Abstract
We introduce a new divide and conquer approach to deal with the problem of de novo genome assembly in the presence of ultra-deep sequencing data (i.e. coverage of 1000x or higher). Our proposed meta-assembler Slicembler partitions the input data into optimal-sized ‘slices’ and uses a standard assembly tool (e.g. Velvet, SPAdes, IDBA_UD and Ray) to assemble each slice individually. Slicembler uses majority voting among the individual assemblies to identify long contigs that can be merged to the consensus assembly. To improve its efficiency, Slicembler uses a generalized suffix tree to identify these frequent contigs (or fraction thereof). Extensive experimental results on real ultra-deep sequencing data (8000x coverage) and simulated data show that Slicembler significantly improves the quality of the assembly compared with the performance of the base assembler. In fact, most of the times, Slicembler generates error-free assemblies. We also show that Slicembler is much more resistant against high sequencing error rate than the base assembler.
Availability and implementation: Slicembler can be accessed at http://slicembler.cs.ucr.edu/.
Contact: hamid.mirebrahim@email.ucr.edu
1 Introduction
Since the early days of DNA sequencing, the problem of de novo genome assembly has been characterized by insufficient and/or uneven depth of sequencing coverage (see e.g. Ekblom et al., 2014). Insufficient sequencing coverage, along with other shortcomings of sequencing instruments (e.g. short read length and sequencing errors) exacerbated the algorithmic challenges in assembling large, complex genome—in particular those with high repetitive content. Some of the third generation of sequencing technology currently on the market, e.g. Pacific Biosciences (Eid et al. 2009) and Oxford Nanopore (Clarke et al. 2009), offers very long reads at a higher cost per base, but sequencing error rate is much higher. As a consequence, long reads are more commonly used for scaffolding contigs created from second generation data, rather than for de novo assembly (English et al. 2012).
Thanks to continuous improvements in sequencing technologies, life scientists can now easily sequence DNA at depth of sequencing coverage in excess of 1000x, especially for smaller genomes like viruses, bacteria or bacterial artificial chromosome (BAC)/YAC clones. ‘Ultra-deep’ sequencing (i.e. 1000 x or higher) has already been used in the literature for detecting rare DNA variants including mutations causing cancer (Campbell et al. 2008), for studing viruses (Beerenwinkel and Zagordi 2011; Widasari et al. 2014), as well as other applications (Ekblom et al. 2014). As it becomes more and more common, ultra-deep sequencing data are expected to create new algorithmic challenges in the analysis pipeline. In this article, we focus on one of these challenges, namely the problem of de novo assembly. We showed recently that modern de novo assemblers SPAdes (Bankevich et al. 2012), IDBA_UD (Peng et al. 2012) and Velvet (Zerbino and Birney 2008) are unable to take advantage of ultra-deep coverage (Lonardi et al. 2015). Even more surprising was the finding that the assembly quality produced by these assemblers starts degrading when the sequencing depth exceeds 500x–1000x (depending on the assembler and the sequencing error rate). By means of simulations on synthetic reads, we also showed in Lonardi et al. (2015) that the likely culprit is the presence of sequencing errors: the assembly quality degradation cannot be observed with error-free reads, whereas higher sequencing error rate intensifies the problem. The ‘message’ of our study (Lonardi et al. 2015) is that when the data are noisy, more data are not necessarily better. Rather, there is an error-rate-dependent optimum.
Independently from us, study (Desai et al. 2013) reached similar conclusions: the authors assembled E. coli (4.6 Mb), S. kudriavzevii (11.18 Mb) and C. elegans (100 Mb) using SOAPdenovo, Velvet, ABySS, Meraculous and IDBA_UD at increasing sequencing depths up to 200x (which is not ultra-deep according to our definition). Their analysis showed an optimum sequencing depth (around 100x) for assembling these genomes, which depends on the specific genome and the assembler.
In addition to sequencing errors, real sequencing data are also plagued by read duplications that contribute to uneven coverage. Read duplication is typically attributed to polymerase chain reaction amplification bias (Aird et al. 2011; Zhou et al. 2014). The presence of highly duplicated reads complicates the task for assemblers when they contain sequencing errors; if unique, it would be easy to detect and remove them. As the coverage increases, the probability of an overlap that involves duplicated reads agreeing to each other due to sequencing errors becomes higher and higher. These new overlaps can induce spurious contigs (typically short) or prevent the creation of longer contigs. In turn, this manifests in a degradation of the assembly quality (N50, number of misassemblies, portion of the target genome covered, etc.). We also suspect that the removal of bubbles/bulges from the de Bruijn graph [for details on bubbles/bulges, see e.g. Zerbino and Birney (2008) or Bankevich et al. (2012)] is significantly harder with ultra-deep sequencing data.
As sequencing errors are the source of the problem, one could attempt to correct them before the assembly. Several stand-alone methods have been proposed in the literature [see Yang et al. (2013) for a recent survey], and several de novo assemblers [e.g. SPAdes (Bankevich et al. 2012)] employ a preprocessing step for correcting errors. Unfortunately, error correction is not very effective for ultra-deep sequencing data. Most error correction tools are based on k-mer spectrum analysis: the underlying assumption is that ‘rare’ k-mers are likely to contain sequencing errors. As the depth of sequencing increases, so does the number of occurrences of any k-mer, including the ones that contain sequencing errors. In Lonardi et al. (2015) and this article, we have collected experimental evidence of the inefficacy of error-correction methods on the assembly of ultra-deep sequencing data.
An alternative approach to deal with excessive sequencing data is down-sampling. The idea of down-sampling is to disregard a fraction of the input reads, according to some predetermined strategy. The simplest approach is to randomly sample the input and only assemble a fraction of the reads. Although coverage reduction has been primarily used for unbalanced data (Brown et al. 2012), we have shown in Lonardi et al. (2015) that in the presence of ultra-deep sequencing data, the assembly of a random sample of the input reads only marginally improves the assembly quality compared with the assembly of entire dataset. Diginorm (Brown et al. 2012) and NeatFreq (McCorrison et al. 2014) are two examples of down-sampling methods aimed to produce a more uniform coverage. They both reduce coverage by selecting representative reads binned by their median k-mer frequency. In general, down-sampling is not a satisfactory technique to deal with large datasets, unless it is expected to remove most of the ‘bad’ reads and none of the ‘good’ reads. Otherwise, it has the undesirable effect of removing ‘critical’ reads, i.e. rare but error-free reads that can help bridge or fill assembly gaps.
In this article, we address the question of how to create high-quality assemblies when an ultra-deep dataset is available. We propose a meta-assembly method called Slicembler that, unlike down-sampling techniques, takes the advantage of the whole input dataset. Slicembler uses a divide-and-conquer approach: it ‘slices’ a large input into smaller sets of reads, assembles each set individually (using a standard assembler) and then merges the individual assemblies. Our experimental results on real and synthetic data show that Slicembler can produce higher quality assemblies than the regular assembly of entire dataset (before or after error correction), as well as better assemblies compared with the assembly of random samples of the reads. The assemblies produced by Slicembler demonstrate that, when an ultra-deep coverage dataset is available, it is possible to create long contigs with no assembly errors. We believe these results can be considered the first step toward making ‘perfect assemblies’. We also show that Slicembler is less sensitive to sequencing error rates, which could make it desirable for third-generation sequencing data.
2 Methods
The availability of ultra-deep sequencing data opens the opportunity to construct assemblies from multiple independent samples of the reads and then compare them with the objective either to (i) merge them or (ii) discover assembly errors and correct them. Slicembler is based on majority voting: if a contig (or a fraction thereof) appears in the majority of the individual assemblies, we assume that it is safe to add that contig to the consensus assembly being built. Slicembler is a meta-assembler for second-generation paired-end short reads, but its framework can be adapted to other type of sequencing data.
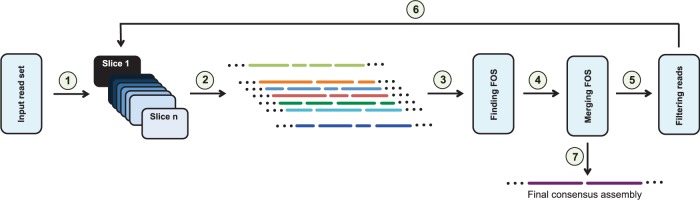
Figure 1 illustrates the proposed iterative algorithm. First, Slicembler partitions the reads into several smaller sets (slices). In the second step, it assembles each set individually using a standard assembler (e.g. Velvet, SPAdes, IDBA_UD or Ray). Third, Slicembler analyzes the individual assemblies and identifies long common contigs (or fractions thereof) supported by a majority of the assemblies. In the fourth step, it merges these common contigs (or fractions thereof) to the partially constructed (consensus) assembly being built. Before repeating steps 2, 3 and 4, any read that maps to the consensus assembly is removed from the input.
Fig. 1.
SLICEMBLER’s pipeline: First, the input reads are partitioned into smaller slices (1). Each slice is assembled individually (2), and the resulting assemblies are merged by a ‘majority voting’ process (3, 4). Before repeating these steps, any read in the input that maps to the consensus assembly is removed (6). When no further merging is possible, the final consensus assembly is produced (7)
2.1 ‘Slicing’ the input
In the first step, the set of input reads is partitioned into n distinct slices. Each paired-end read is assigned to exactly one slice, although it is also possible to assign a read to multiple slices. For simplicity, each slice contains approximately the same number of reads. The number of slices is determined from the desired depth of coverage Ds for each slice. As we discussed in Lonardi et al. (2015), the coverage Ds is a critical parameter for the quality of assembly. To find a good value for Ds, one can run the base assembler (e.g. Velvet, SPAdes, Ray or IDBA_UD) on larger and larger samples of the input and find the coverage that maximizes the chosen assembly statistics (e.g. N50). Once the value of Ds is established, one can determine the number of slices by computing , where Dt is the depth of coverage for the whole input read set. Given the set of input reads, the slice coverage Ds and the average read length, it is straightforward to partition the reads into n slices with the desired coverage.
2.2 Assembling the slices
In the second step, each of the n slices is assembled independently with a standard assembler (e.g. Velvet, SPAdes, Ray or IDBA_UD), possibly with different choices of the k-mer values in each slice. Under the assumption that the number of reads in every slice is sufficient for a complete assembly, the ideal outcome is that each of the n assemblies covers the entire target genome. In practice, each assembly is expected to contain a mixture of ‘good’ and ‘bad’ contigs due to sequencing errors, repetitive regions and imperfections in the assembly algorithms. The objective of the next step is to identify the ‘good portion’ of each contig by taking a majority vote among the assemblies.
2.3 Finding frequently occurring substrings
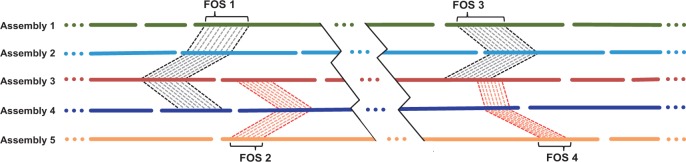
In the third step, Slicembler searches for long substrings that occur exactly in the majority of individual assemblies. The input to this step is a set of n assemblies where each assembly Ai is represented as a set of contigs. Given a string s, we define c(s) as a subset of assemblies in which s appears exactly in at least one contig of each assembly in T. Given a minimum support u and minimum length l, Slicembler identifies all maximal substrings r such that |r| > l and |c(r)| > u, that is, r is longer than l nucleotides and it appears in at least u assemblies. By maximal we mean that if string r was extended by one extra symbol to the left or to the right, then |c(r)| would decrease below threshold u + 1. We call such substrings r, frequently occurring substrings (FOS). Figure 2 illustrates four FOS detected from a set of five assemblies. FOS1 occurs in four assemblies, whereas FOS2 appears in three of them. FOS3 and FOS4 is a pair of overlapping substrings occurring in three assemblies.
Fig. 2.
Examples of frequently occurring substrings (FOS) from five assemblies (FOS can overlap)
To find FOS, we build a generalized suffix tree on the contigs of n assemblies (and their reverse complement), then use a variant of the algorithm proposed in Hui (1992). In this algorithm, each input string is assigned a distinct ‘color’. The algorithm uses the generalized suffix tree to compute for each tree node u the number of distinct colors in the subtree rooted at node u. The algorithm computes the number of colors for each node in linear time in the length of the input strings. Algorithm (Hui 1992), however, does not produce maximal substrings. Once the internal nodes have the color information, to ensure right-maximality our algorithm finds the deepest internal node u (spelling out string r, |r| > l), such that |c(r)| > k. To guarantee left-maximality, we take advantage of suffix links: if node u has a suffix link to node v, and subtrees rooted at u and v have the same number of leaves and colors then the string corresponding to v is not left-maximal and should not be reported. We mark all the nodes corresponding to the loci of strings contained in string r, then the process above is repeated to find the second longest FOS.
As we mentioned earlier, repetitive regions in the genome represent a major challenge for assemblers. Often a FOS includes a repetitive pattern at the end due to disagreements among assemblies on how many times that pattern should be repeated. The ends of each FOS are critical for merging, which requires a prefix–suffix overlap. Any error in these sections may prevent the algorithm from merging overlapping FOS (discussed in Section 2.4 later). To avoid merging errors in later steps, Slicembler checks 20 bp at the ends of each FOS. If a tandem repeat is found at any of the ends, all copies (except one) of the repeated pattern are eliminated. Specifically, any string in the form of αββ+ (where |β| < 10 bp) is replaced with αβ.
2.4 Merging frequently occurring sequences
When detected FOS are overlapping (e.g. FOS3 and FOS4 in Fig. 2), they can be merged to obtain longer FOS (FOS will also be merged to the contigs in the consensus assembly being built). Slicembler identifies any FOS that has an exact suffix–prefix overlap (i.e. no mismatches/indels) with another FOS (or its reverse complement) and determines the number of paired-end reads that connect each pair of such overlapping FOS. A pair of FOS is merged if either (i) the exact overlap is at least 100 bp or (ii) the exact overlap is 50–99 bp and the number of paired end reads connecting them is at least Dt/1000 or (iii) the exact overlap is 20–49 bp and the number of paired end reads connecting them is at least Dt/100. This idea of using paired-end read to increase the confidence of an overlap is similar to the scaffolding step used to order and orient contigs in de novo assemblers or specialized scaffolding tools like (Pop et al. 2004).
2.5 Slicembler algorithm
Slicembler is an iterative meta-assembler. The main steps of slicing/assembling/merging are executed iteratively until a predetermined condition is met. Table 1 presents a sketch of our algorithm. As described in Section 2.1, the number of slices is calculated from the chosen slice coverage (DS). The input read set is partitioned into n slices (line 2). The rest of the algorithm is performed iteratively (lines 3–19) until the total length of the consensus assembly F meets or exceeds the target genome size, or no sufficiently long FOS can be found. At the beginning of a new iteration, Slicembler assembles the reads in each slice individually (lines 4–6). Next, a generalized suffix tree T is created from the contigs in the individual assemblies (both forward and reverse complement) (line 7). Using the suffix tree, Slicembler produces the set of maximal substrings longer than l bases that occur in at least u assemblies (out of n, line 11). The FOS set could contain any number of strings (including none). Then, Slicembler checks whether FOS overlapping with the current consensus assembly meet the conditions described in Section 2.4 and merges them (line 12). The parameter u is set to n initially, so Slicembler first tries to determine whether there is any FOS that appears in all the assemblies. If no new FOS is found, the support u is decreased (by one) and the loop is repeated. The parameter u is decreased until at least one FOS is detected or u becomes smaller than n/2. If u becomes smaller than n/2, the minimum length l is halved and u is initialized again to n. We selected n/2 as the ‘turning point’ because we would not trust any common substring that appears in the minority of the assemblies. The initial value for l is one-fifth of the size of the target; based on our observations, using a larger value for the initial value of l is unlikely to improve the results but makes Slicembler slower. The iterative process stops when l drops below lmin, which is desired minimum contig length in the final assembly (lmin is user-defined, typically 200–500 bp). If l is below lmin and no new FOS have been identified in the current iteration (line 17), Slicembler’s iterative process is terminated and the consensus assembly is reported.
Table 1.
A sketch of Slicembler’s algorithm
| Inputs | Input reads (S), slice coverage (DS), min contig length (lmin), size of the target genome (ltarget) |
|---|---|
| Output | Set of contigs (F) |
| 1 | F ← Ø |
| 2 | Partition S into n slices S1,S2,…, Sn each of which has coverage DS |
| 3 | while (|F| < ltarget) do |
| 4 | A ← Ø |
| 5 | for i ←1 to n do |
| 6 | A ← A ∪ Assemble(Si) |
| 7 | T ← GeneralizedSuffixTree (A, ReverseComplement(A)) |
| 8 | u ← n |
| 9 | l ← ltarget/5 |
| 10 | while (l > lmin) |
| 11 | FOS ← FindFOS(T, u, l) |
| 12 | if (FOS ≠ Ø) then F ← MergeFOS(FOS, F) |
| 13 | break |
| 14 | else if (u > n/2) then u ← u – 1 |
| 15 | else l ← l/2 |
| 16 | u ← n |
| 17 | if (l < = lmin) and (FOS = Ø) then break |
| 18 | for i ← 1 to n do |
| 19 | Si ← FindUnmappedReads(F, Si) |
| 20 | return F |
Before starting a new iteration, all the reads in each slice are mapped to all detected FOS in the current consensus assembly. Each paired end read that maps exactly to any contig in the current assembly is removed (lines 18-19), and only the remaining reads are assembled in the next iteration. Note that we do not repartition the read sets after this step, because although the number of reads decreases, so does the size of the target we are supposed to reconstruct. There is one exception to this strategy of read elimination. Recall that to be able to merge the FOS set with the current assembly, the strings have to overlap a minimum number of bases. To make sure that this will be possible in future iterations, reads that are mapped close to the ends of contigs of the current assembly are not eliminated.
Like any other assembly pipeline, gap filling and scaffolding can be applied at the end of the process to improve the quality of final assembly. In this case, gap filling is easier than usual because of the high quality of contigs produced by Slicembler and the very large number of reads available for filling the gaps. Also, the number of gaps to be filled at the end is relatively small since Slicembler fills some of the gaps during the merging process (see Fig. 4 for an example). The merging step uses small FOS identified in the later iterations to ‘glue’ adjacent contigs.
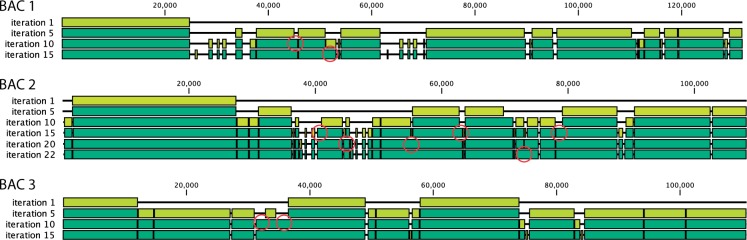
Fig. 4.
An illustration of SLICEMBLER’s progressive construction of the consensus assembly for BACs 1, 2 and 3 (‘snapshots’ are taken every five iterations). Each box represents a perfect alignment between that contig and the reference. Light green boxes indicate a new FOS compared with the previous snapshot. Circles point to gaps closed or contig extended via the merging process (picture created with CLC sequence viewer)
3 Experimental results
We implemented Slicembler in Python. Our tool can be accessed at http://slicembler.cs.ucr.edu/. Slicembler is a meta-assembler; its performance directly depends on the base assembler. In the following experiments, we used Velvet as the base assembler, unless stated otherwise. The performance of Slicembler using other base assemblers (IDBA_UD, Ray and SPAdes) is presented in Section 3.3. The number of slices and the sequencing error rate for the input reads are other factors that critically influence the quality of the final assembly. We study these issues in Section 3.4 and Section 3.5.
Recall that at the end of every iteration, all reads are mapped to the partially constructed assembly to detect bridge reads (to be used later in the merging step) and to eliminate reads that are already represented in the assembly. Slicembler uses BWA (Li and Durbin 2009) to find perfect alignments (no mismatches, no gaps) for this purpose. We used a minimum contig length lmin = 200 (which is the default parameter for Slicembler). We did not use any gap filling or scaffolding tool on the final assemblies. All experiments were carried on a Linux server with 20 computing cores and 194 GB of RAM.
3.1 Ultra-deep sequencing of barley BACs
To carry out experiments on real ultra-deep data, we sequenced a set of 16 BAC genomic clones of barley (Hordeum vulgare L.) on an Illumina HiSeq2000 at UC Riverside at a depth of coverage 8000 x–15 000 x. The average read length was about 88 bases after quality trimming; reads were paired-end with an average insert size of 275 bases. Another set of 52 barley BACs was sequenced by the Department of Energy Joint Genome Institute using Sanger sequencing. As the primary DNA sequences for each of these 52 BACs were assembled in one solid contig [details in Lonardi et al. (2015)], we assumed these Sanger-based assemblies to be the ‘ground truth’ or ‘reference’. Five ultra-deep sequenced BACs had such a reference, so we used them to objectively evaluate the performance of Slicembler. To have an equal-sized input dataset for all BACs, we used only 8000x worth of coverage. These five barley BAC clones, hereafter referred as BAC 1–5 have the following lengths: 131 747 bp, 108 261 bp, 110 772 bp, 111 748 bp and 102 968 bp, respectively. We should remind the reader that the barley genome is highly repetitive. Approximately 84% of the genome consists of mobile elements or other repeat structures (International Barley Genome Sequencing Consortium et al., 2012).
3.2 Quality of Slicembler’s assemblies
Slicembler divided each of the five ultra-deep BAC inputs into 10 slices (Ds = 800 x coverage). We showed in Lonardi et al. (2015) that such coverage is expected to provide a ‘good’ assembly in terms of N50, longest contig, number of misassemblies (i.e. misjoined contigs) and percentage of the target genome covered. We compared the performance of Slicembler to three alternative methods, namely (A) assemble all reads (8000x coverage) with the same assembler used in Slicembler, (B) run error-correction [using Racer (Ilie and Molnar 2013)] on all reads (8000x coverage) then assemble the corrected reads with the same assembler used in Slicembler and (C) assemble each of the slices (800x coverage) individually and consider the average statistics over the 10 slices (down-sampling).
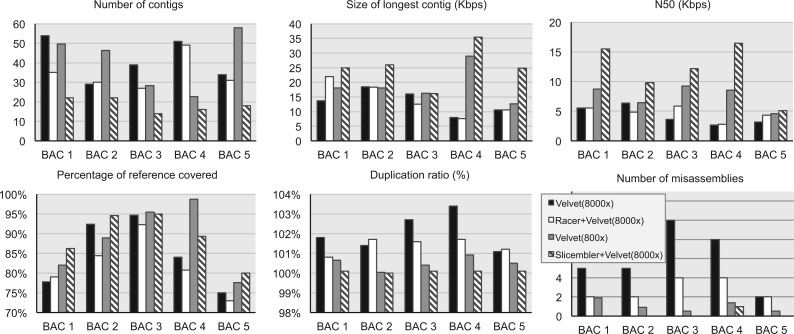
Figure 3 summarizes the assembly statistics collected with QUAST (Gurevich et al. 2013) for Slicembler compared with methods A, B and C described above. The base assembler was Velvet (hash value 69). Several observations on Figure 3 are in order. First, note that for most of the BACs, down-sampling at 800 x leads to better quality assemblies than the assembly of all the reads at 8000 x. This is consistent with our previous results (Lonardi et al. 2015). Second, error correction increases the quality of assemblies for most of the BACs. At the same time, error correction affects negatively other statistics like duplication ratio and N50.
Fig. 3.
Summary of assembly statistics on five barley BACs sequenced at 8000x. We compared Slicembler (using Velvet) with three alternative methods: Velvet on the entire dataset, Racer + Velvet on the entire dataset and the average performance of Velvet on the slices of 800 x each (see legend). Ground truth was based on Sanger-based assemblies. Statistics were collected with QUAST for contigs longer than 500 bp
Finally and more importantly, observe that in the majority of cases, Slicembler generates the highest quality assemblies. Its assemblies are less fragmented, which is reflected by a smaller number of contigs, longer longest contigs and higher N50. Also, Slicembler’s assemblies cover a higher fraction of the target genome and they have a much smaller number of misassembly errors compared with the other approaches. In fact, Slicembler’s assemblies are almost error-free. BAC 4 is the only exception: although Slicembler’s assembly of BAC 4 has fewer misassemblies than the assembly of all the reads before or after error correction, it contains more errors than the average downsampling-based assembly. The slightly higher number of assembly errors for Slicembler is due to the merging step, which could be made more conservative. Slicembler’s contigs also contained less mismatches and indels compared with the other methods (data not shown). Finally, note that Slicembler’s assemblies are less inflated than the other approaches. The assembly of all the reads, with or without error correction, has quite large duplication ratio.
To illustrate the progress during Slicembler’s iterative refinements, Figure 4 shows the status of the consensus assembly created for BACs 1, 2 and 3 every five iterations. Each box represents a perfect alignment of a Slicembler’s contig to the reference genome (no insertion/deletion/mismatches allowed). Observe that in the last iteration 85–95% of the target genome is covered by the error-free contigs. In the first iterations, most of the target genome is covered by large FOS. In later iterations, FOS are smaller but they can connect adjacent contigs or extend them (see red circles). Most of the small gaps between the contigs are composed by repetitive patterns. These gaps are induced by the ‘trimming’ step of the algorithm, which eliminates repetitive patterns from the ends of FOS to avoid false overlaps. A gap-filling tool can easily close these small gaps during the finishing step.
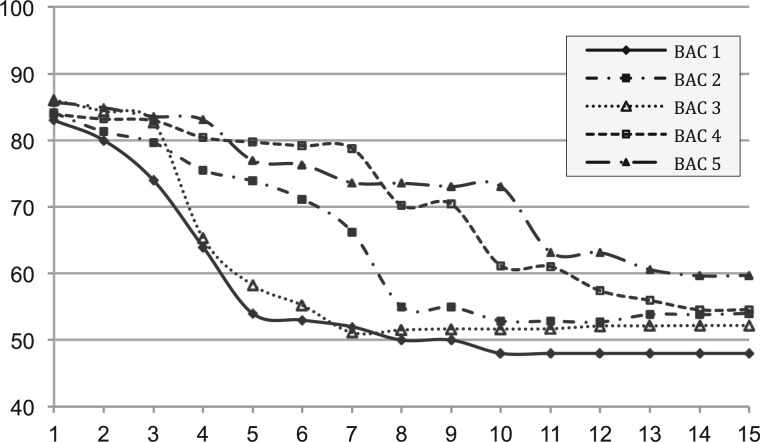
As mentioned above, at the end of each iteration, Slicembler maps the current set of input reads to the consensus assembly: any read that is mapped exactly is discarded. This allows Slicembler (and its base assembler) to ‘focus’ on the parts of the genome/BAC that are still missing from the consensus assembly. Because FOS in early iterations are ‘safer’ to be added to the consensus assembly, the set of reads discarded in early iterations are expected to be of higher quality. To this end, we determined the percentage of reads at each iteration of Slicembler that could be mapped exactly (i.e. no mismatches/indels) to the reference genome. Figure 5 shows these percentages for the first 15 iterations in the assembly of the five BACs. Observe that the percentage of high-quality reads is about 85% in early iterations.
Fig. 5.
The percentage of reads (y axis) at each iteration of Slicembler (x axis) that map exactly (i.e. zero mismatches/indels) to the reference on the five ultra-deep sequenced BACs
As the number of iterations increases, the percentage of high-quality reads in the input monotonically decreases. In the last few iterations, the percentage stays somewhat flat because later FOS are shorter, so the additional number of high-quality reads mapped to these FOS is also small.
3.3 The choice of the base assembler
Recall that Slicembler is a meta-assembler, and its performance depends on the performance on the base assembler. To evaluate the influence of base assembler on the assembly quality, we compared several assemblers, namely Velvet (Zerbino and Birney 2008), SPAdes (Bankevich et al. 2012), Ray (Boisvert et al. 2010) and IDBA_UD (Peng et al. 2012).
Experimental results for BAC 3 are shown below in Table 2. We compared the assembly produced by Velvet, SPAdes, Ray and IDBA_UD on all the reads (8000x) against the assemblies created by Slicembler in conjunction with the corresponding base assembler. Slicembler was run on 10 slices (800x each). The k-mer used was 69 for Velvet and Ray. For IDBA_UD and SPAdes, the reported assembly was based on three different k-mers (29, 49 and 69).
Table 2.
Comparing BAC assemblies produced with IDBA_UD, Velvet, SPAdes and Ray to the assemblies produced by SLICEMBLER in conjunction with the same assembler; the numbers in bold represent the best assembly statistic between SLICEMBLER and the coresponding base assembler
| Methods | Number of contigs | Percent ref covered | Duplication ratio | Misassemblies|mismatches per 100 kb | N50 | Longest contig |
|---|---|---|---|---|---|---|
| IDBA_UD (8000x) | 34 | 97.0% | 1.010 | 0|0.93 | 7335 | 13 889 |
| Slicembler + IDBA (10 slices of 800 x) | 13 | 97.0% | 1.010 | 0|1.1 | 16 121 | 31 161 |
| Velvet (8000x) | 39 | 94.7% | 1.027 | 10|20.0 | 3649 | 16 048 |
| Slicembler + Velvet (10 slices of 800 x) | 14 | 95.1% | 1.001 | 0|0 | 12 178 | 16 128 |
| SPAdes (8000x) | 49 | 95.7% | 1.006 | 0|0.94 | 9129 | 21 872 |
| Slicembler + SPAdes (10 slices of 800 x) | 11 | 96.9% | 1.024 | 0|1.2 | 27 685 | 31 158 |
| Ray (8000x) | 35 | 80.0% | 1.003 | 0|0 | 3996 | 7186 |
| Slicembler + Ray (10 slices of 800 x) | 24 | 88.0% | 1.000 | 0|0 | 7192 | 12 842 |
Statistics were collected with QUAST for contigs longer than 500 bp.
Observe that among the stand-alone assemblers, IDBA_UD and SPAdes created higher quality assemblies compared with Velvet and Ray. However, regardless of the choice of the base assembler, Slicembler improved the quality of the assemblies.
The only ‘negative’ statistics for Slicembler is that it introduced a few more errors in the assemblies created using IDBA_UD and SPAdes. We determined that these additional errors were due to incorrect merging in later iterations. Also, Slicembler had a slightly higher duplication ratio than SPAdes. Other than these, Slicembler significantly improved all other statistics. In fact, similar results were observed on the other four BACs (data not shown). In general, Slicembler created higher quality assemblies when used in conjunction with IDBA_UD and SPAdes
3.4 The choice of depth of coverage for each slice
The depth of coverage in each slice is critical to optimize on the quality of the assemblies. If the depth of coverage is too low, the assembly of each slice will be fragmented, which will be reflected in shorter FOS. On the other hand, more slices can increase the confidence in choosing FOS due to more ‘votes’ available. For this reason, we decided to use simulations to study the tradeoffs of the depth of coverage in each slice. To this end, we used wgsim (https://github.com/lh3/wgsim) to generate synthetic datasets with 500x, 1000x, 2500x, 5000x, 7500x and 10 000x reads at 1% sequencing error rate (no indels) based on the reference sequence of BAC 3. Each dataset was assembled with Slicembler using Velvet as the base assembler by dividing the input into 10 slices, so that the coverage in each slice was 50x, 100x, 250x, 500x, 750x and 1000x.
Table 3 shows the usual quality statistics for the assemblies on simulated reads. Observe that Slicembler’s best performance is observed when slices are in the 100x–500x coverage range. When the slice coverage is lower than 100x, assemblies are more fragmented due to insufficient coverage. When the slice coverage is higher than 500x, we experience the negative effects of ultra-deep sequencing data on the quality of the individual assemblies: FOS become smaller and the final assembly is more fragmented. Note that despite the 1% sequencing error rate, Slicembler was able to create error free contigs for all cases.
Table 3.
Quality statistics for SLICEMBLER's assemblies for simulated reads with different depth of coverage; the number in bold represent the best assembly statistic in each row
| 500x | 1000x | 2500x | 5000x | 7500x | 10 000x | |
|---|---|---|---|---|---|---|
| Number of contigs | 20 | 12 | 11 | 10 | 18 | 38 |
| Longest contig | 27 364 | 31 823 | 31 946 | 31 950 | 21 865 | 9425 |
| N50 | 6707 | 26 275 | 26 288 | 26 267 | 12 428 | 3643 |
| Percent refer. covered | 90.6% | 88.7% | 94% | 93.9% | 92.9% | 84.7% |
| Duplication ratio | 1 | 1 | 1 | 1 | 1 | 1 |
| Misassemblies | 0 | 0 | 0 | 0 | 0 | 0 |
| Mismatches and indels | 0 | 0 | 0 | 0 | 0 | 0 |
We used 10 slices in all experiments (i.e. the coverage for each slice was 50x, 100x, 250x, 500x, 750x and 1000x). Statistics were collected with QUAST for contigs longer than 500 bp.
3.5 Effect of sequencing error rate in the reads
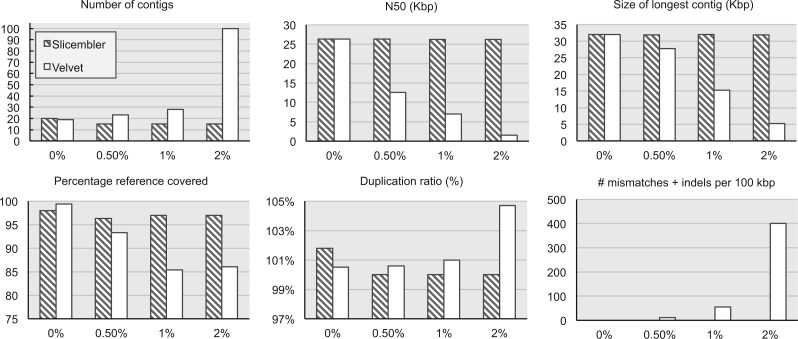
De novo assemblers are quite sensitive to sequencing error rate in the input reads. Even assemblers that have a preprocessing step for error correction (e.g. SPAdes) have difficulties handling errors when the depth of coverage is very high (Lonardi et al. 2015). Because Slicembler relies on majority voting for common contigs in the slice assemblies, we wondered whether it would be more resilient compared with its base assembler. To this end, we used wgsim to generate datasets at 3000x coverage with increasing sequencing error rate, namely 0% (errorless), 0.5%, 1% and 2% error rate based on BAC 3. We assembled each set with Slicembler+Velvet using six slices of 500x coverage each. Results are reported in Figure 6.
Fig. 6.
The effect of increasing sequencing error rates on the quality of assemblies created by Velvet and Slicembler + Velvet. Input paired-end reads were generated using wgsim with a coverage of 3000x using BAC 3 as a reference. For Slicembler, simulated read sets were divided into six slices. Statistics were collected with QUAST for contigs longer than 500 bp
First, note that Slicembler was not able to improve the quality of assembly when the reads are error-free. This is consistent with the results in Lonardi et al. (2015) for error-free reads. Velvet and other de novo assemblers are capable of producing high-quality assemblies when reads are error-free, since there are no imperfections in the de Bruijn graph. More importantly, observe that as the sequencing error rate increases, the performance of Velvet quickly degrades, whereas the performance of Slicembler is unaffected (despite using Velvet as the base assembler). Particularly remarkable is the number of mismatches and indels per 100 kb, which stays at zero for Slicembler for all the tested error rates (up to 2%). There were no misassemblies in the assemblies created by both Velvet and Slicembler.
4 Discussion and conclusion
Advancement in sequencing technologies has been reducing sequencing costs exponentially fast. Ultra-deep sequencing is now feasible, especially for smaller genomes and clones. We expect that in the near future life, scientists will sequence ‘as much as they want’ because the sequencing cost will be a minor component of total project costs. This explosion of data will create new algorithmic challenges. We have shown previously that popular modern de novo assemblers are unable to take advantage of ultra-deep coverage, and the quality of assemblies starts degrading after a certain depth of coverage. Slicembler is an iterative meta-assembler that solves this problem: it takes advantage of the whole dataset and significantly improves the final quality of the assembly. The strength of Slicembler is based on the majority voting scheme: in our experiments, FOS selected by Slicembler from the slice assemblies never contain errors with the exception of FOS belonging to the very ends of the target genome, which are not as reliable because coverage tends to be lower. Slicembler extracts high-quality contigs from the slice assemblies, and it prevents contigs containing mis-joins and calling errors to be included in the final assembly.
Experiments on a set of ultra-deep barley BACs and simulated data show that our proposed method leads to higher quality assemblies than the corresponding base assembler. We also demonstrated that Slicembler is more resilient to high sequencing error rates than its base assembler. Our proposed algorithm is expected to work for genomes of any length, but the current implementation of Slicembler has been tested only on relatively small genomic target sequences for which real ultra-deep coverage is now available. For Slicembler to scale to larger genomes, its efficiency must be improved. Slicembler has to execute the base assemblers tens to hundreds of times (depending on the number or slices and iterations). Obviously, Slicembler is expected to be significantly slower than the base assembler. For example, Slicembler was around 50x slower than Velvet to assemble BAC 1. Most of the computational effort in Slicembler is spent in finding FOS (this required construction of the generalized suffix tree), merging FOS (this requires computing exact prefix–suffix overlaps) and mapping the reads (this requires running BWA) at every iteration. One way to increase the algorithm speed would be to process the slices in parallel. Another possible improvement would be to map the reads to each slice assembly only once and process the alignment file to determine which reads should be passed to the following iteration, instead of mapping the reads to the slice assembly from scratch in every iteration. We are also working on improving the merging step, to prevent mis-joins. More advanced approaches for merging contigs, like methods proposed for merging draft assemblies (Nijkamp et al. 2010; Soueidan et al. 2013; Vicedomini et al. 2013), may improve the quality of Slicembler results. We plan to release soon an improved version of Slicembler implemented in C++.
To conclude, the results presented in this article indicate the possibility of having (almost) perfect assemblies when the depth of coverage is very high. Although there is more work to be done to achieve a perfect assembly, we believe that Slicembler represents a significant step forward in this direction.
Acknowledgements
We thank Weihua Pan (UC Riverside), Hind Alhakami (UC Riverside) and Prof. Pavel Pevzner (UC San Diego) for early comments on this study.
Funding
This work was supported in part by the U.S. National Science Foundation [DBI-1062301] and [IIS-1302134], by the USDA National Institute of Food and Agriculture [2009-65300-05645], by the USAID Feed the Future program [AID-OAA-A-13-00070] and the UC Riverside Agricultural Experiment Station Hatch Project CA-R-BPS-5306-H.
Conflict of Interest: none declared.
References
- Aird D., et al. (2011) Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol., 12, R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., et al. (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol., 19,455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerenwinkel N., Zagordi O. (2011) Ultra-deep sequencing for the analysis of viral populations. Curr. Opin. Virol., 1, 413–418. [DOI] [PubMed] [Google Scholar]
- Boisvert S., et al. (2010) Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J. Comput. Biol., 17, 1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.T., et al. (2012) A reference-free algorithm for computational normalization of shotgun sequencing data. arXiv:1203.4802.
- Campbell P.J., et al. (2008) Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc. Natl. Acad. Sci. USA, 105, 13081–13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui L. (1992) Color set size problem with applications to string matching. In: Apostolico A., et al. (eds.), Combinatorial Pattern Matching. Springer, Berlin Heidelberg, pp. 230–243. [Google Scholar]
- Clarke J., et al. (2009) Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol., 4, 265–270. [DOI] [PubMed] [Google Scholar]
- Desai A., et al. (2013) Identification of optimum sequencing depth especially for de novo genome assembly of small genomes using next generation sequencing data. PLoS One, 8, e60204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid J., et al. (2009) Real-time DNA sequencing from single polymerase molecules. Science, 323, 133–138. [DOI] [PubMed] [Google Scholar]
- Ekblom R., et al. (2014) Patterns of sequencing coverage bias revealed by ultra-deep sequencing of vertebrate mitochondria. BMC Genomics, 15, 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English A.C., et al. (2012) Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One, 7,e47768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A., et al. (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics, 29, 1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilie L., Molnar M. (2013) RACER: rapid and accurate correction of errors in reads. Bioinformatics, 29, 2490–2493. [DOI] [PubMed] [Google Scholar]
- International Barley Genome Sequencing Consortium. et al.. (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature, 491, 711–716. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonardi S., et al. (2015) When less is more: “slicing” sequencing data improves read decoding accuracy and de novo assembly quality. Bioinformatics, doi: 10.1093/bioinformatics/btv311 (in press).
- McCorrison J.M., et al. (2014) NeatFreq: reference-free data reduction and coverage normalization for de novo sequence assembly. BMC Bioinformatics, 15, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijkamp J., et al. (2010) Integrating genome assemblies with MAIA. Bioinformatics, 26, i433–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., et al. (2012) IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics, 28, 1420–1428. [DOI] [PubMed] [Google Scholar]
- Pop M., et al. (2004) Hierarchical scaffolding with Bambus. Genome Res., 14, 149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soueidan H., et al. (2013) Finishing bacterial genome assemblies with Mix. BMC Bioinformatics, 14, S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicedomini R., et al. (2013) GAM-NGS: genomic assemblies merger for next generation sequencing. BMC Bioinformatics, 14, S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widasari D.I., et al. (2014) A deep-sequencing method detects drug-resistant mutations in the hepatitis B virus in indonesians. Intervirology, 57, 384–392. [DOI] [PubMed] [Google Scholar]
- Yang X., et al. (2013) A survey of error-correction methods for next-generation sequencing. Brief. Bioinform., 14, 56–66. [DOI] [PubMed] [Google Scholar]
- Zerbino D.R., Birney E. (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res., 18, 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., et al. (2014) Bias from removing read duplication in ultra-deep sequencing experiments. Bioinformatics, 30, 1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]