Abstract
The CFTR inhibitory factor (Cif) is an epoxide hydrolase (EH) virulence factor secreted by the bacterium Pseudomonas aeruginosa. Sequence alignments reveal a pattern of Cif-like substitutions that proved to be characteristic of a new subfamily of bacterial EHs. At the same time, crystallographic and mutagenetic data suggest that EH activity is required for virulence and that Cif’s active site remains generally compatible with a canonical two-step EH mechanism. A hallmark of this mechanism is the formation of a covalent hydroxyalkyl-enzyme intermediate by nucleophilic attack. In several well-studied EHs, this intermediate has been captured at near stoichiometric levels, presumably reflecting rate-limiting hydrolysis. Here we show by mass spectrometry that only minimal levels of the expected intermediate can be trapped with WT Cif. In contrast, substantial amounts of intermediate are recovered from an active-site mutant (Cif-E153Q) that selectively targets the second, hydrolytic release step. Utilizing Cif-E153Q and a previously reported nucleophile mutant (Cif-D129S), we then captured Cif in the substrate-bound, hydroxyalkyl-intermediate, and product-bound states for 1,2-epoxyhexane, yielding the first crystallographic snapshots of an EH at these key stages along the reaction coordinate. Taken together, our data illuminate the proposed two-step hydrolytic mechanism of a new class of bacterial virulence factor. They also suggest that the failure of WT Cif to accumulate a covalent hydroxyalkyl-enzyme intermediate reflects an active-site chemistry in which hydrolysis is no longer the rate-limiting step, a non-canonical kinetic regime that may explain similar observations with a number of other EHs.
Graphical Abstract
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen responsible for severe infections at sites including the airway, eyes, and burn wounds. The bacterium possesses an inherent resistance to many antibiotics and can form biofilms that further enhance drug resistance and impede immune clearance (1–3). P. aeruginosa also deploys an arsenal of toxins and virulence factors, whose expression and secretion are adaptively modulated during the progression from colonization to early and ultimately chronic infection (4). One of these is the CFTR inhibitory factor (Cif), which is expressed by both early and late clinical isolates (5). Cif is a secreted epoxide hydrolase (EH) that can enter human epithelial cells and prevent deubiquitination of the cystic fibrosis (CF) transmembrane conductance regulator (CFTR) following endocytic uptake (6).
As a result, Cif blocks endocytic recycling of CFTR, redirecting the protein towards lysosomal degradation and leading to a rapid decline in cell-surface abundance (7, 8). CFTR plays a key role in airway mucociliary clearance, and CFTR loss-of-function mutations cause CF. By reducing net CFTR channel activity, Cif could thus allow P. aeruginosa to induce a CF-like phenotype in airway cells at the site of colonization or infection. Furthermore, although there is no precedent for epoxide regulation of protein deubiquitination, and no in vivo substrates have been reported, EH activity is strictly required for Cif’s deleterious effect: active-site mutations and an enzyme inhibitor each block Cif-mediated degradation of apical CFTR (9).
Cif is thus the first example of an EH that serves as a bacterial virulence factor. Its active site exhibits multiple sequence and structural differences compared to canonical EH enzymes, such as the mammalian soluble (sEH) and microsomal (mEH) epoxide hydrolases (10). Indeed, these distinguishing sequence motifs are shared among a family of secreted proteins from related Gram-negative opportunistic pathogens such as Acinetobacter nosocomialis (10, 11). Nevertheless, based on based on structural and mechanistic analogy to the canonical EH enyzmes (12), we were able to predict key active-site residues (9).
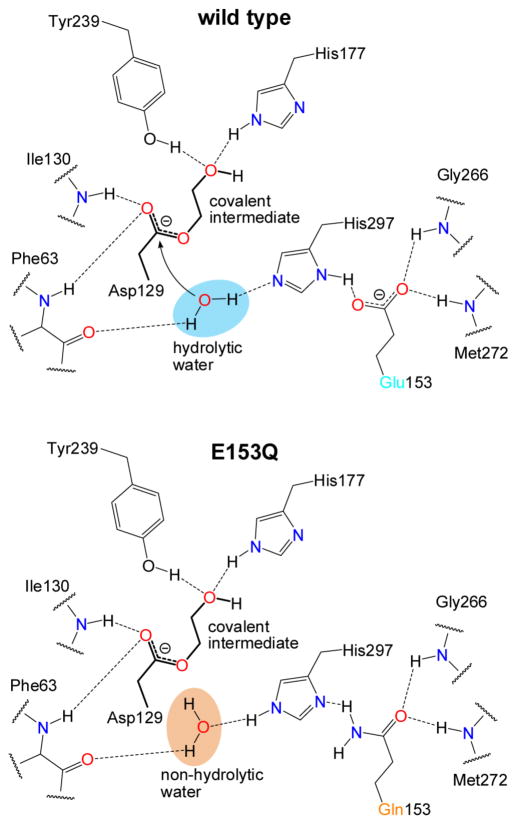
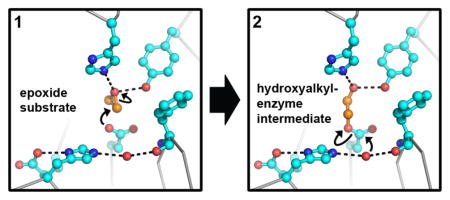
According to the resulting mechanistic paradigm, a tyrosine and histidine form an oxyanion hole, donating hydrogen bonds that polarize the epoxide ring. Together with the steric constraints of the active site, these interactions also orient an oxirane carbon for nucleophilic attack by an aspartate side chain, opening the ring and forming a covalent hydroxyalkyl enzyme intermediate (13). To release the product, a charge-relay histidine-acid pair activates a water molecule (Figure 1) for nucleophilic attack on the covalent intermediate, producing a vicinal diol. This mechanism has been inferred by structural studies of apo or inhibitor-bound EHs and similarities to the well-studied haloacid dehalogenases (HADs) (14). In addition, kinetic analysis of mammalian EHs has shown that the formation of the covalent intermediate is quicker than its hydrolysis, leading to its accumulation and permitting its biochemical isolation (12, 15–17).
Figure 1. Chemical illustration comparing the proposed hydrogen bond networks of the charge-relay pair in the WT and E153Q Cif active site.
The nucleophile Asp129 is shown covalently bound to a epoxide hydrolysis intermediate (chemical bonds shown in bold). In the WT enzyme (top), the carboxylic acid moiety of Glu153 accepts hydrogen bonds from the backbone amide nitrogens of Gly266 and Met272 and from the imidazole ring of the charge-relay His297. In the Cif-E153Q mutant (bottom), the hydrogen-bonding pattern suggests that the Gln153 carbonyl oxygen continues to accept the main-chain amide hydrogen bonds, positioning the side-chain amide nitrogen to donate a hydrogen bond to His297, in contrast to the WT interaction. Along with the predicted pKa shift of the His297, this flipping of hydrogen bond directionality has cascading effects on the orientation of the nucleophilic water molecule, impairing its activation for hydrolysis.
In this study, we proposed to exploit the formation of covalent intermediates to visualize the interactions of substrate adducts with the Cif active site. However, mass spectrometry showed that there is little accumulation of adduct in the WT enzyme. As a result, we used further mutagenesis to probe the role of the charge-relay pair in Cif enzyme activity, highlighting the unusual kinetics of this non-canonical EH and confirming the role of adduct formation in hydrolysis. Finally, our expanded set of Cif mutants enabled us to determine the first crystallographic structures of the active-site geometry at critical steps along the reaction coordinate shared by the EH family of enzymes.
MATERIALS AND METHODS
Mutagenesis and protein purification
The E153Q mutation was generated using yeast recombineering as previously described (18, 19). Carboxy-terminal hexa-histidine tagged Cif protein was expressed in Top10 Escherichia coli (Invitrogen) cells and purified by immobilized metal affinity chromatography as described previously in detail (20). Unless otherwise specified, purified Cif protein was prepared and assayed in the following buffer: 100 mM NaCl, 20 mM sodium phosphate (pH 7.4).
Enzyme labeling
Capture of the enzyme-substrate hydroxyalkyl intermediate was performed as described previously by Müller et al. (38) In summary, 2 μg of Cif protein was incubated with 1 μmol of EBH for 15 sec at 37 °C in a 50 μL volume. Earlier EH trapping experiments were performed at pH 7.0–7.4. WT Cif shows little (< 25%) variation in EH activity from pH 5 – 8, and the protein exhibits reduced solubility at or below pH 6. Furthermore, neutral or slightly basic conditions significantly reduce baseline chemical hydrolysis of the epoxide substrates, compared to an acidic environment. Thus, we selected the physiologically relevant pH of 7.4 for all labeling experiments. The reaction was halted by addition of 450 μL of 1 mM HCl in acetone at −20 °C. Following a 3 hr incubation at −20 °C, the solution was centrifuged at 13,000×g, and the protein pellet was washed twice with 1 mM HCl in acetone. Following evaporation of residual acetone at −20 °C, protein precipitate was dissolved in non-reducing SDS sample buffer.
Liquid chromatography and mass spectrometry
Coomassie-stained SDS-PAGE gel bands corresponding to Cif-WT and Cif-E153Q proteins were destained to clarity in 50 mM ammonium bicarbonate/40% acetonitrile solution, dehydrated, and subjected to chymotryptic proteolysis using sequencing grade chymotrypsin (Roche Life Sciences; 15 ng/μl) overnight at room temperature. The resulting peptide digests were collected by extraction and dried by vacuum centrifugation, then loaded on to an in-house fabricated microcapillary HPLC column and resolved across a 50-minute gradient from 3% acetonitrile/0.125% formic acid to 35% acetonitrile/0.125% formic acid for online LTQ-Orbitrap data-dependent analysis essentially as described using a “top-10” method (21–23). The resulting MS/MS scans were data-searched using a modified Sequest algorithm (24) with no enzyme specificity against a single-protein FASTA containing the Cif-WT and Cif-E153Q sequences. Ion chromatograms were extracted to a mass measurement accuracy of +/− 1.5 parts-per-million from the calculated theoretical exact masses (http://physics.nist.gov). Annotated MS/MS spectral peaks were considered as matches within a 1 m/z tolerance from theoretical average masses.
Crystallization, data collection and processing, structure refinement, and analysis
Cif protein crystals were obtained by vapor diffusion against 400 μl of reservoir solution in a 4 μl hanging drop at 291 K (20, 25). Cif protein was prepared in 100 mM NaCl, 20 mM 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (Cif-WT and Cif-E153Q) or sodium phosphate (Cif-D129S) at pH 7.4 for crystallization trials, and drops were set up by mixing well solution with Cif protein in a 1:1 ratio. Crystallization well solutions contained polyethylene glycol (PEG) 8000, 125 mM CaCl2, and were buffered with 100mM sodium acetate, pH 5.0. Epoxide or diol compounds were included if appropriate. PEG8000 (w/v) and ligand concentrations were as follows: 14.0% for apo Cif-E153Q; 13.0% for Cif-D129S with 20 mM 1,2-epoxyhexane; 14.0% for Cif-E153Q with 10 mM EBH; 12.5% for Cif-E153Q with 10 mM 1,2-epoxyhexane; and 14.0% for Cif-WT with 20 mM 1,2-hexanediol). Final Cif protein concentration was 5.0 mg/mL for all trials, except 8.1 mg/mL for the WT complex with 1,2-hexanediol. Upon harvesting for data collection, crystals were soaked in a cryoprotectant solution consisting of the well solution supplemented with 20% (w/v) glycerol. The crystals were then flash cooled either in the nitrogen stream of an Oxford Cryostream 700 operating at 100 K or by rapid plunging into a liquid nitrogen bath. Oscillation data were collected at 100 K at the X6A beamline of the National Synchrotron Light Source at Brookhaven National Laboratory. Diffraction images were processed and scaled with the XDS package (26). Molecular replacement and iterative rounds of automated refinement were performed using Phenix (27, 28). Once clear electron density could be resolved for ligand or adduct molecules, they were added to the model. The asymmetric unit contains two Cif dimers. To estimate the extent of saturation, the occupancy of ligand molecules was allowed to refine in parallel with B-factors. WinCoot (29) was used for manual adjustment of the model, and PyMOL (30) was used to generate images of the final model.
Circular dichroism spectroscopy
Circular dichroism (CD) spectra were determined for 10 μM solutions of Cif protein in the following buffer: 100 mM NaCl, 20 mM sodium phosphate (pH 7.4). Variable wavelength CD spectra were first measured at 20 °C. Next, the thermal denaturation of each protein was monitored by observing the CD signals at 208, 215, and 222 nm wavelengths and increasing the temperature of the protein by 1 °C per measurement.
Assay for enzyme activity
Epoxide hydrolase enzyme activity was determined for the reporter substrate epibromohydrin (Sigma) using an adrenochrome reporter assay as described previously (25, 31, 32). Lipoprotein lipase from Pseudomonas spp. (Sigma) was used as a negative protein control, and a standard curve was generated using 3-bromo-1,2-propanediol (Sigma). The enzyme reaction was carried out using 20 μM protein incubated with 10 mM substrate at 37 °C for 30 min. We define 1 U as 1 μmol of substrate hydrolyzed per minute.
RESULTS AND DISCUSSION
A low-abundance covalent intermediate is observed for wild-type Cif
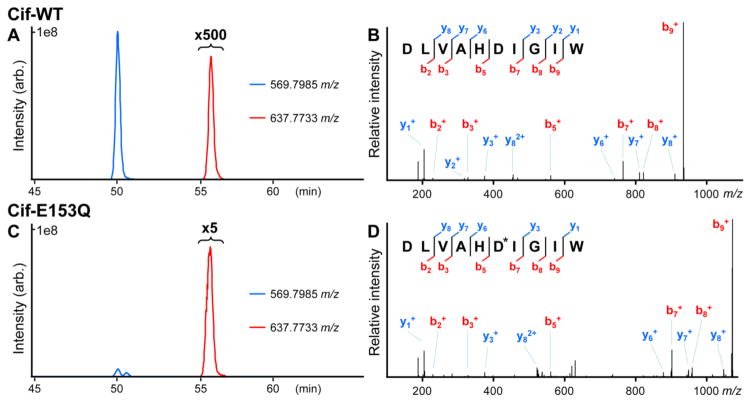
Traditionally, radiolabeling has been used to detect EH covalent intermediates (12, 16, 17). Although highly sensitive, this approach requires custom radiolabeling for each candidate substrate. To circumvent this requirement, we explored a mass-spectrometry (MS) approach for Cif. WT Cif was incubated with the reporter substrate epibromohydrin (EBH), and then precipitated with acidic acetone to terminate the reaction and trap existing adducts, using protocols developed with other EHs (30). The band containing the Cif protein was resolved by SDS-PAGE and digested with chymotrypsin, which is predicted to generate a peptide spanning residues 124–133 and including the catalytic nucleophile Asp129. The chymotryptic peptides were separated by online reverse-phase chromatography mass spectrometry to assess substrate adduct formation with residue-specific resolution not possible using radiolabeling assays. Surprisingly, analysis of reactions containing Cif-WT and EBH reveals a dominant signal corresponding to the nucleophile-spanning, native peptide ion DLVAH(D129)IGIW (Figure 2A, blue chromatogram), which generates the expected fragment ions in tandem MS/MS spectra (Figure 2B). Although a peak corresponding to the predicted adduct is observed at a later elution timepoint, it is present only at extremely low levels (Figure 2A, red chromatogram).
Figure 2. Substrate capture and observation by tandem LC-MS.
(A) Extracted ion chromatograms from Cif-WT + EBH in vitro reaction digests with chymotrypsin for the mass-to-charge values corresponding to the doubly-charged native (blue) and epibromohydrin-adducted (red) DLVAH(D129)IGIW to a mass measurement precision of +/− 1.5 parts-per-million from theoretical. The red trace across the adducted peptide elution profile is expanded 500-fold. (B) Ion-trap CID MS/MS spectrum of the doubly-charged precursor ion corresponding to the native sequence DLVAHDIGIW. Ion fragment assignments are shown. (C) Extracted ion chromatograms from Cif-E153Q in vitro reaction digests with chymotrypsin as in (A). Note that the region corresponding to the adducted peptide elution profile is expanded only 5-fold. The maximum absolute intensity of 1e8 arbitrary units is consistent between both chromatograms. (D) Ion-trap CID MS/MS spectrum of the doubly-charged precursor ion corresponding to the same peptide adducted with the reaction intermediate. Note the selective shift in fragment ions spanning D129, denoted as adducted with an asterisk. Both precursor ions were observed at less than 1.5 parts-per-million mass deviation from theoretical.
The observation of an adduct is consistent with a canonical two-step hydrolysis, but the levels of intermediate (≤1%) were much lower than expected based on previous studies, which in general yielded >60% adduction (16, 17). Substantial accumulation of the intermediate for the human soluble and microsomal EH enzymes has typically been interpreted as evidence that the rate constant of nucleophilic attack (k2 in ref. (14)) is much faster than the rate constant of hydrolysis (k3), a prediction that has also been confirmed by direct kinetic determination (15, 33). One explanation for the divergent outcome with Cif is that this relationship may have shifted, such that the intermediate can be released on a time-scale faster than or comparable to its formation, leading to low steady-state levels. If so, a mutation selectively impairing the hydrolysis step should permit much greater accumulation of the covalent intermediate. In the case of Cif, the Glu153-His297 acid-relay system is a logical target: according to our model (Figure 1, WT panel), Asp129 does not require activation for nucleophilic attack, but the hydrolytic water does.
Neutralization of the charge-relay acid blocks hydrolysis
Although an aspartic acid substitution for the WT charge-relay glutamic acid markedly increased activity for the rat mEH (34), the equivalent substitution in Cif has only a modest impact on EH activity (9). We reasoned that the nearly isosteric replacement with glutamine would also be well-tolerated structurally, but would eliminate the carboxylate functionality that is required to shift the pKa of the adjacent histidine. We were able to express and purify the Cif-E153Q mutant at the single-milligram scale using our previously established protocols (20). While the total yield is somewhat reduced (roughly 30% of the yield for WT protein), size-exclusion chromatography revealed an elution volume consistent with the WT dimer (25).
This ability to express Cif-E153Q recombinantly contrasts with earlier active-site mutagenesis experiments, in which replacement of the charge-relay His297 led to a loss of expression, likely due to protein misfolding (25). To test for possible destabilization of the protein fold, we performed circular dichroism spectroscopy with the mutant protein and monitored wavelengths associated with secondary structure content during thermal denaturation (Figure 3A). The melting temperature of Cif-E153Q differs by less than 1 °C from that previously reported for the WT protein (9). Taken together, these data suggest that the mutation has not fundamentally affected the overall structural integrity of the protein.
Figure 3. Characterization of Cif-E153Q.
(A) Circular dichroism spectroscopy coupled with thermal denaturation of Cif-E153Q reveals cooperative unfolding with a melting temperature of 57°C. (B) Hydrolysis of the epoxide substrate epibromohydrin in the presence of the E153Q mutant is indistinguishable from that observed in a negative protein control.
To determine the impact of the carboxamide substitution on catalysis, we then quantified the ability of the mutant enzyme to hydrolyze the epoxide group of EBH using the well-characterized adrenochrome-reporter assay. As a positive control, we observed robust product formation with the WT enzyme. However, the amount of product formed in the presence of Cif-E153Q was indistinguishable from the negative control (Figure 3B), confirming a loss of EH activity.
Cif-E153Q is a sterically conservative mutation
To establish the structural basis for this effect, we crystallized Cif-E153Q and determined its structure by X-ray crystallography to a resolution of 1.66 Å (Table 1). Crystals form under precipitant conditions comparable to those originally identified for the WT protein and exhibit a very similar morphology. Phases were determined by molecular replacement, using the WT structure (PDB entry 3KD2) as a search model. We refined the structure using procedures designed to minimize phase bias, yielding a model with excellent geometry and agreement with the experimental diffraction data (Table 1).
Table 1.
Data collection and refinement statistics
| Cif-E153Q | Cif-E153Q + epibromohydrin | Cif-D129S + epoxyhexane | Cif-E153Q + epoxyhexane | Cif-WT + 1,2-hexanediol | |
|---|---|---|---|---|---|
| Data Collection | |||||
| Wavelength (Å) | 1.0000 | 0.8984 | 0.9795 | 1.2006 | 1.0000 |
| Space Group | C2 | C2 | C2 | C2 | C2 |
| Unit cell dimensions: | |||||
| a,b,c (Å) | 168.4, 83.9, 89.5 | 168.1, 84.0, 89.5 | 167.4, 83.9, 88.7 | 168.1, 83.7, 89.2 | 169.8, 84.2, 90.1 |
| α,β,γ (°) | 90, 100.4, 90 | 90, 100.3, 90 | 90.0, 100.3, 90.0 | 90, 100.5, 90 | 90, 100.3, 90 |
| Resolutiona (Å) | 36.27-1.66 (1.70-1.66) | 46.14-1.80 (1.97-1.80) | 43.4-1.85 (1.90-1.85) | 46.01-1.95 (2.00-1.95) | 46.45-1.65 (1.70-1.65) |
| Rsymb (%) | 9.0 (56.4) | 9.0 (27.8) | 7.2 (39.2) | 9.9 (39.4) | 5.8 (37.7) |
| Rmrgd-Fc (%) | 8.4 (38.5) | 10.9 (30.8) | 8.5 (39.1) | 12.0 (49.0) | 8.4 (42.3) |
| I/σI | 16.5 (4.1) | 11.9 (4.8) | 18.1 (4.6) | 11.7 (3.5) | 15.6 (3.6) |
| Completeness (%) | 97.1 (95.8) | 99.4 (99.6) | 96.9 (95.8) | 99.7 (98.5) | 99.9 (99.9) |
| Redundancy | 7.5 (7.5) | 4.1 (4.2) | 5.8 (5.8) | 4.8 (3.8) | 4.1 (4.2) |
| Refinement | |||||
| Total number of reflections | 140462 | 112809 | 99957 | 88434 | 149955 |
| Reflections in the test set | 7020 | 5797 | 4990 | 4411 | 7520 |
| Rworke/Rfreef (%) | 18.1/20.8 | 18.9/22.4 | 17.7/21.4 | 17.3/22.5 | 16.9/19.7 |
| Number of atoms: | |||||
| Protein | 9496 | 9509 | 9651 | 9496 | 9463 |
| Solvent | 870 | 829 | 1059 | 995 | 1334 |
| Ligand | 0 | 20 | 28 | 28 | 16 |
| Ramachandran plotg (%) | 91.3/8.3/0.4/0 | 91.1/8.5/0.4/0 | 90.4/9.2/0.4/0 | 90.8/8.8/0.4/0 | 91.2/8.4/0.4/0 |
| Bav (Å2) | |||||
| Protein | 14.4 | 12.6 | 17.5 | 18.9 | 15.2 |
| Solvent | 24.5 | 22.0 | 28.8 | 28.8 | 29.5 |
| Ligand | - | 24.0 | 27.5 | 16.2 | 24.9 |
| Bond length RMSD | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 |
| Bond angle RMSD | 1.028 | 1.012 | 1.036 | 1.006 | 1.061 |
| PDB ID | 4DMC | 4DNF | 4EHB | 4DNO | 4EUS |
Values in parentheses are for data in the highest-resolution shell.
Rsym = ΣhΣi |I(h) − Ii(h)|/ΣhΣi Ii(h), where Ii(h) and I(h) values are the i-th and mean measurements of the intensity of reflection h.
Rmrgd-F is a robust indicator of the agreement of structure factors of symmetry-related reflections and is described in detail by Diederichs & Karplus (52).
SigAno = <(|F(+) − F(−)|/σΔ)>.
Rwork = Σh |Fobs(h) − Fcalc(h)|/Σh Fobs(h), h ∈ {working set}.
Rfree = Σh |Fobs(h) − Fcalc(h)|/Σh Fobs(h), h ∈ {test set}.
Core/allowed/generously allowed/disallowed.
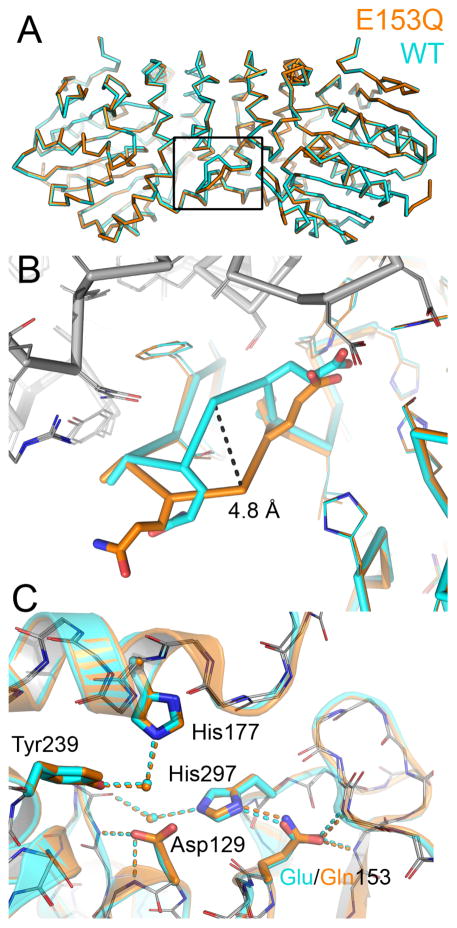
The WT and E153Q dimer models align with an all-atom root-mean-square difference (RMSD) of 0.18 Å (4,190 atoms, Figure 4A), which is less than the maximum-likelihood based coordinate error estimate of 0.21 Å. Aside from surface-exposed side chains, the only notable structural difference we observed between the WT and E153Q models is a shift in the small loop containing the β7 hairpin in one protomer of the Cif dimer. This results in displacement of residues 170–172, with a maximal Cα shift of ≈ 5 Å occurring at Gly171 (Figure 4B). This alternate conformation has been observed in some previous Cif mutant structures (e.g., Cif-H207A; PDB ID 4DMH). Thus far, all Cif crystals have grown under similar conditions, in the C2 space group, and with two Cif dimers comprising the asymmetric unit. The Cif protomers within each dimer are not perfectly symmetrical, and the loop shift is only observed on equivalent chains A and C of each dimer. The β7 hairpin loops of these two chains interact with protomers of neighboring asymmetric unit, while the equivalent loops on chains B and D do not. Therefore, the conformational variation is most likely influenced by subtle changes in packing within the crystal lattice.
Figure 4. Structural characterization of Cif-E153Q.
(A) The WT (cyan) and E153Q (orange) Cif dimer models are shown as Cα traces and are nearly superimposable, except for residues 170–172 of chain A. The boxed area is illustrated in (B), where the differing side chain atoms of chain A are shown as bold sticks, both models of chain A have colored carbon atoms, and side chains for aligning residues are shown as thin lines. Chain B for both WT and E153Q are shown with the Cα trace and side chain carbon atoms colored gray. The Cα of Gly171 exhibits the largest positional shift, which is indicated by a dashed line (C) Mutant residue Gln153 is located at the same position as the WT Glu residue, where it accepts hydrogen bonds from the backbone amides of Gly266 and Met272. A semi-transparent cartoon rendering denotes secondary structure, and the main chain is shown with carbon atoms colored gray for both models.
Within the active site, the position of the nucleophile and ring-opening residues are essentially unchanged. Furthermore, the impaired catalytic triad of Cif-E153Q retains the WT configuration (Figure 4C). Gln153 occupies the same position within the active site as the WT Glu residue, and forms equivalent hydrogen bonds to backbone amides (Figure 4C).
Although we cannot determine the carboxamide orientation directly from our X-ray diffraction data, we can assign the most likely orientation based on the hydrogen bonding profile of this residue (Figure 1), and this orientation is corroborated computationally by NQ-Flipper (35). As a carboxamide nitrogen cannot accept hydrogen bonds, we have oriented the carboxamide oxygen of Gln153 to accept the hydrogen bonds from the two backbone amides. As a result, the carboxamide nitrogen, which can only donate hydrogen bonds, is oriented to hydrogen bond with the charge-relay side chain of His297. In the WT protein, the charge-relay acid accepts a hydrogen bond from His297 (Figure 1). Thus, it appears that the E153Q substitution reverses the hydrogen bond orientation between residues 153 and 297, in turn reversing the hydrogen bond orientation between His297 and the water molecule that mediates hydrolysis of the covalent intermediate in the WT enzyme. Taken together, these data suggest that the E153Q mutation blocks Cif EH enzyme activity by preventing activation of the water molecule necessary for release of the covalent enzyme-substrate intermediate.
Blockade of hydrolysis increases abundance of the covalent intermediate
Based on our proposed mechanism, the Cif-E153Q mutant is unable to hydrolyze EBH because it cannot release the covalently attached hydroxyalkyl intermediate. By setting krelease = 0, the mutation should create an extreme example of the kinetic regime (kattack ≫ krelease) that previously permitted capture of covalent intermediates for canonical EH enzymes (36–38). As a result, substrates should be converted to suicide inhibitors, and tandem MS should detect a substantial fraction of the 124–133 peptide in adducted form.
To test this prediction, we repeated our MS analysis using Cif-E153Q incubated with EBH. Importantly, following precipitation, separation, and chymotryptic proteolysis, the spectra obtained with the mutant protein show a significant (>80%) reduction of the native peptide ion intensity, and a corresponding ~100-fold increase in intensity of the hydroxyalkyl adduct (Figure 2C) relative to the levels observed with Cif-WT (Figure 2A). In this case, manual assignment of the fragment ions in tandem MS/MS spectra further confirmed the site of adduction to the Asp129-Ile130 dipeptide (Figure 2D), consistent with our mechanistic model.
The high sensitivity of our LC-MS/MS platform also allows us to explore alternative catalytic activities. Cif is a member of the α/β-hydrolase superfamily, and for some other members of the family, multiple catalytic activities have been observed (39). In our analysis of Cif’s phylogenetic relationships, we noted that the His/Tyr pair is characteristic of the HADs, whereas a Tyr/Tyr pair is found in canonical EHs (10). To test the possibility of alternate HAD activity, we exploited that fact that EBH possesses a halogen in addition to an epoxide moiety, and interrogated our LC-MS/MS data for the production of an adduct representing the covalent intermediate expected for dehalogenase activity. Surprisingly, we were able to observe a minor peak corresponding to a nucleophile-spanning peptide adducted with a dehalogenated EBH intermediate at < 10% of the intensity of the halogenated intermediate (data not shown). The EBH substrate was provided at ≈ 1,000-fold molar excess, and the active-site substitution may have affecte the dehalogenating activity of the mutant. Nevertheless, although other HAD active-site motifs are missing in Cif, a low level of dehalogenating activity is clearly observed. These data underscore the sensitivity of our MS assay, which allows identification of intermediates at low substoichiometric levels. It is therefore well suited to the tracking of alternative activities in this broad family of enzymes (40).
Crystallographic observation of the EH catalytic mechanism
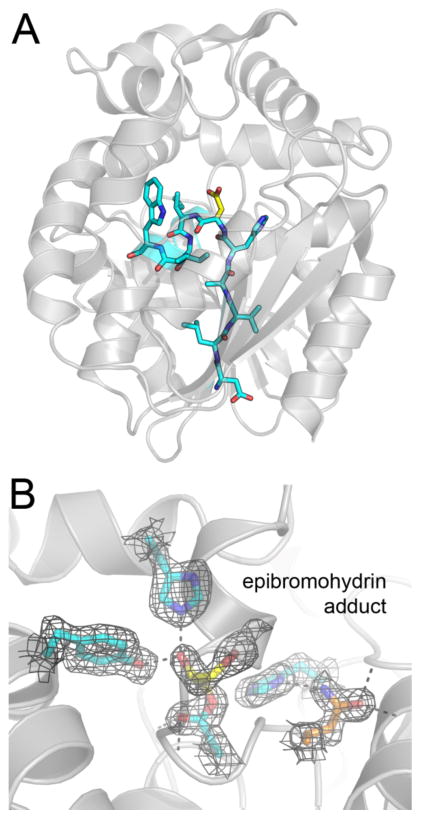
The near-quantitative loss of the native peptide detected by MS suggested we should be able to observe the corresponding adduct by X-ray crystallography. To test this hypothesis, we crystallized Cif-E153Q in the presence of excess EBH and determined the structure to 1.80 Å resolution (Table 1). We do not observe any major differences between the protein residues in the apo-E153Q and EBH-adducted E153Q structures, and the dimer models align with an all-atom RMSD of 0.13 Å (4,146 atoms), which is less than the maximum-likelihood based coordinate error estimate of 0.22 Å. We do not see electron density corresponding to the covalent intermediate associated with the alternative debromination reaction, consistent with the low level of the signal obtained by MS. In contrast, we do observe clear electron density for the adduct of the Asp129 nucleophile in ~60% of the molecules, as expected for the EH activity of the enzyme (Figure 5B). This structure suggests that Cif preferentially selects the terminal carbon on the EBH substrate for nucleophilic attack. Furthermore, hydrogen bonding to ring-opening residues His177 and Tyr239 stabilizes the oxygen derived from the epoxide ring, as previously suggested by biochemical dissection of the EH catalytic mechanism (9, 10). This represents the first direct structural observation of a hydroxyalkyl intermediate for any EH enzyme.
Figure 5. A covalent intermediate detected by mass spectrometry and X-ray crystallography.
(A) A Cif protomer is shown as a gray cartoon, with residues 124–133 shown as bold sticks with carbon atoms colored cyan. This peptide is produced by digesting Cif with chymotrypsin, and it spans the nucleophilic Asp129, highlighted by yellow colored carbon atoms. (B) Co-crystallization of Cif-E153Q with EBH results in an observable hydroxyalkyl-enzyme intermediate with ≥ 60% occupancy. The refined 2Fo-Fc map is shown as black mesh contoured to 1 σ. Side chains of catalytic active site residues and covalent adduct are shown with side chains as bold sticks; WT residues have carbon atoms colored cyan, mutant Gln153 is shown with carbon atoms colored orange, and the adduct has carbon atoms colored yellow.
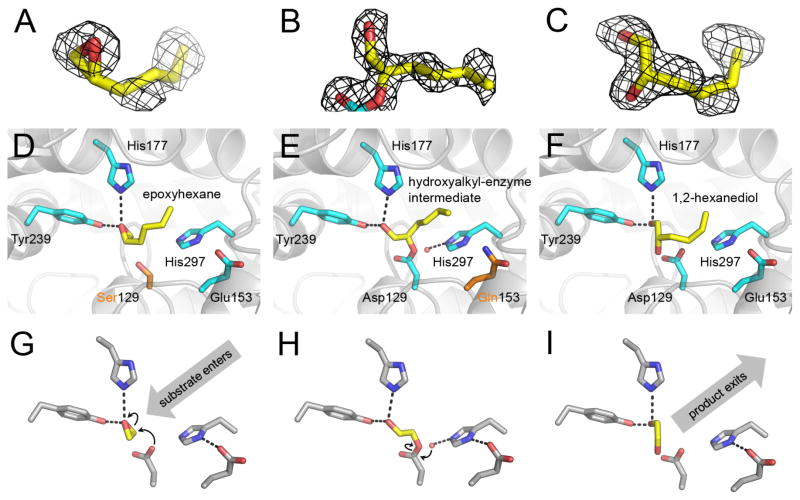
Next, we sought to determine crystal structures capturing each step of the catalytic process: 1) substrate bound in the Michaelis complex prior to nucleophilic attack, 2) the ester-linked hydroxyalkyl-enzyme intermediate, and 3) post-hydrolysis product bound in the active site. To capture and observe the pre-attack substrate bound within the Cif active site, we utilized the previously described Cif-D129S mutation (9), which abrogates enzyme activity by mutation of the nucleophile. To observe the product-bound state, we added a large molar excess of product during crystallization, in an attempt to leverage mass-action to drive formation of a potentially low affinity complex. Unfortunately, we were unable to determine co-crystal structures with EBH or its hydrolysis product 3-bromo-1,2-propanediol. Despite repeated attempts, no ligand electron density was observed in the active site (data not shown). To circumvent this issue, we utilized a different epoxide substrate: 1,2-epoxyhexane, which has a higher specific activity than EBH (Hvorecny et al., manuscript in preparation). This strategy was successful, and we were able to determine structures of Cif-D129S bound to epoxyhexane to 1.85 Å resolution, Cif-E153Q bound to the 1,2-epoxyhexane hydrolysis intermediate to 1.95 Å resolution, and Cif-WT bound to the hydrolysis product 1,2-hexanediol to 1.65 Å resolution (Table 1). The captured or trapped ligands in this reaction series refined with occupancies ≥90%, ≥88%, and ≥74%, respectively. We again observe no significant structural rearrangements within the protein induced by either mutation or small molecule binding, and each dimer model aligns to the apo-WT dimer (PDB ID 3KD2) with an all-atom RMSD ≤ 0.19 Å. The maximum-likelihood based coordinate error estimates of these models range from 0.22 to 0.28 Å.
In the Cif-D129S Michaelis complex for epoxyhexane (Figure 6A), we observe the substrate bound and coordinated within the active site. The aliphatic carbon tail is pointed towards the protein surface, occupying the tunnel that was previously hypothesized to be the pathway by which substrates access the catalytic machinery of Cif (9, 25). The epoxide oxygen is stabilized by hydrogen bonds to residues His177 and Tyr239, which in the WT enzyme position the terminal epoxide carbon for nucleophilic attack by the Asp129 carboxylate and form an oxyanion hole.
Figure 6. The catalytic mechanism of Cif-mediated epoxide hydrolysis is observed crystallographically in three distinct states.
(A–C) 2Fo-Fc electron density maps were calculated before inclusion of ligand or adduct atoms in the phase model. These ‘omit’ maps are shown as a black mesh contoured at 1 σ for (A) epoxyhexane bound to Cif-D129S, (B) the Cif-E153Q hydroxyalkyl-enzyme intermediate of epoxyhexane hydrolysis, and (C) 1,2-hexanediol bound to Cif-WT, along with stick figures showing the refined models (yellow carbons). (D–F) Refined crystallographic structures are shown for complexes that correspond to a step along the reaction coordinate. Refined models are shown as stick figures (WT Cif residues, cyan carbons; mutant Cif residues, orange carbons; substrate- or product-associated residues, yellow carbons). (D) First, the D129S mutation at the nucleophile allows observation of the substrate epoxyhexane bound in the pre-attack Michaelis complex. (E) Second, the E153Q mutation at the charge-relay acid allows observation of the hydroxyalkyl-enzyme intermediate with Asp129, as formed during epoxyhexane hydrolysis. (F) Third, the wild-type enzyme is observed with the product 1,2-hexanediol bound in active site. (G–I) Corresponding steps in the proposed hydrolysis mechanism are exhibited below panels D-F, with stick figures showing the wild-type active-site residues (grey carbons) and a generic epoxide substrate (yellow carbons). Non-carbon atoms are colored by atom type (N = blue; O = red).
The structure of the covalently bound intermediate clearly identifies the structure of the modified side chain (Figure 6B). In contrast to the E153Q-EBH hydrolysis intermediate, we observe covalent substrate-adduction following nucleophilic attack on the secondary epoxide carbon rather than the terminal carbon, suggesting that there may be some variability in the positioning of the epoxide carbons relative to the attacking nucleophile, depending on the steric and chemical characteristics of individual substrates. As seen in the EBH hydrolysis intermediate structure, the oxygen from the epoxide ring remains coordinated by hydrogen bonding to residues His177 and Tyr239. As for EBH, the aliphatic carbon tail also points toward the protein surface.
Finally, in the product-bound structure with Cif-WT (Figure 6C), we again see coordination by His177 and Tyr239 of the 2-OH oxygen, which corresponds to the former epoxide oxygen. The hydrolysis and release of the covalent intermediate occurs by nucleophilic attack via a water molecule activated by the His297-Glu153 charge-relay pair. The oxygen from this water molecule regenerates the nucleophile, and the former nucleophilic oxygen atom is retained by the hydrolysis product. The aliphatic tail remains in the same overall orientation in all models, suggesting that any perturbations associated with the active-site mutations are modest. Thus, we have obtained first atomic-resolution snapshots of the structures formed following substrate binding, nucleophilic attack, and hydrolysis for a member of the EH family of α/β-hydrolases. These crystallographic observations corroborate and reinforce previous predictictions made based on biochemical analysis of EH mechanism and inferences from apo-structures.
Concluding remarks
This study confirms the mechanistic proposal in which the catalytic triad coordinates successive nucleophilic attacks by Asp129 and then by a hydrolytic water. The first step generates a hydroxyalkyl-enzyme intermediate. Mutation of the charge-relay acid side chain (Glu153) in the catalytic triad traps this intermediate in a form that is susceptible to crystallization and detailed structural analysis. This should enable us to delineate more clearly the stereo- and regiospecificity of Cif. Several EHs have relatively accommodating active sites (41–43), but particularly among the bacterial EHs, there are also examples of high selectivity (44–46). In the case of Cif, comparison of the intermediates formed by EBH and 1,2-epoxyhexane shows a switch in attack from the primary to the secondary epoxide carbon. Ultimately, a more detailed understanding of these preferences may help to narrow the universe of candidate substrates that may mediate Cif’s virulence activity.
While our study confirms that Cif utilizes a canonical two-step EH mechanism, it also suggests a non-canonical kinetic regime. For most EHs studied to date, hydrolysis is the rate-limiting step, facilitating the identification of the covalent intermediate, validating a two-step mechanism and permitting detailed kinetic analysis (12, 14, 47, 48). In some cases, however, the expected intermediate has not been observed, and a one-step hydrolytic attack by water has been proposed as an alternative. This may be the case, particularly for enzymes that do not share the α/β-hydrolase fold (49–51). However, our data suggest that in other cases, the enzymes may exhibit Cif-like kinetics, with release relatively fast compared to attack. In the present case, the failure of the covalent intermediate to accumulate significantly complicates kinetic analysis. As a result, further study will be required to determine experimental rate constants and thus establish whether this is due to slowing of attack or acceleration of release. Furthermore, the high sensitivity of our MS approach suggests that it can provide a rapid test for covalent intermediates in kinetic regimes that frustrate traditional labeling strategies and in side reactions that may help to illuminate structure-function relationships among the α/β-hydrolases (40).
Acknowledgments
FUNDING SOURCE STATEMENT
Funding support was provided by NIH grants R01-AI091699, P30-GM106394, R01-ES002710, R01-CA155260, P42-ES004699, T32-AI007519, T32-DK007301, and T32-GM008704, as well as Cystic Fibrosis Foundation grants MADDEN08G0 and STANTO15R0.
We would like to thank Dr. Jessica D. St. Laurent for technical assistance, Dr. Vivian Stojanoff and Dr. Jean Jakoncic at NSLS/Brookhaven for assistance with data collection, Dr. Daniel P. MacEachran for assistance with yeast recombineering, and Dr. Yash R. Patankar for assistance with crystallization.
ABBREVIATIONS
- CFTR
cystic fibrosis transmembrane conductance regulator
- Cif
CFTR inhibitory factor
- EBH
epibromohydrin
- EH
epoxide hydrolase
- HAD
haloacid dehalogenase
- WT
wild-type
References
- 1.Davies JC, Bilton D. Bugs, biofilms, and resistance in cystic fibrosis. Respir Care. 2009;54:628–640. doi: 10.4187/aarc0492. [DOI] [PubMed] [Google Scholar]
- 2.El Solh AA, Akinnusi ME, Wiener-Kronish JP, Lynch SV, Pineda LA, Szarpa K. Persistent infection with Pseudomonas aeruginosa in ventilator-associated pneumonia. Am J Respir Crit Care Med. 2008;178:513–519. doi: 10.1164/rccm.200802-239OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy TF, Brauer AL, Eschberger K, Lobbins P, Grove L, Cai X, Sethi S. Pseudomonas aeruginosa in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;177:853–860. doi: 10.1164/rccm.200709-1413OC. [DOI] [PubMed] [Google Scholar]
- 4.Sousa AM, Pereira MO. Pseudomonas aeruginosa Diversification during Infection Development in Cystic Fibrosis Lungs-A Review. Pathogens. 2014;3:680–703. doi: 10.3390/pathogens3030680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballok AE, Bahl CD, Dolben EL, Lindsay AK, St Laurent JD, Hogan DA, Madden DR, O’Toole GA. Epoxide-mediated CifR repression of cif gene expression utilizes two binding sites in Pseudomonas aeruginosa. J Bacteriol. 2012;194:5315–5324. doi: 10.1128/JB.00984-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bomberger JM, Ye S, MacEachran DP, Koeppen K, Barnaby RL, O’Toole GA, Stanton BA. A Pseudomonas aeruginosa toxin that hijacks the host ubiquitin proteolytic system. PLoS Pathog. 2011;7:e1001325. doi: 10.1371/journal.ppat.1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bomberger JM, Barnaby RL, Stanton BA. The deubiquitinating enzyme USP10 regulates the post-endocytic sorting of cystic fibrosis transmembrane conductance regulator in airway epithelial cells. J Biol Chem. 2009;284:18778–18789. doi: 10.1074/jbc.M109.001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swiatecka-Urban A, Moreau-Marquis S, MacEachran DP, Connolly JP, Stanton CR, Su JR, Barnaby R, O’Toole GA, Stanton BA. Pseudomonas aeruginosa inhibits endocytic recycling of CFTR in polarized human airway epithelial cells. Am J Physiol Cell Physiol. 2006;290:C862–872. doi: 10.1152/ajpcell.00108.2005. [DOI] [PubMed] [Google Scholar]
- 9.Bahl CD, Hvorecny KL, Bomberger JM, Stanton BA, Hammock BD, Morisseau C, Madden DR. Inhibiting an Epoxide Hydrolase Virulence Factor from Pseudomonas aeruginosa Protects CFTR. Angew Chem Int Ed Engl. 2015;54:9881–9885. doi: 10.1002/anie.201503983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bahl CD, Madden DR. Pseudomonas aeruginosa Cif Defines a Distinct Class of α/β Epoxide Hydrolases Utilizing a His/Tyr Ring-Opening Pair. Protein Pept Lett. 2012;19:186–193. doi: 10.2174/092986612799080392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bahl CD, Hvorecny KL, Bridges AA, Ballok AE, Bomberger JM, Cady KC, O’Toole GA, Madden DR. Signature motifs identify an Acinetobacter Cif virulence factor with epoxide hydrolase activity. J Biol Chem. 2014;289:7460–7469. doi: 10.1074/jbc.M113.518092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Widersten M, Gurell A, Lindberg D. Structure-function relationships of epoxide hydrolases and their potential use in biocatalysis. Biochim Biophys Acta. 2010;1800:316–326. doi: 10.1016/j.bbagen.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 13.Pinot F, Grant DF, Beetham JK, Parker AG, Borhan B, Landt S, Jones AD, Hammock BD. Molecular and biochemical evidence for the involvement of the Asp-333-His-523 pair in the catalytic mechanism of soluble epoxide hydrolase. J Biol Chem. 1995;270:7968–7974. doi: 10.1074/jbc.270.14.7968. [DOI] [PubMed] [Google Scholar]
- 14.Morisseau C, Hammock BD. Epoxide hydrolases: mechanisms, inhibitor designs, and biological roles. Annu Rev Pharmacol Toxicol. 2005;45:311–333. doi: 10.1146/annurev.pharmtox.45.120403.095920. [DOI] [PubMed] [Google Scholar]
- 15.Tzeng H-F, Laughlin LT, Armstrong RN. Semifunctional Site-Specific Mutants Affecting the Hydrolytic Half-Reaction of Microsomal Epoxide Hydrolase. Biochemistry. 1998;37:2905–2911. doi: 10.1021/bi9727388. [DOI] [PubMed] [Google Scholar]
- 16.Lacourciere GM, Armstrong RN. The catalytic mechanism of microsomal epoxide hydrolase involves an ester intermediate. Journal of the American Chemical Society. 1993;115:10466–10467. [Google Scholar]
- 17.Morisseau C, Du G, Newman JW, Hammock BD. Mechanism of mammalian soluble epoxide hydrolase inhibition by chalcone oxide derivatives. Arch Biochem Biophys. 1998;356:214–228. doi: 10.1006/abbi.1998.0756. [DOI] [PubMed] [Google Scholar]
- 18.MacEachran DP, Ye S, Bomberger JM, Hogan DA, Swiatecka-Urban A, Stanton BA, O’Toole GA. The Pseudomonas aeruginosa secreted protein PA2934 decreases apical membrane expression of the cystic fibrosis transmembrane conductance regulator. Infect Immun. 2007;75:3902–3912. doi: 10.1128/IAI.00338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O’Toole GA. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol. 2006;72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bahl CD, MacEachran DP, O’Toole GA, Madden DR. Purification, crystallization and preliminary X-ray diffraction analysis of Cif, a virulence factor secreted by Pseudomonas aeruginosa. Acta Crystallogr. 2010;F66:26–28. doi: 10.1107/S1744309109047599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chhabra ES, Ramabhadran V, Gerber SA, Higgs HN. INF2 is an endoplasmic reticulum-associated formin protein. J Cell Sci. 2009;122:1430–1440. doi: 10.1242/jcs.040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kettenbach AN, Schweppe DK, Faherty BK, Pechenick D, Pletnev AA, Gerber SA. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal. 2011;4:rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sano H, Peck GR, Kettenbach AN, Gerber SA, Lienhard GE. Insulin-stimulated GLUT4 protein translocation in adipocytes requires the Rab10 guanine nucleotide exchange factor Dennd4C. J Biol Chem. 2011;286:16541–16545. doi: 10.1074/jbc.C111.228908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faherty BK, Gerber SA. MacroSEQUEST: efficient candidate-centric searching and high-resolution correlation analysis for large-scale proteomics data sets. Anal Chem. 2010;82:6821–6829. doi: 10.1021/ac100783x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bahl CD, Morisseau C, Bomberger JM, Stanton BA, Hammock BD, O’Toole GA, Madden DR. Crystal structure of the Cystic Fibrosis Transmembrane Conductance Regulator Inhibitory Factor Cif reveals novel active-site features of an epoxide hydrolase virulence factor. J Bacteriol. 2010;192:1785–1795. doi: 10.1128/JB.01348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- 27.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific LLC; Palo Alto, CA, USA: 2008. http://www.pymol.org. [Google Scholar]
- 31.Cedrone F, Bhatnagar T, Baratti JC. Colorimetric assays for quantitative analysis and screening of epoxide hydrolase activity. Biotechnol Lett. 2005;27:1921–1927. doi: 10.1007/s10529-005-3904-1. [DOI] [PubMed] [Google Scholar]
- 32.MacEachran DP, Stanton BA, O’Toole GA. Cif is negatively regulated by the TetR family repressor CifR. Infect Immun. 2008;76:3197–3206. doi: 10.1128/IAI.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laughlin LT, Tzeng HF, Lin S, Armstrong RN. Mechanism of Microsomal Epoxide Hydrolase. Semifunctional Site-Specific Mutants Affecting the Alkylation Half-Reaction. Biochemistry. 1998;37:2897–2904. doi: 10.1021/bi972737f. [DOI] [PubMed] [Google Scholar]
- 34.Arand M, Muller F, Mecky A, Hinz W, Urban P, Pompon D, Kellner R, Oesch F. Catalytic triad of microsomal epoxide hydrolase: replacement of Glu404 with Asp leads to a strongly increased turnover rate. Biochem J. 1999;337(Pt 1):37–43. [PMC free article] [PubMed] [Google Scholar]
- 35.Weichenberger CX, Sippl MJ. NQ-Flipper: validation and correction of asparagine/glutamine amide rotamers in protein crystal structures. Bioinformatics. 2006;22:1397–1398. doi: 10.1093/bioinformatics/btl128. [DOI] [PubMed] [Google Scholar]
- 36.Arand M, Wagner H, Oesch F. Asp333, Asp495, and His523 form the catalytic triad of rat soluble epoxide hydrolase. J Biol Chem. 1996;271:4223–4229. doi: 10.1074/jbc.271.8.4223. [DOI] [PubMed] [Google Scholar]
- 37.Hammock BD, Pinot F, Beetham JK, Grant DF, Arand ME, Oesch F. Isolation of a putative hydroxyacyl enzyme intermediate of an epoxide hydrolase. Biochem Biophys Res Commun. 1994;198:850–856. doi: 10.1006/bbrc.1994.1121. [DOI] [PubMed] [Google Scholar]
- 38.Muller F, Arand M, Frank H, Seidel A, Hinz W, Winkler L, Hanel K, Blee E, Beetham JK, Hammock BD, Oesch F. Visualization of a covalent intermediate between microsomal epoxide hydrolase, but not cholesterol epoxide hydrolase, and their substrates. Eur J Biochem. 1997;245:490–496. doi: 10.1111/j.1432-1033.1997.00490.x. [DOI] [PubMed] [Google Scholar]
- 39.Marchot P, Chatonnet A. Enzymatic activity and protein interactions in alpha/beta hydrolase fold proteins: moonlighting versus promiscuity. Protein Pept Lett. 2012;19:132–143. doi: 10.2174/092986612799080284. [DOI] [PubMed] [Google Scholar]
- 40.Lenfant N, Hotelier T, Velluet E, Bourne Y, Marchot P, Chatonnet A. ESTHER, the database of the alpha/beta-hydrolase fold superfamily of proteins: tools to explore diversity of functions. Nucleic Acids Res. 2013;41:D423–429. doi: 10.1093/nar/gks1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arand M, Cronin A, Oesch F, Mowbray SL, Jones TA. The telltale structures of epoxide hydrolases. Drug Metab Rev. 2003;35:365–383. doi: 10.1081/dmr-120026498. [DOI] [PubMed] [Google Scholar]
- 42.Decker M, Arand M, Cronin A. Mammalian epoxide hydrolases in xenobiotic metabolism and signalling. Arch Toxicol. 2009;83:297–318. doi: 10.1007/s00204-009-0416-0. [DOI] [PubMed] [Google Scholar]
- 43.Newman JW, Morisseau C, Hammock BD. Epoxide hydrolases: their roles and interactions with lipid metabolism. Prog Lipid Res. 2005;44:1–51. doi: 10.1016/j.plipres.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Okamura-Ikeda K, Hosaka H, Yoshimura M, Yamashita E, Toma S, Nakagawa A, Fujiwara K, Motokawa Y, Taniguchi H. Crystal structure of human T-protein of glycine cleavage system at 2.0 A resolution and its implication for understanding non-ketotic hyperglycinemia. J Mol Biol. 2005;351:1146–1159. doi: 10.1016/j.jmb.2005.06.056. [DOI] [PubMed] [Google Scholar]
- 45.Steinreiber A, Faber K. Microbial epoxide hydrolases for preparative biotransformations. Curr Opin Biotechnol. 2001;12:552–558. doi: 10.1016/s0958-1669(01)00262-2. [DOI] [PubMed] [Google Scholar]
- 46.van Loo B, Kingma J, Arand M, Wubbolts MG, Janssen DB. Diversity and biocatalytic potential of epoxide hydrolases identified by genome analysis. Appl Environ Microbiol. 2006;72:2905–2917. doi: 10.1128/AEM.72.4.2905-2917.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Armstrong RN. Kinetic and chemical mechanism of epoxide hydrolase. Drug Metab Rev. 1999;31:71–86. doi: 10.1081/dmr-100101908. [DOI] [PubMed] [Google Scholar]
- 48.Armstrong RN, Cassidy CS. New structural and chemical insight into the catalytic mechanism of epoxide hydrolases. Drug Metab Rev. 2000;32:327–338. doi: 10.1081/dmr-100102337. [DOI] [PubMed] [Google Scholar]
- 49.de Medina P, Paillasse MR, Segala G, Poirot M, Silvente-Poirot S. Identification and pharmacological characterization of cholesterol-5,6-epoxide hydrolase as a target for tamoxifen and AEBS ligands. Proc Natl Acad Sci U S A. 2010;107:13520–13525. doi: 10.1073/pnas.1002922107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hopmann KH, Hallberg BM, Himo F. Catalytic mechanism of limonene epoxide hydrolase, a theoretical study. J Am Chem Soc. 2005;127:14339–14347. doi: 10.1021/ja050940p. [DOI] [PubMed] [Google Scholar]
- 51.Thunnissen MM, Nordlund P, Haeggstrom JZ. Crystal structure of human leukotriene A(4) hydrolase, a bifunctional enzyme in inflammation. Nat Struct Biol. 2001;8:131–135. doi: 10.1038/84117. [DOI] [PubMed] [Google Scholar]
- 52.Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]