabolic requirements than normal, differentiated cells and that metabolic reprogramming can provide a selective advantage that drives tumor growth. In support of this hypothesis, most oncogenes and tumor suppressors, including the PI3K/Akt, Ras, Myc, and p53 pathways, directly regulate cellular metabolism (for reviews, see [22, 23]).
A common feature of oncogenic metabolic reprogramming is the redirection of metabolic intermediates towards anabolic pathways. In general, increased glycolytic flux enables various glycolytic intermediates to enter biosynthetic pathways, generating the building blocks that are critical for lipid, protein, and nucleotide biosynthesis (Figure 2) [24]. Consequently, metabolic alterations that promote accumulation of glycolytic intermediates—whether by increasing glucose uptake and glycolytic enzyme expression or slowing the final step in glycolysis—will favor the shunting of glycolytic intermediates towards anabolic pathways [22]. Indeed, proliferating cancer cells preferentially express a less-active (and readily inhibited) isoform of pyruvate kinase (PKM2), the enzyme that catalyzes the final, ATP producing step in glycolysis [25]. With this isoform, less ATP is produced during glycolysis, and upstream glycolytic intermediates are shunted toward macromolecule synthesis. Thus, while Warburg postulated that the major purpose of increased glycolysis is the generation of ATP, current evidence suggests that cancer cells may hijack glycolytic metabolism to produce key biosynthetic intermediates rather than ATP itself [24].
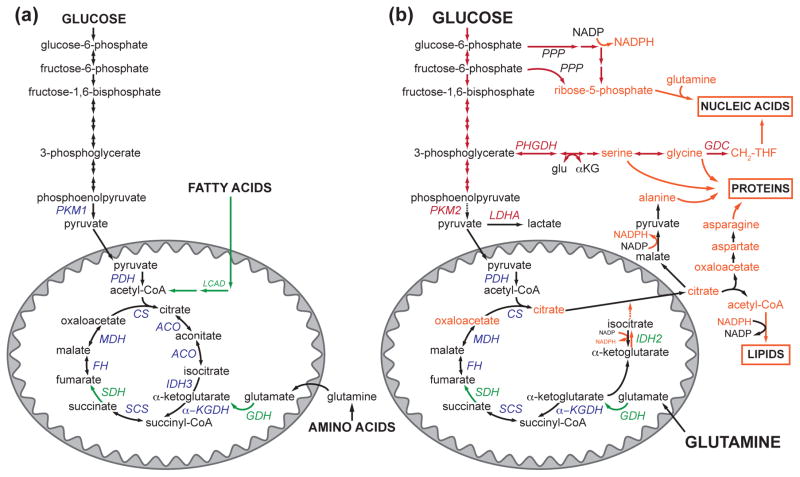
Figure 2. Metabolic pathways are rewired to support proliferation.
(a) Quiescent cells funnel glucose, amino acids, and fatty acids into the mitochondrial TCA cycle for energy production. They also express the PKM1 isoform of pyruvate kinase, which efficiently catalyzes the last step in glycolysis. SIRT3 can promote nutrient catabolism by targeting enzymes in fatty acid oxidation (LCAD), amino acid catabolism (GDH), and the TCA cycle itself (SDH). At the same time, SIRT3 activates OXPHOS to harness the products of the TCA cycle and convert them to ATP. (b) Proliferating cells rewire their metabolic pathways to support biomass generation and can take up high levels of nutrients, including glucose and glutamine. Often, proliferating cells convert pyruvate to lactate at a high rate through the action of lactate dehydrogenase, LDHA, a hallmark of the Warburg effect. Many proliferating cells express the PKM2 isoform of pyruvate kinase, which slows the final step of glycolysis. Glycolytic intermediates are shunted to anabolic pathways, promoting the generation of the carbon units and reducing potential required to synthesize proteins, lipids, and nucleic acids. Mitochondrial intermediates, particularly citrate, are critical for proliferation, and the mitochondrial TCA cycle is reconfigured to maintain metabolite efflux. Although SIRT3 can target several of the enzymes that play a role in mitochondrial anaplerosis and cataplerosis (i.e., IDH2 and GDH), the role of SIRT3 in supporting these processes is largely unknown. Glu, glutamate.
Recent studies illustrate how hyperactivation of enzymes that regulate biosynthetic pathways can support tumor growth. For example, elevated expression of glycine decarboxylase (GDC), which generates the methylene-tetrahydrofolate that is critical for nucleotide biosynthesis, is necessary and sufficient for cellular transformation and tumorigenesis [26]. Similarly, 3-phosphoglycerate dehydrogenase (PHGDH), which catalyzes the first step in the serine synthesis pathway that branches from glycolysis (Figure 2), is amplified in several human cancers, and increased flux into the serine synthesis pathway can support cancer cell proliferation [27, 28]. These studies demonstrate that increased flux through anabolic pathways is in some cases sufficient to support tumor growth.
Proliferating cells rely on mitochondrial metabolic pathways
Whereas glycolysis is an important source of many biosynthetic intermediates, several mitochondrial metabolic pathways play critical roles in generating key anabolic metabolites. The macromolecules needed to support cell proliferation are derived from metabolic building blocks provided by mitochondria [29]. For example, citrate is produced in the mitochondria by the condensation of acetyl-CoA and oxaolacetate, a reaction that is catalyzed by citrate synthase in the first step of the mitochondrial TCA cycle (Figure 2). Citrate can be exported to the cytosol through mitochondrial citrate transporters and converted back to acetyl-CoA by ATP-citrate lyase [30]. This acetyl-CoA is the major carbon source for fatty acid synthesis, generating the lipids that will make the membranes needed for cell replication. Similarly, oxaloacetate and α-ketoglutarate provide the carbon backbone for nonessential amino acids, which are used for protein and nucleic acid synthesis. Thus, while quiescent cells mainly utilize the TCA cycle to oxidize nutrients, generating NADH and FADH2 to fuel ATP production through the mitochondrial electron transport chain, proliferating cells use the TCA cycle to provide the building blocks necessary to support cell growth. In this manner, mitochondrial pathways are rewired to support proliferation.
Consequently, metabolic rearrangements that support mitochondrial metabolic networks play important roles in cancer growth. Notably, Possemato et al. demonstrated that PHGDH amplification did not promote cell growth by increasing intracellular serine levels, but rather, the serine biosynthesis pathway provided a critical source of glutamine-derived α-ketoglutarate, a key intermediate in the mitochondrial TCA cycle [28]. Indeed, generating α-ketoglutarate from glutamine is critical to the survival and growth of many cancer cells. To maintain efflux of TCA cycle metabolites for anabolic purposes, cells must replenish TCA cycle intermediates in a process known as anaplerosis. Glutamine-derived α-ketoglutarate is a major source of anaplerosis in proliferating cells; not surprisingly, many cancer cells take up high levels of glutamine [31]. Despite the importance of anaplerosis in supporting cell proliferation, little is known about the regulation of anaplerotic flux or the activity of mitochondrial metabolic networks [29].
These insights into the role of TCA cycle metabolites in proliferating cells emphasize the active role that mitochondria play in the metabolic reprogramming of cancer cells. Our revised understanding of the role of mitochondria in cancer metabolism increases the importance of elucidating how mitochondrial metabolism is regulated during both normal and pathological conditions.
Mitochondrial acetylation is dynamic
The sheer amount of acetylation of mitochondrial proteins suggests that acetylation may provide an important layer of regulation over mitochondrial metabolic pathways. Furthermore, acetylation of mitochondrial proteins is dynamically regulated by changes in nutrient status. Several studies have elegantly detailed how changes in availability of specific nutrients can influence the acetylation of individual proteins in cultured cells (for examples, see [11, 32]). Strikingly, calorie restriction, high fat diet, ethanol feeding, as well as fasting and refeeding can all cause dramatic shifts in global mitochondrial protein acetylation in vivo [33–37]. Similarly, SIRT3 expression is regulated by conditions that alter cellular nutrient balance. Conditions that increase demand for mitochondrial energy production, such as fasting, exercise, and calorie restriction, can induce SIRT3 gene expression in both liver and skeletal muscle [38–42]. Conversely, aging and high fat diet reduce SIRT3 expression [41, 43]. Such changes in SIRT3 expression indicate that acetylation may rewire mitochondrial metabolic pathways in response to changing nutrient status, and it will be interesting for future studies to examine the tissue-specific responses of SIRT3 expression to various metabolic stresses.
The growing list of SIRT3 targets supports the hypothesis that acetylation can direct mitochondrial metabolism. In general, SIRT3 appears to help mitochondria adjust to periods of real or perceived energy limitation by activating enzymes involved in fuel catabolism and energy production (see below). Consequently, SIRT3 activity may provide an avenue for cells to toggle mitochondrial metabolism between oxidative, catabolic states that enable survival during periods of energy limitation and reductive, anabolic states that promote cell growth.
SIRT3 regulates multiple mitochondrial oxidative pathways
SIRT3 deacetylates enzymes involved in diverse mitochondrial functions, including energy production and antioxidant defense, indicating that SIRT3 activity may ensure coordinated regulation of several distinct mitochondrial functions (Figure 3). In particular, SIRT3 targets enzymes involved in multiple mitochondrial oxidative pathways, with the cumulative effect of promoting nutrient oxidation and energy production.
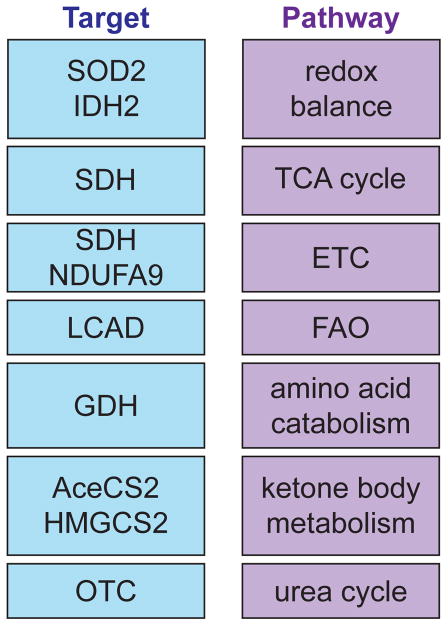
Figure 3. SIRT3 targets proteins in multiple mitochondrial pathways.
A list of selected proteins that are directly deacetylated by SIRT3 (target) and the mitochondrial pathways they influence reveals that SIRT3 can coordinate disparate mitochondrial processes. ETC, electron transport chain; FAO, fatty acid oxidation; SOD2, manganese superoxide dismutase; IDH2, isocitrate dehydrogenase 2; SDH, succinate dehydrogenase; NDUFA9, subunit of complex I; LCAD, long chain acyl-CoA dehydrogenase; GDH, glutamate dehydrogenase; AceCS2, acetyl-CoA synthetase 2; HMGCS2, 3-hydroxy-3-methylglutaryl CoA synthase 2; OTC, ornithine transcarbamoylase.
Oxidative phosphorylation
Oxidative phosphorylation (OXPHOS) is the process by which mitochondria generate ATP. High-energy electrons derived from the oxidation of nutrients by the TCA cycle are passed through a series of protein complexes (I–IV) embedded in the inner-mitochondrial membrane. The energy of the electrons is used to pump protons from the mitochondrial matrix into the inner-membrane space, generating a proton gradient. Protons then flow down their energy gradient through ATP synthase (or complex V), which harnesses their energy to form ATP from ADP. Finally, the electrons combine with oxygen to form water in a reaction catalyzed by subunits of complex IV. Inevitable by-products of this process are reactive oxygen species (ROS) that form when electrons leak out of the ETC at complex I or complex III and combine with oxygen to form the highly reactive superoxide radical [20].
Each of the OXHPOS complexes (I–V) contains multiple subunits, many of which are acetylated [15]. Several lines of evidence indicate that SIRT3 may influence multiple steps of OXPHOS. First, the activities of complexes I–III are reduced in SIRT3 null cells [44–46], with increased ROS leakage out of complex III [47] and lowered cellular ATP levels [45] both associated with loss of SIRT3. These observations suggest that efficient electron transport through the ETC may be slowed in SIRT3 null cells, and it will be important for future studies to elucidate the mechanisms by which SIRT3 influences OXPHOS function. Currently, it is known that SIRT3 physically associates with complex I, II and V; SIRT3 can deacetylate subunits of complex I and II, and subunits of complexes I–III and V are hyperacetylated in SIRT3 KO cells [45, 46, 48, 49]. A combination of biochemistry and structural biology may be required to elucidate how acetylation of specific residues can influence electron transfer through the ETC. Given that proteins in macromolecular complexes are more likely to be acetylated [50] and that acetylation can influence the physical association of two proteins [32], one intriguing hypothesis is that acetylation of OXPHOS subunits may regulate OXPHOS complex assembly or subunit interactions.
TCA cycle
Every enzyme in the TCA cycle can be acetylated [11], but to date, only succinate dehydrogenase (SDH) is known to be a target of SIRT3 [46, 48]. SIRT3 can directly deacetylate the flavoprotein subunit of SDH (SDHA), and SIRT3 loss in mouse embryonic fibroblasts (MEFs) and murine brown adipose tissue (BAT) results in reduced SDH activity [48]. While future studies will likely show that SIRT3 regulates multiple TCA cycle enzymes, our current knowledge that SDH is regulated by SIRT3 underscores the potential of SDH as an important regulatory node. SDH (also known as complex II) participates in both the TCA cycle and OXPHOS and is thus poised to regulate the activity of both pathways. By modulating SDH activity, SIRT3 could ensure coordination of fuel oxidation through the TCA cycle and substrate delivery to the electron transport chain to reduce the likelihood of overloading the electron transport chain and generating ROS.
SIRT3 also targets isocitrate dehydrogenase 2 (IDH2) [42, 51]. Although isocitrate dehydrogenase activity—the oxidation of isocitrate to α-ketoglutarate—is a key step of the TCA cycle, the NAD-dependent enzyme IDH3 is likely the major IDH isoform active in the TCA cycle. IDH2 is NADP-dependent, and because NADP is not an electron carrier for the ETC, IDH2 is not traditionally considered to be a TCA cycle enzyme. Nevertheless, IDH2 carries out several functions that are particularly relevant to a discussion of cancer metabolism. First, by reducing NADP to NADPH, IDH2 has an important role providing reducing equivalents that could be used to combat oxidative stress or promote anabolic processes (Figure 2) [22, 42].
Second, IDH2 can also provide a critical source of citrate during periods of hypoxia that characterize the tumor microenvironment [52]. Normal cells use pyruvate and glutamine to generate citrate, which provides carbon for fatty acid and cholesterol synthesis and can generate cytosolic NAPDH for anabolic processes. Both the conversion of pyruvate to acetyl-CoA and glutamine to oxaloacetate via the TCA cycle require oxidized NAD, which can be limiting under hypoxic conditions. Several elegant studies demonstrated that during conditions of hypoxia or mitochondrial dysfunction, both cytosolic and mitochondrial NADP-dependent isocitrate dehydrogenases can catalyze reductive carboxylation of glutamine-derived α-ketoglutarate to form isocitrate, which can be converted to citrate [52–54]. Thus, reductive carboxylation catalyzed by IDH2 may be a key mechanism through which tumors maintain anabolic processes during periods of hypoxia.
SIRT3 directly deacetylates IDH2, promoting its forward activity—oxidative decarboxylation—and generating NADPH [42]. This NAPDH supports glutathione antioxidant systems and protects against oxidative stress. It will be interesting for future studies to examine whether SIRT3 actively suppresses the reverse, reductive carboxylation activity of IDH2, which would have important implications for tumor cell growth.
Other mitochondrial pathways
An increasing number of mitochondrial proteins are targeted by SIRT3 and a common theme is emerging: SIRT3 generally activates a program of mitochondrial oxidative metabolism (Figure 3). For example, SIRT3 stimulates fatty acid oxidation by deacetylating long chain acyl-CoA dehydrogenase (LCAD) [40]. Similarly, SIRT3 activates glutamate dehydrogenase (GDH) to increase amino acid oxidation via the TCA cycle [51]. In addition to the energy-generating pathways listed above, SIRT3 targets mitochondrial pathways that are important for maintaining mitochondrial homeostasis during conditions of nutrient deprivation. In response to fasting, SIRT3 deacetylates and activates 3-hydroxy-3-methylglutaryl CoA synthase 2 (HMGCS2), the rate-limiting enzyme in the synthesis of the ketone body β-hydroxybutyrate [55]. Moreover, SIRT3 promotes energy generation from ketone bodies through acetyl-CoA synthase 2 (AceCS2), which converts acetate to acetyl-CoA for metabolism by the TCA cycle [56, 57]. During conditions such as fasting or CR, elevated amino acid catabolism requires a concomitant increase in the activity of the urea cycle. SIRT3 coordinates these two pathways by deacetylating ornithine transcarbamoylase (OTC), facilitating flux through the urea cycle during CR [39]. Taken together, these studies demonstrate that SIRT3 functions to reprogram mitochondrial metabolism during nutrient stress. SIRT3 funnels fatty acids, amino acids and ketone bodies into the TCA cycle to fuel OXPHOS and sustain energy production. At the same time, SIRT3 stimulates the urea cycle and ketone body synthesis in order to preserve metabolic homeostasis.
SIRT3 influences cellular signaling pathways that promote tumorigenesis
SIRT3 also has an important role maintaining metabolic homeostasis by regulating mitochondrial redox balance. Several groups have reported stress- and diet-inducible roles for SIRT3 in the regulation of manganese superoxide dismutase (SOD2), the major mitochondrial superoxide detoxifier [58, 59]. Activation of IDH2 by SIRT3 similarly protects against oxidative stress-induced cell death [42]. It is interesting to speculate that the antioxidant program could be an integral part of SIRT3-mediated metabolic reprogramming. Because increased OXPHOS amplifies mitochondrial ROS production, SIRT3 must also improve antioxidant defenses to maintain an appropriate redox state.
Elevated ROS production can also promote tumorigenesis through many mechanisms and is a common feature of human cancers [60]. ROS activate signaling pathways that regulate cell growth and proliferation, differentiation, survival, inflammation, and metabolism [60]. Furthermore, oxidative damage to proteins, lipids, and DNA can harm cellular processes and cause mutations that further promote tumor progression.
Several groups have recently found that SIRT3 can function as a tumor suppressor by regulating ROS levels [44, 47, 59, 61]. Cells with reduced SIRT3 expression had elevated ROS levels, stemming from increased electron leakage at complex III or reduced MnSOD detoxifying activity [47, 59]. In turn, SIRT3 null MEFs displayed increased genomic instability and could be transformed by expression of a single oncogene [44, 59]. Consequently, SIRT3 null cells formed larger tumors in xenograft assays [44, 47, 61], and tumor growth could be reversed by treatment with antioxidants or induction of SIRT3 expression [47]. A variety of human tumors have reduced SIRT3 expression, supporting the hypothesis that SIRT3 acts a tumor suppressor in humans [44, 61]. Breast cancers, in particular, have striking reductions in SIRT3 levels: 40% of a panel of human breast tumors demonstrated loss of at least one SIRT3 allele and 20% had no detectable SIRT3 protein [61].
SIRT3 is a tumor suppressor that opposes cancer-associated metabolic reprogramming
Given the prominent role of SIRT3 in regulating mitochondrial metabolic processes, we hypothesized that metabolic regulation by SIRT3 might contribute to its ability to act as a tumor suppressor. SIRT3 activity promotes substrate oxidation and ATP production by directly deacetylating numerous mitochondrial targets, and conversely, SIRT3 loss triggers increased glucose uptake and lactate production, hallmarks of elevated glycolysis [44, 61]. Metabolomic analyses revealed that SIRT3 null cells had a metabolic profile similar to proliferating cancer cells, including accumulation of glycolytic intermediates, and this elevated glycolytic metabolism supported increased cell proliferation [61]. These metabolic shifts were not merely the result of dysfunctional mitochondria in the absence of SIRT3 expression but also active signaling downstream of elevated ROS.
The transcription factor hypoxia-inducible factor 1α (HIF1α) regulates cell metabolism in response to changes in ROS or oxygen concentration [62]. Under normal conditions, HIF1α is hydroxylated by prolyl hydroxylases (PHDs), targeting it for polyubiquitination and proteasomal degradation. Low oxygen availability or elevated ROS serve as a signals to reduce PHD activity, enabling HIF1α stabilization [63]. Together with its binding partner HIF1β, HIF1α activates transcription of genes crucial for the cellular response to hypoxia, including many enzymes that promote anaerobic metabolism of glucose through glycolysis [64]. By regulating ROS, SIRT3 can directly influence HIF1α stability and target gene expression [47, 61]. Furthermore, reduced SIRT3 expression in human breast cancers correlates with increased HIF1α target gene expression, suggesting a metabolic mechanism for tumor suppression by SIRT3 [61].
Conclusions
Warburg’s vision of cancer cell metabolism, which sidelined mitochondria as dysfunctional bystanders, must be revised to include the critical contribution of mitochondria to supporting tumor growth. Mitochondrial pathways that are used largely to produce ATP in nonproliferating cells are rewired to serve as biosynthetic factories in rapidly proliferating cells, and SIRT3 is emerging as a critical regulator of the balance between mitochondrial, oxidative metabolism and glycolytic, anabolic pathways. In nutrient-deprived conditions, SIRT3 deacetylates numerous mitochondrial proteins to promote nutrient oxidation and ATP production. Conversely, low SIRT3 activity increases ROS production, which signals through HIF1α to increase glycolytic metabolism and cellular proliferation. Increasing evidence indicates that SIRT3 plays a unique regulatory role, integrating mitochondrial regulation with intracellular signaling cascades. By targeting more than a half a dozen key metabolic enzymes, SIRT3 is perfectly positioned to orchestrate coordinated shifts in mitochondrial metabolism, with potential implications for diseases that rely on mitochondrial metabolic activities. It will be important for future studies to increase our understanding of how mammalian sirtuins integrate metabolism with signaling cascades by post-translational modification of diverse substrates, and how modulation of sirtuin activity can influence health and disease (see Box).
BOX. Outstanding questions.
How is SIRT3 activity regulated? Although SIRT3 expression is known to be regulated by conditions that alter cellular nutrient balance, little is known about factors that acutely influence SIRT3 activity. As SIRT3 is a NAD-dependent deacetylase, it is tempting to speculate that SIRT3 could sense flux through the ETC. For example, reduced ETC flux—whether because of dysfunction or reduced ADP availability—would decrease NAD levels and inhibit SIRT3 activity (Figure 1). Subsequent increases in ROS would stabilize HIF1α and redirect metabolism away from mitochondrial oxidative processes. Conversely, low membrane potential and high ADP levels would promote NADH oxidation, increasing NAD levels and SIRT3 activity, this promoting mitochondrial substrate oxidation to further supply the ETC. Alternatively, nutrient sensitive modifications such as acetylation or phosphorylation may influence SIRT3 activity, as has been shown for SIRT1 [65].
What factors promote mitochondrial aceyltation? Because SIRT3 can function as a tumor suppressor, a mitochondrial acetyltransferase could be an oncogene. Although no mitochondrially-localized acetyltransferase has been identified, the acetylation of a subunit of complex V that is encoded by mitochondrial DNA suggests that acetylation—whether enzymatic or not—may take place in the mitochondria itself [15]. Furthermore, it is not clear if changes in the mitochondrial acetyl-CoA pool can underlie shifts in mitochondrial acetylation analogous to how intracellular acetyl-CoA can drive histone acetylation [66]. Given that altering mitochondrial enzyme acetylation appears to affect multiple metabolic processes, it will be important for future studies to address these questions.
Can SIRT3 function as a tumor suppressor and an oncogene, depending on the context? Several studies suggest that in certain circumstances (for example, oral squamous cell carcinoma), SIRT3 can function as an oncogene [67, 68]. One potential explanation for this discrepancy is the context-specific metabolic programs that can support tumor development. For example, many cancer cells rely on fatty acid synthesis for growth, and enzymes involved in fatty acid synthesis have been suggested as therapeutic targets [69]. However, fatty acid oxidation can promote survival in matrix-detached cells, providing an oncogenic advantage [70]. Similarly, ROS can be oncogenic in some contexts and tumor suppressive in others; these apparent contradictions may depend upon ROS levels in the cell. Whereas moderate ROS can promote oncogenic signaling and mutagenesis, excessive ROS can trigger senescence and cell death [22]. Thus, the influence of SIRT3 on various metabolic pathways and ROS production may have opposite effects on tumor cell growth in different contexts.
Is altered metabolism a driver or a passenger in cancer progression? This is one of the largest questions currently facing the field. Certainly, common oncogenes that can transform cells can also reprogram metabolism to increase nutrient uptake, glycolysis and glutaminolysis, and promote anabolic pathways [22]. Furthermore, these metabolic alterations are apparently required for cell proliferation, as many transformed cells display heightened susceptibility to nutrient withdrawal or metabolic pathway inhibition [69]. This observation has fueled much research into the therapeutic opportunities provided by altered cancer metabolism, and several drugs that target the specific metabolic properties of cancer cells are in development [69]. Nevertheless, evidence that altered metabolism could by itself promote tumorigenesis has, until recently, remained elusive. The first evidence that expression of a single metabolic enzyme could transform cells was provided by Zhang et al, who demonstrated that glycine decarboxylase (GDC) can transform multiple cell lines [26]. GDC expression triggers increases in several metabolic intermediates, including metabolites that support pyrimidine synthesis, and it will be important for future studies to elucidate the specific metabolites that contribute to oncogenesis. We are increasingly recognizing the ability of metabolic intermediates to serve as signaling molecules that regulate multiple cellular processes, including signaling cascades and gene expression. In general, oncogenic signaling and metabolic pathways are so entwined that it will be difficult to separate the two hypotheses. Regardless of whether altered metabolism is a driver or a passenger in tumorigenesis, it is increasingly clear that to understand how tumors grow we must understand how—and why—they reprogram their metabolism. Perhaps this understanding will lead to new, productive therapeutic avenues.
Acknowledgments
We thank Pat Ward for helpful discussion and Natalie German for critical reading of the manuscript. M.H. is supported by funding from the Paul F. Glenn Foundation for Medical Research, Ellison Medical Foundation, American Cancer Society, and NIH-AG032375.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annual review of pathology. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du J, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng C, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Molecular & cellular proteomics : MCP. 2011;10 doi: 10.1074/mcp.M111.012658. M111 012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith BC, Denu JM. Acetyl-lysine analog peptides as mechanistic probes of protein deacetylases. The Journal of biological chemistry. 2007;282:37256–37265. doi: 10.1074/jbc.M707878200. [DOI] [PubMed] [Google Scholar]
- 6.Garrity J, et al. N-lysine propionylation controls the activity of propionyl-CoA synthetase. The Journal of biological chemistry. 2007;282:30239–30245. doi: 10.1074/jbc.M704409200. [DOI] [PubMed] [Google Scholar]
- 7.Thao S, Escalante-Semerena JC. Control of protein function by reversible Nvarepsilon-lysine acetylation in bacteria. Current opinion in microbiology. 2011;14:200–204. doi: 10.1016/j.mib.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Molecular & cellular proteomics : MCP. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan KL, Xiong Y. Regulation of intermediary metabolism by protein acetylation. Trends in biochemical sciences. 2011;36:108–116. doi: 10.1016/j.tibs.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao S, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canto C, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen D, et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes & development. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong L, Denu JM. Function and metabolism of sirtuin metabolite O-acetyl-ADP-ribose. Biochimica et biophysica acta. 2010;1804:1617–1625. doi: 10.1016/j.bbapap.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SC, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Molecular cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 16.Scott I, et al. Identification of a molecular component of the mitochondrial acetyltransferase programme: a novel role for GCN5L1. The Biochemical journal. 2012;443:655–661. doi: 10.1042/BJ20120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakin RE, Jung MO. Cytoplasmic sequestration of HDAC7 from mitochondrial and nuclear compartments upon initiation of apoptosis. The Journal of biological chemistry. 2004;279:51218–51225. doi: 10.1074/jbc.M409271200. [DOI] [PubMed] [Google Scholar]
- 18.Verdin E, et al. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends in biochemical sciences. 2010;35:669–675. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lombard DB, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Molecular and cellular biology. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annual review of genetics. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 22.Cairns RA, et al. Regulation of cancer cell metabolism. Nature reviews. Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 23.Vander Heiden MG, et al. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Locasale JW, Cantley LC. Metabolic flux and the regulation of mammalian cell growth. Cell metabolism. 2011;14:443–451. doi: 10.1016/j.cmet.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 26.Zhang WC, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 27.Locasale JW, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nature genetics. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeBerardinis RJ, et al. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell metabolism. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Srere PA. The citrate cleavage enzyme. I. Distribution and purification. The Journal of biological chemistry. 1959;234:2544–2547. [PubMed] [Google Scholar]
- 31.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends in biochemical sciences. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin YY, et al. Functional dissection of lysine deacetylases reveals that HDAC1 and p300 regulate AMPK. Nature. 2012;482:251–255. doi: 10.1038/nature10804. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Schwer B, et al. Calorie restriction alters mitochondrial protein acetylation. Aging cell. 2009;8:604–606. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirschey MD, et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Molecular cell. 2011;44:177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kendrick AA, et al. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. The Biochemical journal. 2011;433:505–514. doi: 10.1042/BJ20100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Picklo MJ., Sr Ethanol intoxication increases hepatic N-lysyl protein acetylation. Biochemical and biophysical research communications. 2008;376:615–619. doi: 10.1016/j.bbrc.2008.09.039. [DOI] [PubMed] [Google Scholar]
- 37.Yang L, et al. The fasted/fed mouse metabolic acetylome: N6-acetylation differences suggest acetylation coordinates organ-specific fuel switching. Journal of proteome research. 2011;10:4134–4149. doi: 10.1021/pr200313x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barger JL, et al. A low dose of dietary resveratrol partially mimics caloric restriction and retards aging parameters in mice. PloS one. 2008;3:e2264. doi: 10.1371/journal.pone.0002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hallows WC, et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Molecular cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirschey MD, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palacios OM, et al. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1alpha in skeletal muscle. Aging. 2009;1:771–783. doi: 10.18632/aging.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Someya S, et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lanza IR, et al. Endurance exercise as a countermeasure for aging. Diabetes. 2008;57:2933–2942. doi: 10.2337/db08-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim HS, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahn BH, et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cimen H, et al. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry. 2010;49:304–311. doi: 10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bell EL, et al. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finley LW, et al. Succinate dehydrogenase is a direct target of sirtuin 3 deacetylase activity. PloS one. 2011;6:e23295. doi: 10.1371/journal.pone.0023295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jing E, et al. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14608–14613. doi: 10.1073/pnas.1111308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 51.Schlicker C, et al. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. Journal of molecular biology. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 52.Wise DR, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shimazu T, et al. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell metabolism. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hallows WC, et al. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwer B, et al. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qiu X, et al. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell metabolism. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 59.Tao R, et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Molecular cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liou GY, Storz P. Reactive oxygen species in cancer. Free radical research. 2010;44:479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finley LW, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Semenza GL. Hypoxia-inducible factor 1: regulator of mitochondrial metabolism and mediator of ischemic preconditioning. Biochimica et biophysica acta. 2011;1813:1263–1268. doi: 10.1016/j.bbamcr.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Molecular cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 64.Majmundar AJ, et al. Hypoxia-inducible factors and the response to hypoxic stress. Molecular cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gerhart-Hines Z, et al. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+) Molecular cell. 2011;44:851–863. doi: 10.1016/j.molcel.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cai L, et al. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Molecular cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alhazzazi TY, et al. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer. 2011;117:1670–1678. doi: 10.1002/cncr.25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alhazzazi TY, et al. SIRT3 and cancer: tumor promoter or suppressor? Biochimica et biophysica acta. 2011;1816:80–88. doi: 10.1016/j.bbcan.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nature reviews. Drug discovery. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 70.Schafer ZT, et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]