Summary
The transition from vegetative to reproductive development in the temperate cereals is mainly regulated by seasonal cues including vernalization (determined mainly by VRN1 and VRN2 genes) and photoperiod (determined mainly by PPD1 and CO2 genes). The wheat VRN3 gene, which is similar to Arabidopsis FT, plays a central role in the integration of the competing signals from these two pathways. Under long days, VRN3 transcription is down-regulated by VRN2, a unique flowering repressor in cereals, and up-regulated by CO2. Overexpression of VRN3 overcomes VRN2 repression and promotes VRN1 transcription and flowering initiation. Using yeast two- and three-hybrid assays we show here that the CCT domains present in VRN2 and CO2 proteins interact with the same subset of eight NF-Y proteins, and that these CCT proteins compete with NF-YA for interactions with NF-YB proteins. We have confirmed all these interactions in vitro, and the interactions between VRN2 and two of the three NF-YB proteins were further confirmed in planta. In addition, we show that mutations in the CCT domain of VRN2 that eliminate the vernalization requirement in winter wheat also reduce the strength of the interactions between VRN2 and NF-Y proteins, and the ability of VRN2 to compete with CO2. Taken together, our results suggest that the interactions between CCT and NF-Y proteins play an important role in the integration of the vernalization and photoperiod seasonal signals, and provide a flexible combinatorial system to integrate multiple developmental and environmental signals in the regulation of flowering initiation in the temperate cereals.
Keywords: wheat flowering, NF-Y, VRN2, CO, VRN3 regulation
Introduction
One of the most important points in the regulation of plant development is the transition from vegetative to reproductive development. The precise regulation of this transition, which is critical for maximizing grain production in annual temperate cereals such as wheat and barley, is determined mainly by seasonal changes in day length (photoperiod) and temperature. Winter cereals require an extended exposure to low temperatures to accelerate flowering, a process known as vernalization. In wheat and barley, natural allelic variation in vernalization requirement is mainly determined by vernalization genes VRN1, VRN2, VRN3 and VRN4 (Yan et al., 2003; 2004; 2006; Yoshida et al., 2010). The first three genes have been cloned, and their functions and interactions have been summarized in two recent reviews (Trevaskis et al., 2007; Distelfeld et al., 2009b).
The VRN1 gene encodes a MADS-box transcription factor closely related to the duplicated Arabidopsis genes APETALA1, CAULIFLOWER and FRUITFULL (Yan et al., 2003). The VRN2 locus from temperate grasses includes two tandemly duplicated genes ZCCT1 and ZCCT2; both involved in flowering repression (Yan et al., 2004; Distelfeld et al., 2009a). VRN2 has no close homologues in Arabidopsis, but plays a similar role to FLOWERING LOCUS C (FLC). Both ZCCT1 and ZCCT2 proteins have a putative Zinc finger in the N-terminus, and a CCT domain in the C-terminus (Yan et al., 2004). The CCT domain is a 43-amino acid region, first described in the Arabidopsis proteins CONSTANS (CO), CONSTANS-like (COL) and TIMING OF CAB1 (TOC1) (Strayer et al., 2000). Deletions and mutations involving the CCT domain of ZCCT1 and ZCCT2 proteins are associated with recessive alleles for spring growth habit in both wheat and barley (Yan et al., 2004; Dubcovsky et al., 2005; Hemming et al., 2009; Distelfeld et al., 2009a).
VRN3 encodes a RAF kinase inhibitor–like protein with high homology to Arabidopsis FLOWERING LOCUS T (FT) (Yan et al., 2006). The FT protein acts as a long-distance flowering signal (florigen) that moves from leaves to apices through the phloem and promotes flowering in a wide range of plant species (Corbesier et al., 2007; Tamaki et al., 2007). In wheat, the VRN3 protein interacts with the bZIP protein FDL2, which binds to the VRN1 promoter (Li and Dubcovsky, 2008). Overexpression of VRN3 in winter wheat results in the up-regulation of VRN1 and a spring growth habit (Li and Dubcovsky, 2008). In the fall, prior to vernalization, VRN3 is repressed by VRN2 (Yan et al., 2006; Hemming et al., 2008; Distelfeld et al., 2009b). Vernalization releases VRN1 chromatin from an inactive state and results in a gradual increase in its transcript levels (Oliver et al., 2009). The up-regulation of VRN1 is associated with the down-regulation of VRN2, which allows the long-day induction of VRN3. The up-regulation of VRN3 promotes further increases in VRN1 transcripts, generating a positive feedback loop that up-regulates VRN1 transcripts until a threshold level required to initiate flowering is reached (Loukoianov et al., 2006; Distelfeld et al., 2009b).
The VRN3 gene functions as an integrator of the vernalization and photoperiod pathways in temperate cereals (Yan et al., 2006; Hemming et al., 2008). The down-regulation of VRN3 by VRN2 described above is counterbalanced by the up-regulation of VRN3 by the photoperiod pathway (Turner et al., 2005). In photoperiod-sensitive wheat and barley varieties, the CCT domain protein CO up-regulates VRN3 and accelerates flowering under long days. Two closely related CO genes, CO1 and CO2, are present in the temperate cereals as a result of a duplication that occurred after the wheat-rice divergence. The wheat CO2 gene is known to complement mutations in the rice Hd1 gene (= CO) suggesting that CO2 is the functional homologue of CO in wheat (Nemoto et al., 2003).
Recent studies indicate that several members of the NUCLEAR FACTOR-Y (NF-Y) transcription factor family, also known as HEME ACTIVATOR PROTEIN (HAP), are involved in the regulation of plant flowering through interactions with CCT domain proteins CO and COL (Ben-Naim et al., 2006; Wenkel et al., 2006; Cai et al., 2007; Chen et al., 2007; Kumimoto et al., 2008; 2010). In yeast and mammals, NF-Y proteins form a complex that consists of three subunits: NF-YA, NF-YB and NF-YC, each encoded by a single gene (Mantovani, 1999). In plants, however, each of the NF-Y subunits is encoded by gene families with more than 10 members (Edwards et al., 1998; Yang et al., 2005). A total of 35 NF-Y genes (10 NF-YA, 11 NF-YB, 14 NF-YC) have been identified in Triticum aestivum (Stepheson et al., 2007, 2010, 2011).
The Arabidopsis AtNF-YB2 (AtHAP3b) protein and its closest homologous protein AtNF-YB3 promote the expression of FT through interactions with CCT domain proteins CO and COL (Cai et al., 2007; Kumimoto et al., 2008). Promoter GUS fusions for AtNF-YB2 and AtNF-YB3 show clear vascular expression patterns in leaves (Siefers et al., 2009), which is important because the role of CO in floral induction is known to be phloem-specific (An et al., 2004). In contrast to the promoting effects of the AtNF-YB2 and AtNF-YB3 genes on flowering, overexpression of AtNF-YA1 (HAP2a) or AtNF-YB1 (HAP3a) genes under a phloem specific promoter delays flowering and significantly reduces FT transcript levels without affecting CO transcript levels (Wenkel et al., 2006). These results suggest that different NF-Y proteins may promote or delay flowering depending on the other subunits in the NF-Y complex and also on which CCT domain proteins they interact with.
The NF-YA (HAP2) protein shares significant sequence homology (18 similar amino acids) with the CCT domain (Wenkel et al., 2006). Natural and induced mutations at six of these conserved amino acids in the CCT domains of ZCCT1, ZCCT2, PPD1 and CO are associated with differences in flowering time (Wenkel et al., 2006; Distelfeld et al., 2009a). In the NF-YA protein, these conserved positions are all located in a sub-domain involved in both the interactions with NF-YB and NF-YC subunits, and the recognition of the CCAAT cis regulatory elements in the promoters of many eukaryotic genes. Based on the interactions observed between the CCT domain proteins (CO and COL) and NF-YB and NF-YC subunits in Arabidopsis, and the NF-Y complex’s role in transcriptional regulation in mammals and yeast, a model has been proposed in which CCT proteins act by replacing the NF-YA subunits of the NF-YA/B/C complexes, altering the transcriptional regulation ability of the complexes to their target genes (Wenkel et al., 2006). A more recent study has shown that NF-Y proteins can interact with a membrane-associated basic domain/leucine zipper (bZIP28) transcription factor, and interact with compound CCAAT-N10-CACG sequences (ERSE-I) present in the promoters of some stress related genes (Liu and Howell, 2010). CO alone can bind to the TGTG-N2-3-ATG sequence (CORE1) present in the promoter of FT (Tiwari et al., 2010). The presence of additional CCAAT boxes potentiates CO-mediated activation, but is not essential for CO activity as a transcription factor (Tiwari et al., 2010).
In this study, we identified interactions between wheat VRN2 (ZCCT1 and ZCCT2) and eight different NF-Y proteins and showed that the same eight NF-Y proteins also interact with CO2. We also demonstrate that VRN2 and CO2 proteins compete for interactions with their common NF-Y protein partners using a yeast three-hybrid system. Finally, we show that mutations in the CCT domain of VRN2 that eliminate its ability to repress flowering also reduce the strength of the interactions between VRN2 and NF-Y proteins and its ability to compete with CO2 for NF-Y interactions. Taken together these results suggest that the CCT-NF-Y protein interactions play an important role in the integration of the photoperiod and vernalization signals in the temperate cereals.
Results
Wheat ZCCT1 and CO2 proteins interact with the same set of NF-Y proteins
cDNA fragments of 13 NF-Y genes were PCR amplified and cloned in frame to the GAL4 activation domain of the yeast prey vector pGADT7 (primers and GenBank accession numbers are listed in Table S1). The cloned cDNA fragments encompass most of the coding regions (85–95%) including the complete NF-Y core domains, which are known to be critical for protein-protein interactions. The ZCCT1 full-length cDNA was cloned into pGBKT7 vector as bait (primers in Table S1). Interactions between ZCCT1 and these 13 NF-Y proteins were tested using yeast two-hybrid assays (Y2H). Two NF-YA, 3 NF-YB and 3 NF-YC proteins showed strong interactions with ZCCT1 (Table 1).
Table 1.
Interactions of CCT domain proteins with NF-Y factors.
| Prey
|
NF-YA
|
NF-YB
|
NF-YC
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bait | 7 | 9 | 1 | 2 | 3 | 4 | 5 | 9 | 11 | 12 | 7 | 10 | 11 |
| ZCCT1 | + | + | + | - | + | - | - | + | - | - | + | + | + |
| CO2-CCT | + | + | + | - | + | - | - | + | - | - | + | + | + |
| CO1-CCT | - | - | - | - | + | - | - | - | - | - | - | - | - |
| PPD1-CCT | - | - | - | - | - | - | - | - | - | - | + | - | - |
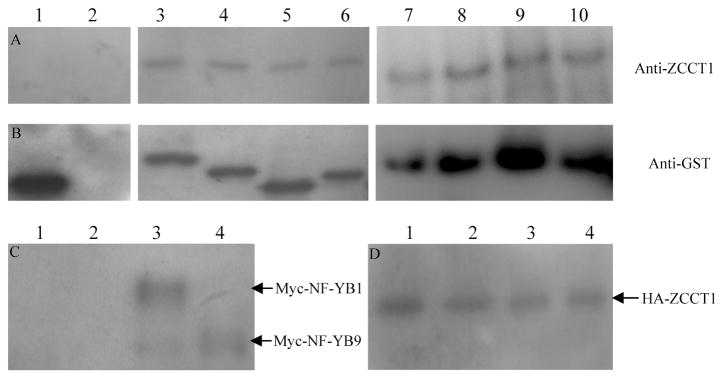
The interactions between ZCCT1 and 8 NF-Yproteins were further confirmed by in vitro Co-immunoprecipitation (Co-IP) assays using purified tagged proteins (Supplemental methods S3). When ZCCT1-MBP was incubated together with the GST tag (Fig. 1A, lane 1) or used alone (Fig. 1A, lane 2) in the in vitro interaction assays, no ZCCT1 protein was detected in the co-immunoprecipitated samples. On the contrary, ZCCT1-MBP protein was co-precipitated with the NF-Y proteins (Fig. 1A, lane 3–10). The same membranes were then stripped and re-hybridized with GST antibody (Fig. 1B).
Figure 1. Validation of the interactions between ZCCT1 and NF-Y proteins by in vitro and in planta Co-IPs.
GST antibody was used to immunoprecipitate protein complexes in the in vitro analysis, and Anti-HA Affinity Matrix was used to precipitate protein complexes in the in planta analysis. A&B) Western blot analyses of the in vitro Co-IPs, A) ZCCT1 antibody was used to detect the presence of ZCCT1 protein in the immunoprecipitates, B) the same membranes were stripped and re-hybridized with GST antibody. The proteins included in the in vitro assays are: 1) ZCCT1-MBP and GST tag; 2) ZCCT1-MBP protein alone; 3) ZCCT1-MBP and NF-YB1-GST; 4) ZCCT1-MBP and NF-YB9-GST; 5) ZCCT1-MBP and NF-YC11-GST; 6) ZCCT1-MBP and NF-YC7-GST; 7) ZCCT1-MBP and NF-YB3-GST; 8) ZCCT1-MBP and NF-YA7-GST; 9) ZCCT1-MBP and NF-YA9-GST; 10) ZCCT1-MBP and NF-YC10-GST. C&D) Western blot analyses of the in planta Co-IPs, C) Anti-Myc antibody was used to detect Myc-NF-YB fusions, D) the same membrane were stripped and re-hybridized with Anti-HA antibody. Protein extracts of the following infiltrations were used in the in planta Co-IPs: 1) Infiltration of HA-ZCCT1 alone; 2) Co-infiltration of HA-ZCCT1 and Myc-NF-YB3; 3) Co-infiltration of HA-ZCCT1 and Myc-NF-YB1; 4) Co-infiltration of HA-ZCCT1 and Myc-NF-YB9.
We also tested whether ZCCT1 interacts with NF-Y proteins in plant cells as shown in yeast and in vitro using a Nicotiana benthamiana transient expression system. ZCCT1 was expressed as an HA fusion and three NF-YB proteins NF-YB1, 3 and 9 were expressed as Myc fusions. All fusion proteins were driven under the control of 35S promoter. Anti-HA Affinity Matrix was used to precipitate protein complexes, Anti-Myc and anti-HA antibodies were used in the western blot analyses. As shown in Fig. 1C, NF-YB1 and NF-YB9 were co-precipitated with HA-ZCCT1 protein, whereas NF-YB3 was not detected. The same membrane was stripped and re-hybridized with anti-HA antibody as the loading control for equal protein input (Fig. 1D) A ZCCT1 protein truncation lacking the putative 5’ terminal zinc finger (ZCCT1-CCT, Fig. S1) was generated using primers ZCCT1-F3 and ZCCT1-R1 (Table S1) to test the protein region required for the interactions with NF-Y proteins. ZCCT1-CCT was able to interact with all eight NF-Y proteins in the absence of the zinc finger (Fig. S2), confirming that the CCT domain region is required for the ZCCT1-NF-Y protein interaction.
cDNA fragments including the complete CCT domains of CO1 (CO1-CCT, Fig. S1), CO2 (CO2-CCT, Fig. S1), and the pseudo-response regulator PPD1 (PPD1-CCT, Fig. S1) were also cloned from T. monococcum accession G3116 (primers in Table S1) and used as baits to test their interactions with the 13 NF-Y proteins. CO2-CCT showed strong interactions with the same eight NF-Y proteins that interacted with ZCCT1 (Table 1) suggesting that CO2 and ZCCT1 proteins may compete for these common NF-Y interactors. CO1-CCT interacted only with NF-YB3, suggesting sub-functionalization of CO1 and CO2 proteins. PPD1-CCT interacted only with NF-YC7 (Table1), further confirming the specificity of the CCT-NF-Y protein interactions. None of the three yeast HAP (NF-Y) proteins was able to interact with the wheat ZCCT1 (Fig. S3B), CO1 or CO2 (Fig. S3E) proteins, suggesting that the Y2H interactions between wheat CCT domain and NF-Y proteins were not mediated by endogenous yeast proteins.
The CCT domain is involved in interactions among CCT-domain proteins
In addition to their interactions with the NF-Y proteins, ZCCT1 and CO2 proteins were also able to self-dimerize as well as to interact with each other and with CO1 in Y2H assays (Table 2). However, PPD1-CCT did not interact with any of the CCT domain proteins we tested (Table 2) confirming the specificity of the CCT-CCT interactions.
Table 2.
Protein interactions among CCT domain proteins.
| Prey
|
ZCCT1 | CO1-BB- CCT | CO1-CCT | CO1-BB | CO2-BB- CCT | CO2-CCT | CO2-BB |
|---|---|---|---|---|---|---|---|
| Bait | |||||||
| ZCCT1 | + | + | + | - | + | + | - |
| ZCCT1-CCT | + | + | + | - | + | + | - |
| CO2-CCT | + | + | + | - | + | + | - |
| CO1-CCT | + | - | - | - | + | + | - |
| PPD1-CCT | - | - | - | - | - | - | - |
The CO1-BB-CCT and CO2-BB-CCT proteins (Fig. S1), which contain the two N-terminal B-boxes and the C- terminal CCT domain, showed strong auto-activation when used as baits, therefore, were only used as preys in the study (Table 2). In addition to the CO1-CCT and CO2-CCT truncations described above, we generated two protein truncations including only the B-boxes region (CO1-BB and CO2-BB, Fig. S2) to further define the region involved in the CO1 and CO2 protein interactions. Interactions were detected only between the complete and truncated proteins that include the CCT domain (Table 2), confirming that this domain is required for the interactions among CCT domain proteins.
Mutations in three conserved arginine (R) residues within the CCT domain of VRN2 reduce the strength of protein interactions
Mutations in conserved arginine (R) amino acids at positions 16 (R16C), 35 (R35W) or 39 (R39C) of the 43- amino acid CCT domain have been shown to result in nonfunctional vrn2 alleles and spring growth habit in our previous genetic studies (Yan et al., 2004, Distelfeld et al., 2009a). The effect of these mutations on the strength of interactions between four ZCCT mutants and both NF-Y and other CCT proteins was quantified and compared to the respective wild type ZCCT proteins using the α-gal quantitative assay.
Effect of mutations on CCT-NF-Y interactions
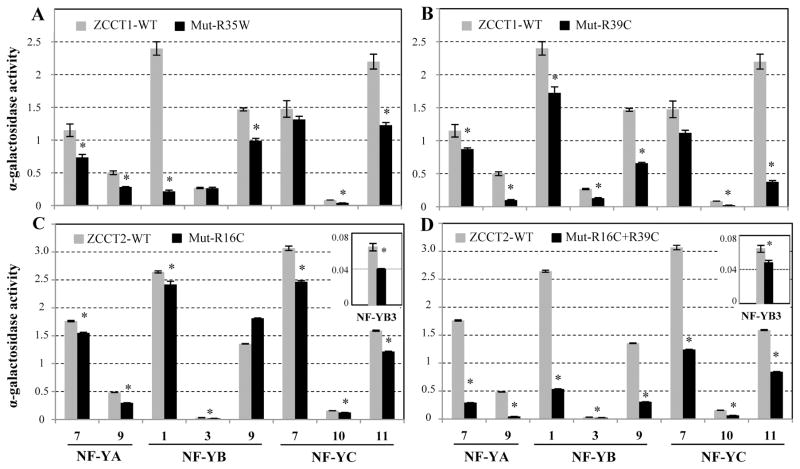
Relative to the wild type ZCCT1 protein, the ZCCT1- R35W mutant protein from T. monococcum DV92 showed significantly weaker interactions (P<0.01) with six out of the eight NF-Y proteins (Fig. 2A). The same six proteins, plus NF-YB3, also showed significantly weaker interactions (P<0.01) with the ZCCT1-R39C mutant protein from tetraploid wheat Langdon (A genome) than with the ZCCT1 wild type control (Fig. 2B).
Figure 2. Effect of mutations in the CCT domain on the strength of protein interactions between ZCCT and NF-Y proteins.
The mutations include A) ZCCT1-R35W and B) ZCCT1-R39C, C) ZCCT2-R16C, and D) ZCCT2-R16C+ R39C. The ZCCT wild type (grey bars) and mutant (black bars) proteins were expressed as DNA-binding domain fusions and the NF-Y proteins as activation domain fusions. The α-gal assays were used to quantify the strength of the interactions. Relative α-gal activity values for each interaction are the average of 10-16 replicates (error bars = s.e.m. and * = P< 0.01). The insets show the α-gal activity level for ZCCT2-NF-YB3 interaction using a different scale.
Since all the ZCCT2 natural alleles identified so far in T. monococcum and in the A genome of tetraploid wheat carry one or two mutations, a ZCCT2 protein with a wild type CCT domain was generated by reverting the R16C mutation in the ZCCT2 protein from G3116 by site-directed mutagenesis (Supplemental Method S2). This engineered non-mutant ZCCT2 protein (ZCCT2-WT), showed significantly stronger interactions (P<0.01) with all the NF-Y protein tested except NF-YB9 (Fig. 2C) relative to the ZCCT2- R16C mutant protein. The ZCCT2 protein from DV92, which has both R16C and R39C mutations (ZCCT2- R16C+R39C) showed even more severe reductions in the strength of its interactions with all eight NF-Y proteins than the single R16C mutant (P<0.01, Fig. 2D).
Effect of mutations on CCT-CCT interactions
Since the CCT domain is also required for protein interactions within the CCT protein family, we also quantified the effect of the R mutations on interactions between ZCCT and CO proteins using the α-gal quantitative assay. All four mutants significantly reduced the strength of protein interactions between ZCCT and CO1 (Fig. S4 A & B). However, these mutations have a different impact on interactions between ZCCT and CO2 proteins. R35W and R39C mutations had almost no effect on ZCCT1-CO2 interaction strength (Fig. S4C). Single mutant R16C and double mutant R16C+R39C caused similar reduction level on the strength of ZCCT2-CO2 interactions (P<0.01, Fig. S4D), confirming the lack of effect of the R39C mutation on ZCCT-CO2 interactions.
ZCCT1 and CO2 compete for interaction with NF-Y proteins in yeast
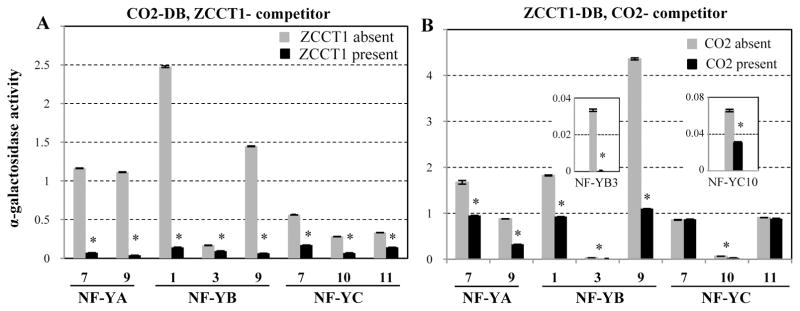
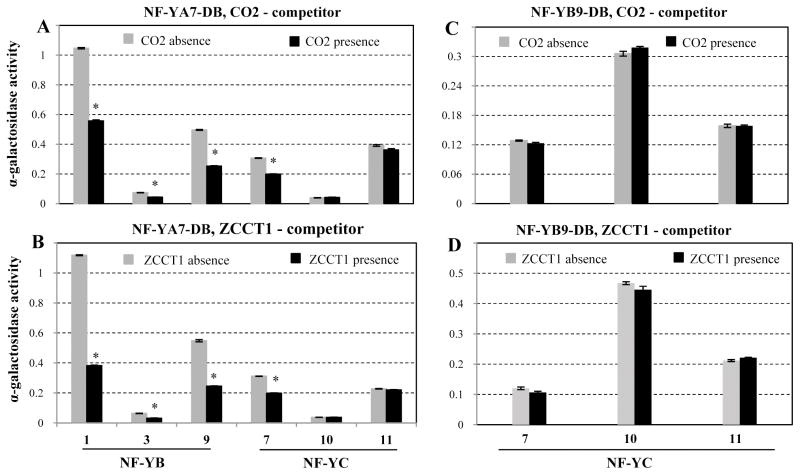
The pBridge yeast three-hybrid (Y3H) system was used to test the hypothesis that ZCCT1 and CO2 proteins compete for interactions with their eight common interacting NF-Y proteins. In the first experiment, CO2 was expressed as the DNA-binding domain fusion, and ZCCT1 was conditionally expressed as the competitor protein under the control of the MET25 promoter (repressed by the presence of methionine in the medium). The quantitative α-gal assay was used to compare the strength of protein interactions between CO2 and NF-Y proteins (expressed as activation domain fusions) in the presence or absence of ZCCT1. The interactions between CO2 and all eight NF-Y proteins were significantly reduced in the presence of ZCCT1 (P<0.01, Fig. 3A), indicating that ZCCT1 competes with CO2 for interactions with these NF-Y proteins.
Figure 3. Competition of ZCCT1 and CO2 proteins for interactions with NF-Y proteins.
Yeast three-hybrid assays were used to test A) the effect of ZCCT1 as competitor when CO2 was expressed as the DNA-binding domain fusion, and B) the effect of CO2 as competitor when ZCCT1 was expressed as the DNA-binding domain fusion (NF-Y proteins were expressed as activation domain fusions). The α-galactosidase activity of the protein interactions in the absence of the competitor is shown in grey bars, and in the presence of the competitor in black bars. Relative α-gal activity values for each interaction are the average of 10-12 replicates (error bars= s.e.m. and * = P< 0.01). The insets show the α-gal activity for ZCCT1-NF-YB3 and ZCCT1-NF-YC10 interactions using a different scale.
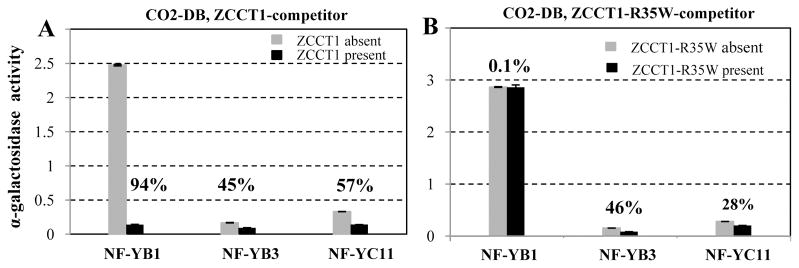
In the second experiment, ZCCT1 was expressed as the DNA-binding domain fusion, and CO2 as the inducible competitor. The interaction between ZCCT1 and NF-YB3 was completely eliminated when CO2 was expressed (Fig. 3B), suggesting CO2 was a stronger interactor for NF-YB3 than ZCCT1. However, the presence of CO2 did not result in significant reduction for interactions between ZCCT1 and NF-YC7 or NF-YC11 (Fig. 3B), indicating that ZCCT1 was a stronger interactor for these two proteins than CO2. The interactions between ZCCT1 and five other NF-Y proteins including NF-YA7, NF-YA9, NF-YB1, NF-YB9 and NF-YC10 were also significantly reduced by the presence of CO2 (Fig. 3B), but less than the reciprocal interactions using ZCCT1 as the competing protein (Fig. 3A). The stronger ability of ZCCT1 to compete with CO2 in most of the NF-Y interactions parallels the dominance of the vernalization requirement over the long day induction of flowering in un-vernalized winter wheat accessions. Since ZCCT1 interacts with both CO2 and the NF-Y proteins, the ZCCT1 competition with the CO2-NF-Y interactions in the Y3H assays can be the result of its interaction with CO2, NF-Y, or both. To differentiate between these alternatives, we took advantage of the unique characteristics of the ZCCT1-R35W mutant, which reduces the strength of the ZCCT1 interaction with several NF-Y proteins (Fig. 2), but has almost no effect on the ZCCT1-CO2 interaction (Fig. S4C). We selected three NF-Y proteins whose interactions with ZCCT1 were differentially affected by the R35W mutation: NF-YB1 (strong reduction), NF-YB3 (not affected), or NC-YC11 (mild reduction) (Fig. 2). The ZCCT1-R35W mutant, which has very limited interaction with NF-YB1 (Fig. 2A), has a significantly reduced ability to compete with the CO2-NF-YB1 interaction (Fig. 4B). The CO2-NF-YB1 interaction is reduced by 94% in the presence of the wild type ZCCT1 protein (Fig. 4A) but only 0.1% in the presence of the mutant ZCCT1-R35W protein (Fig. 4B). In contrast, both ZCCT1 and ZCCT1-R35W showed the same ability to compete with the CO2-NF-YB3 interaction (Fig. 4A &4B), as expected from the inability of this mutation to affect the ZCCT1-NF-YB3 interaction (Fig. 2). Finally, the competing strength of ZCCT1 with the CO2-NF-YC11 interaction was partially affected by the R35W mutation (57 % reduction with ZCCT1 vs. 28% reduction with ZCCT1-R35W), reflecting the intermediate effect of this mutation on the strength of the ZCCT1-NF-YC11 interaction (Fig. 2A and 4). In summary, the effects of the R35W mutation on the ZCCT1-NF-Y interactions in the Y2H assays (Fig. 2A) were well correlated with the effect of the mutation on the ability of ZCCT1 to compete with the CO2-NF-Y interactions in the Y3H assays, but not with its effect on the ZCCT1-CO2 interactions. These results suggest that the ZCCT1 competition was the result of its interactions with the NF-Y proteins rather than with CO2.
Figure 4. Effect of the CCT domain mutation R35W on ZCCT1-NF-Y interactions and on competition between ZCCT1 with CO2.
The α-gal assays were used to quantify the relative strength of the protein interactions. Relative α-gal activity values for each interaction are the average of 10-16 replicates. Percentages indicate the percent reduction in α-gal activity. A) Effect of the ZCCT1-wild type protein expressed as a competitor in Y3H assays on the strength of the interactions between CO2 (DNA-binding domain fusion) and NF-YB1, NF-YB3 and NF-YC11 (activation domain fusions ), B) Effect of ZCCT1-R35W mutant protein expressed as a competitor in Y3H assays on the strength of the interaction between CO2 (DNA-binding domain fusion ) and NF-YB1, NF-YB3 and NF-YC11 (activation domain fusions).
ZCCT1 and CO2 proteins show stronger competition with NF-YA than with the other two subunits in the NF-Y protein complex in yeast
In yeast and mammals, NF-Y proteins are known to function as a trimeric complex. The NF-YB and NF-YC subunits first form a tight heterodimer in the cytoplasm that is then translocated to the nucleus where it interacts with NF-YA (Mantovani, 1999). Since the CCT domain has several conserved amino acids with the NF-YA subunit it has been proposed that CCT domain proteins may compete with the NF-YA subunits (Wenkel et al., 2006). We used the Y3H system to test this hypothesis.
First, we expressed NF-YA7 as a DNA-binding domain fusion and tested how the presence of CO2 or ZCCT1 affected the interactions between NF-YA and NF-YB, and between NF-YA and NF-YC. The strength of the interactions between NF-YA7 and the three tested NF-YB proteins (NF-YB1, NF-YB3 and NF-YB9) was significantly reduced by the introduction of the competing CO2 (40 to 50% reduction, Fig. 5A) or ZCCT1 (50 to 65% reduction, Fig. 5B) proteins. Only one of the NF-YA - NF-YC interactions (NF-YA7 – NF-YC7) was affected by the presence of CO2 (35% reduction, Fig. 5A) or ZCCT1 (36% reduction, Fig. 5B) proteins.
Figure 5. Competition of ZCCT1 and CO2 proteins for interactions with NF-Y subunits.
Yeast three-hybrid assays were used to test the effect of A) CO2 and B) ZCCT1 as competitors on the interactions between NF-YA7 (expressed as DNA-binding domain fusion) and different NF-YB and NF-YC proteins (expressed as activation domain fusions); and the effect of C) CO2 and D) ZCCT1 as competitors on the interactions between NF-YB9 (expressed as DNA-binding domain fusion) and different NF-YC proteins (expressed as activation domain fusion). The α-galactosidase activity generated by the protein interactions in the absence of the competitor is shown in grey bars, and in the presence of the competitor in black bars. Relative α-gal activity values for each interaction are the average of 10-12 replicates (error bars= s.e.m. and * = P< 0.01).
We then expressed NF-YB9 as the DNA-binding domain fusion and tested whether CO2 or ZCCT1 can affect the NF-YB – NF-YC interactions. None of the NF-YB – NF-YC interactions was affected by the presence of CO2 or ZCCT1 (Fig. 5C & 5D). Taken together, these results suggest that ZCCT1, CO2 and NF-YA proteins compete for interactions with NF-YB proteins NF-YB1, NF-YB3 and NF-YB9.
Discussion
Complexity of NF-Y –CCT interactions
The 10-fold expansion of the three NF-Y gene subfamilies in the plant kingdom can potentially generate a large number (>1,000) of different trimeric NF-Y complexes (Siefers et al., 2009).The additional interactions between NF-Y subunits and CCT domain proteins, which include flowering and circadian clock proteins (Griffiths et al., 2003), greatly increase the number of potential combinations. The plant-specific CCT domain shares 18 similar amino acids with NF-YA (Wenkel et al., 2006) and mediates interactions between CCT domain proteins and NF-Y proteins Fig. S2). In Arabidopsis, the interactions between the CCT domain protein CO and NF-YB and NF-YC proteins have been demonstrated in yeast, in vitro, and in planta (Wenkel et al., 2006).
Here, we show similar interactions between NF-Y subunits and the CO homologues in wheat and also report interactions with the flowering repressors ZCCT1 and ZCCT2 (Table 1). Moreover, we show that wheat CO2 and ZCCT1 proteins interact with the same subset of NF-Y subunits (Table 1), and that they compete with each other and with NF-YA subunits for interactions with NF-YB in Y3H assays (Fig. 3 and 5). We also show that mutations in the CCT domain of ZCCT proteins that eliminate the vernalization requirement (Yan et al., 2004; Distelfeld et al., 2009a) also reduce the strength of ZCCT - NF-Y proteins interactions (Fig. 2) and the ability of ZCCT1 to compete with CO2 (Fig. 3). This correlation provides an indirect link between the biochemical interactions described in this study and their effect on flowering time.
In the temperate cereals, the unique presence of ZCCT1 and ZCCT2, the duplication of the CO1 and CO2 flowering promoters, and their complex competitive interactions provide an additional layer of complexity to the interactions between CCT domain and NF-Y complexes. These interactions might be further complicated by other CCT proteins. Compared with ZCCT and CO2 proteins, CO1 and PPD1 seem to be more selective in their NF-Y protein interactions. Among the 13 NF-Y proteins we have tested, CO1 interacted only with NF-YB3, and PPD1 interacted only with NF-YC7. Since we have not tested the ability of CO1 and PPD1 to compete with VRN2 for these common NF-Y interactors, we cannot rule out the existence of additional competitive interactions among those proteins.
Protein interactions in planta
Since the interactions between CO and NF-Y proteins have already been validated in planta (Wenkel et al., 2006), in this study we only tested whether ZCCT1 protein interacts with NF-Y proteins in plant cells. Among the three NF-YB proteins NF-YB1, NF-YB3 and NF-YB9 we tested, we were able to confirm the interactions between ZCCT1 and NF-YB1 and NF-YB9 in Nicotiana benthamiana (Fig 1C). The failure to detect the NF-YB3 – ZCCT1 interaction in the co-immunoprecipitated sample (Fig 1C) can be explained either by the lower strength of this interaction compared with the ZCCT1 interactions with NF-YB1 and NF-YB9 (6–10 fold lower in the α-gal quantitative assays, Fig 2), or by the presence of a competing CO protein from N. benthamiana.
The CCT-NF-Y interaction in planta is also supported by their expression in the leaf vascular tissues. Although in Arabidopsis CO is expressed in several cell types, only its phloem-specific expression is sufficient to induce FT transcription (An et al., 2004). VRN2 has a similar spatial transcription profile in wheat to that of CO. VRN2 transcripts accumulate in the shoot apical meristem and in all tissues of young un-vernalized leaves (Kane et al., 2007). We confirmed the nuclear localization of VRN2 and its expression in wheat leaf mesophyll and phloem cells by in situ RT-PCR (A. Jedrzejuk, J. Jernstedt and J. Dubcovsky unpublished results). Promoter GUS fusion analyses for all 36 Arabidopsis NF-Y genes also show vascular expression patterns in leaves for 5 NF-YA, 4 NF-YB and 7 NF-YC genes including AtNF-YB2 and AtNF-YB3 (Siefers et al., 2009). In summary, the expression of these genes in the leaf vascular tissues may facilitate their interactions in planta, and might be critical for the regulation of FT, which travels through the phloem from the leaves to the apical meristem (Corbesier et al., 2007; Tamaki et al., 2007).
NF-Y protein interactions and flowering models
Genetic and transgenic experiments have shown that the transcriptional level of VRN3 (= FT) is up-regulated by PPD-1 and CO2 (Turner et al., 2005) and down-regulated by VRN2 (Yan et al., 2004; Hemming et al., 2008) under long days. Similarly, changes in the expression of Arabidopsis NF-YB2 and NF-YB3 genes trigger changes in the expression of FT and in flowering time only under long days (Kumimoto et al., 2008). Taken together, these results support our current combinatorial model in which the competition between the photoperiod (CO2) and vernalization (VRN2) proteins for common NF-YB interactors results in the integration of the seasonal cues in the transcriptional regulation of VRN3 and, thus, flowering time (Fig. 6).
Fig. 6. Combinatorial model of flowering.
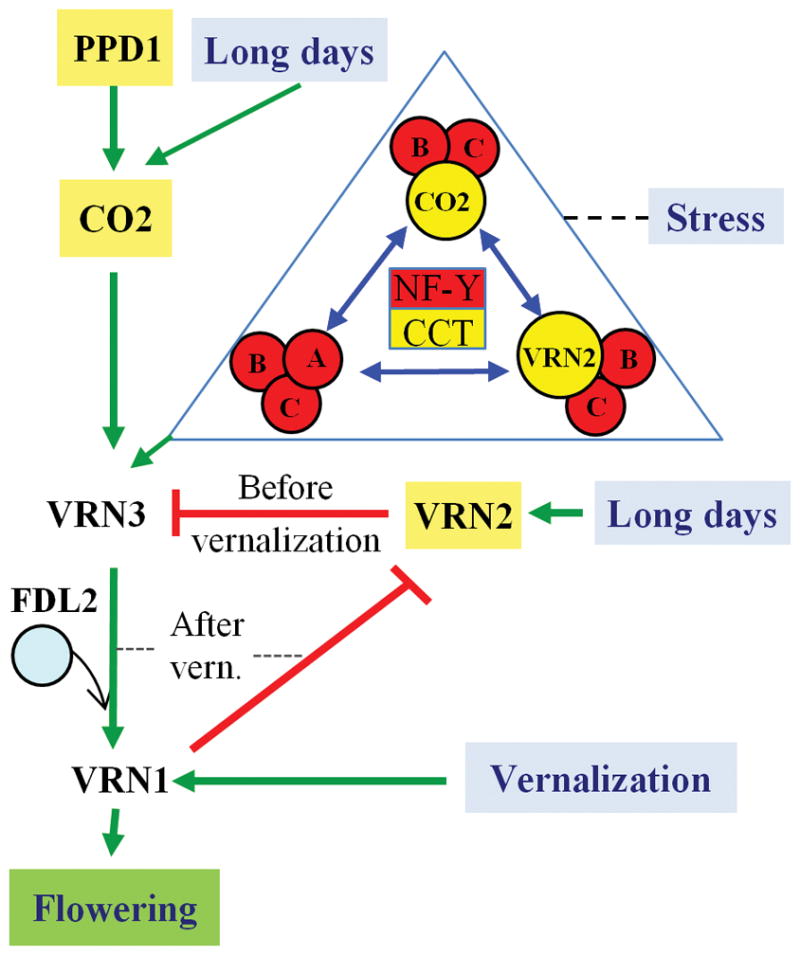
VRN3 integrates signals from the photoperiod (through CO2) and vernalization (through VRN2) pathways. Vernalization induces VRN1 transcription which is associated with the transcriptional repression of VRN2. VRN2 and CO2 compete with each other and with NF-YA subunits for interactions with NF-YB1, NF-YB3 and NF-YB9, generating complexes of different promoting or repressing strengths that converge in the transcriptional regulation of VRN3. VRN3 interacts with FDL2 to induce higher levels of VRN1 transcription and initiate flowering. The regulation of several wheat NF-Y genes by abiotic stresses provides a putative link between the stress and seasonal cues in the regulation of flowering. The expansion of CCT-domain and NF-Y proteins in plants generates a large number of possible combinations with the potential to accommodate the multiple environmental signals that converge in the regulation of flowering. Green arrows indicate promotion, red lines repression and blue double-head arrows competition. Yellow highlights indicate CCT domain proteins.
In contrast to wheat and Arabidopsis, rice is a short day plant, and under long days, CO (Hd1 in rice nomenclature) represses FT (Hd3a in rice nomenclature) and delays flowering (reviewed in Hayama and Coupland, 2004). Flowering under long days is also delayed by GHD7, the closest homologue of wheat VRN2 in rice (Yan et al., 2004; Xue et al., 2008). Therefore, GHD7 and CO have a redundant function as long day flowering repressors in rice, rather than a competitive one as observed in the temperate cereals. This redundant function might be also affected by NF-Y interactions, since DTH8 (OsHAP3H), an NF-YB gene related to wheat NF-YB3, reduces FT transcript levels and delays flowering under long days but not under short days (Wei et al., 2010).
In the temperate cereals the competing forces of the vernalization and photoperiod pathways in the regulation of flowering are reflected in the competition of VRN2 and CO2 proteins for the same set of NF-Y interactors. Since these two proteins interact with each other, it is also possible that one is sequestering the other from the NF-Y complexes. We were able to differentiate between the competition and sequestration hypotheses by using the ZCCT1-R35W mutation, which has very limited effect on the ZCCT1-CO2 interaction but reduces the strength of the interactions between ZCCT1 and different NF-Y subunits to different levels. In the Y3H assays using ZCCT1-R35W as the bridge protein, the strength of the CO2-NF-YB interactions was reduced proportionally to the strength of the ZCCT1-R35W -NF-YB interaction, suggesting that ZCCT1 competed with CO2 for the NF-Y subunits rather than sequestered CO2 from the NF-Y complex. We do not have a similar tool for the CCT-NF-YA interactions yet; therefore, we currently do not know if the ability of CO2 and ZCCT1 to reduce the strength of the NF-A/NF-YB interactions in the Y3H experiments (Fig. 5) is the result of direct competition or sequestration of NF-YA from the NF-Y complexes.
Another area that requires further research is the identification of the DNA binding sites of the different CCT and NF-Y complexes in wheat. In Arabidopsis, CO alone has been shown to bind a CO-responsive element (CORE, TGTG-N2-3-ATG), a motif duplicated in tandem in the FT promoter. CO-dependent FT expression is further enhanced by the presence of CCAAT sequences (Tiwari et al., 2010). In the FT promoters of wheat (e.g. GenBank DQ890162, DQ890164, and DQ890165) and barley (e.g. GenBank DQ898515 to DQ898520) we found 1 and 2 conserved TGTG-N3-ATG motifs respectively, approximately 1.5 kb up-stream from the translational initiation site, and 3-4 conserved CCAAT motifs further up-stream of the TGTG-N3-ATG motifs. The determination of the DNA binding sites of CCT proteins, NF-Y, and NF-Y-CCT complexes will require additional experiments.
Recent studies have shown that NF-Y complexes have the ability to interact with bZIP transcription factors bZIP67 (Yamamoto et al., 2009) and bZIP28 (Liu and Howell, 2010). Arabidopsis bZIP67 is seed-specific and has the ability to activate the CRC and SUS promoters through interactions with NF-Y proteins independent of the CCAAT motifs (Yamamoto et al., 2009). Arabidopsis bZIP28 transcription factor forms complexes with NF-Y proteins that bind to the endoplasmatic reticulum (ER) stress responsive element CCAAT-N10-CACG (ERS-I, Liu and Howell, 2010). None of the CCAAT boxes present in the barley and wheat FT promoter sequences mentioned above is part of a CCAAT-N10-CACG motif suggesting that either this mechanism is not relevant for the regulation of FT in the temperate cereals or that putative NF-Y/bZIP complexes in these species have slightly different recognition sequences.
Several NF-Y proteins are involved in plant responses to various environmental conditions, such as drought and osmotic stresses. For example, the Arabidopsis HAP3b (NF-YB2) gene is transcriptionally regulated by water stress (Nelson et al., 2007), and differences in flowering time between hap3b mutant and wild type alleles are larger under water stress than under water sufficient conditions (Chen et al., 2007). Arabidopsis NF-YA5 and nine wheat NF-Y genes have also been shown to be transcriptionally regulated by abiotic stresses (Stephenson et al., 2007; Li et al., 2008). The combinatorial model of flowering presented in Fig. 6 describes a flexible system in which the NF-Y – CCT interactions may provide a central hub to integrate the seasonal cues with multiple stress signals to fine-tune the regulation of flowering time.
Materials and Methods
Plant Materials and growing conditions
Triticum monococcum winter accession G3116 was used to clone NF-Y, ZCCT1, ZCCT2-R16C, CO1, CO2, and PPD1; spring accession DV92 was used to clone ZCCT1-R35W and ZCCT2-R16C+R39C; and tetraploid wheat Langdon was used to clone ZCCT1-R39C. All plants were grown in a greenhouse at 20°C–25°C under long-day photoperiod (8h of dark/ 16h of light) for 4 weeks before leaves were harvested for RNA extraction.
Yeast two-hybrid assays
Yeast vectors pGBKT7 (DNA binding domain, BD) and pGADT7 (activation domain, AD) and yeast strain AH109 were used in the two-hybrid assays (Clontech). Table S1 lists the primers used to generate yeast bait and prey constructs. The lithium acetate method was used for yeast transformation. Transformants were selected on SD medium lacking leucine (Leu) and tryptophan (Trp) plates and re-plated on SD medium lacking Leu, Trp, histidine (His) and adenine (Ade) to test the interactions.
Quantitative α-galactosidase assays
The yeast strain AH109 used in our two-hybrid assays is a MEL1 gene carrier, therefore, the αGal quantitative assay was used to measure the extracellular α-galactosidase (the product of MEL1 expression) activity produced during the culture growth. Cell populations containing different pairs of interacting proteins were grown to a density of 2~5 × 106 cells ml−1 in SD medium at 30 °C. Cells were pelleted using a microcentrifuge, 200 μl medium aliquot was mixed with 600 μl Assay Buffer (0.33 M sodium acetate pH 4.5; 33 mM p-nitrophenyl-α-D-galactopyranoside) and incubated at 30 °C for 12~24 hours. Reactions were stopped by the addition of 200 μl 2 M Na2CO3. Activity was calculated as the optical density at 410 nm (OD410).
Yeast three-hybrid assays
The pBridge yeast three-hybrid system (Clontech) was used to determine ternary protein interactions. The pBridge system allows the expression of two proteins: a DNA-binding fusion, and a second protein that positively or negatively affects the BD–AD interactions (the AD fusion protein is expressed in the pGADT7 vector). The second protein (Bridge protein) is conditionally expressed from the MET25 promoter in the absence of methioine (Met), and it is repressed in the presence of 1mM methionine. Gold Yeast strain with Aureobasidin A (AbA) reporter (Clontech) was used in the Y3H assays. All the prey constructs used in the Y3H assays were the same as in Y2H assays. Protein interactions were tested on SD-Leu-Trp + 125 ng/ml AbA (Bridge protein repressed by the methionine present in the growth medium) or SD-Leu-Trp-Met +125 ng/ml AbA (Bridge protein expressed). The quantitative α-galactosidase assay was used to compare protein interaction strength in the presence or absence of the Bridge protein.
Validation of interactions between ZCCT1 and NF-YB proteins using Nicotiana benthamiana transient expression system
Gateway entry vector pDONR/Zeo (Invitrogen) was used to generate entry constructs for ZCCT1, NF-YB1, NF-YB3 and NF-YB9 (primers in Table S4). ZCCT1 was then switched by recombination to gateway binary vector pGWB15, which has a 35S promoter and an N-terminal 3XHA tag. NF-YB1, NF-YB3 and NF-YB9 were switched to binary vector pGWB18, which has a 35S promoter and an N-terminal 4XMyc tag.
Constructs were transformed into Agrobacterium tumefaciens strain C58C1. Single Agrobacterium colonies containing each of the HA-ZCCT1, Myc-NF-YB1, Myc-NF-YB3 and Myc-NF-YB9 constructs were cultured at 28 °C for 16–24 h. Bacteria were pelleted and resuspended in infiltration media as described before (Popescu et al., 2007). (For co-infiltration, equal volumes of HA-ZCCT1 and one of the Myc-NF-YB cultures were mixed and infiltrated into the adaxial surface of Nicotiana benthamiana leaves using a 1 ml needleless syringe. The infiltrated leaf regions were collected 3 days after infiltration and stored at −80°C.
Leaf samples were ground in liquid nitrogen and total protein was extracted as described (Liu et al., 2010). One ml of protein extract was incubated with 50 μl of Anti-HA Affinity Matrix (Roche, prewashed twice with extraction buffer) at 4°C on a rocker for 2~ 4 hours to allow the co-precipitation of protein complexes. The Affinity Matrix was pelleted at 13,000 rpm in a microcentrifuge for 30s. The pelleted matrix was then washed with 1 ml of cold extraction buffer for a total of three times. SDS-PAGE sample buffer (50 μl) was added to the final washed matrix pellet and boiled for 5 min. The Affinity Matrix was pelleted again and the supernatant fraction (Co-IP sample) was transferred to a new tube for western blot analysis.
Co-IP samples were separated by SDS-PAGE with 12% acrylamide and transferred to HybondTM-P PVDF membrane (GE Healthcare). Anti-c-Myc monoclonal antibody (sc-40, Santa Cruz Biotechnology) was used to detect the presence of Myc-NF-YB proteins. The same membrane was then stripped in stripping buffer (2 % SDS, 100 mM beta-mercaptoethanol, 50 mM Tris PH 6.8) at 50 °C for 30 min, and re-hybridized with Anti-HA monoclonal antibody (HA.11, Covance).
Supplementary Material
S1. Generation of a monoclonal antibody for ZCCT1 protein
S2. Generation of a ZCCT2 protein carrying the wild type CCT domain by site-directed mutagenesis
S3. Validation of interactions between ZCCT1 and NF-Y proteins by in vitro Co-Immunoprecipitation (Co-IP)
Table S1. PCR Primers used for yeast two-hybrid constructs (restriction sites are underlined).
Table S2. Primers used to generate GST and MAP-tagged fusion proteins (restriction sites are underlined).
Table S3. Primers used in yeast three-hybrid constructs (restriction sites are underlined)
Table S4. Primers used in the generation of gateway constructs
Figure S1. Schematic diagrams of wheat ZCCT1, CO1 and CO2 protein regions used in the yeast two-hybrid constructs.
Figure S2. Yeast two-hybrid analysis of interactions between ZCCT1-CCT truncation and wheat NF-Y proteins.
Figure S3. Yeast two-hybrid analysis of interactions between yeast HAP (NF-Y) and wheat CCT-domain proteins.
Figure S4. Effect of mutations in Arginine (R) amino acids within the CCT domain on the strength of the interactions between ZCCT and CO.
Acknowledgments
We thank Dr. Peter Langridge of the Australian Centre for Plant Functional Genomics, University of Adelaide for his collaboration in the generation of the VRN2 monoclonal antibody. We thank Dr. Andrew Chen for the NF-YB3 gene sequences from diploid wheat and Meiyee Lau and Francine Paraiso for their excellent technical support. This project was supported by the National Research Initiative grants numbers 2007-35301-17737 and 2007-35301-18188 from the USDA National Institute of Food and Agriculture.
References
- An HL, Roussot C, Suarez-Lopez P, Corbesler L, Vincent C, Pineiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, Coupland G. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development. 2004;131:3615–3626. doi: 10.1242/dev.01231. [DOI] [PubMed] [Google Scholar]
- Ben-Naim O, Eshed R, Parnis A, Teper-Bamnolker P, Shalit A, Coupland G, Samach A, Lifschitz E. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 2006;46:462–476. doi: 10.1111/j.1365-313X.2006.02706.x. [DOI] [PubMed] [Google Scholar]
- Cai XN, Ballif J, Endo S, Davis E, Liang M, Chen D, DeWald D, Kreps J, Zhu T, Wu Y. A putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiol. 2007;145:98–105. doi: 10.1104/pp.107.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen NZ, Zhang XQ, Wei PC, Chen QJ, Ren F, Chen J, Wang XC. AtHAP3b plays a crucial role in the regulation of flowering time in Arabidopsis during osmotic stress. J Biochem Mol Biol. 2007;40:1083–1089. doi: 10.5483/bmbrep.2007.40.6.1083. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Distelfeld A, Tranquilli G, Li C, Yan L, Dubcovsky J. Genetic and molecular characterization of the VRN2 loci in tetraploid wheat. Plant Physiol. 2009a;149:245–257. doi: 10.1104/pp.108.129353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A, Li C, Dubcovsky J. Regulation of flowering in temperate cereals. Curr Opin Plant Biol. 2009b;12:178–184. doi: 10.1016/j.pbi.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Chen C, Yan L. Molecular characterization of the allelic variation at the VRN-H2 vernalization locus in barley. Mol Breed. 2005;15:395–407. [Google Scholar]
- Edwards D, Murray JAH, Smith AG. Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis. Plant Physiol. 1998;117:1015–1022. doi: 10.1104/pp.117.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S, Dunford RP, Coupland G, Laurie DA. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 2003;131:1855–1867. doi: 10.1104/pp.102.016188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Coupland G. The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Phys. 2004;135:677–684. doi: 10.1104/pp.104.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming MN, Fieg S, Peacock WJ, Dennis ES, Trevaskis B. Regions associated with repression of the barley (Hordeum vulgare) VERNALIZATION1 gene are not required for cold induction. Mol Gen Genomics. 2009;282:107–117. doi: 10.1007/s00438-009-0449-3. [DOI] [PubMed] [Google Scholar]
- Hemming MN, Peacock WJ, Dennis ES, Trevaskis B. Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiol. 2008;147:355–366. doi: 10.1104/pp.108.116418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane NA, Agharbaoui Z, Diallo AO, Adam H, Tominaga Y, Quellet F, Sarhan F. TaVRT2 represses transcription of the wheat vernalization gene VRN1. Plant J. 2007;51:670–680. doi: 10.1111/j.1365-313X.2007.03172.x. [DOI] [PubMed] [Google Scholar]
- Kumimoto RW, Adam L, Hymus GJ, Repetti PP, Reuber TL, Marion CM, Hempel FD, Ratcliffe OJ. The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta. 2008;228:709–723. doi: 10.1007/s00425-008-0773-6. [DOI] [PubMed] [Google Scholar]
- Kumimoto RW, Zhang Y, Siefers N, Holt BF. NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 2010;63:379–391. doi: 10.1111/j.1365-313X.2010.04247.x. [DOI] [PubMed] [Google Scholar]
- Li C, Dubcovsky J. Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 2008;55:543–554. doi: 10.1111/j.1365-313X.2008.03526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell. 2008;20:2238–2251. doi: 10.1105/tpc.108.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell. 2010;22:782–796. doi: 10.1105/tpc.109.072173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J. Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiol. 2005;138:2364–2373. doi: 10.1104/pp.105.064287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. Development of Series of Gateway Binary Vectors, pGWBs, for Realizing Efficient Construction of Fusion Genes for Plant Transformation. J Biosci Boeng. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J, Warner DC, Anstrom DC, Bensen RJ, Castiglioni PP, Donnarummo MG, Hinchey BS, Kumimoto RW, Maszle DR, Canales RD, Krolikowski KA, Dotson SB, Gutterson N, Ratcliffe OJ, Heard JE. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci USA. 2007;104:16450–16455. doi: 10.1073/pnas.0707193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto Y, Kisaka M, Fuse T, Yano M, Ogihara Y. Characterization and functional analysis of three wheat genes with homology to the CONSTANS flowering time gene in transgenic rice. Plant J. 2003;36:82–93. doi: 10.1046/j.1365-313x.2003.01859.x. [DOI] [PubMed] [Google Scholar]
- Oliver SN, Finnegan EJ, Dennis ES, Peacock WJ, Trevaskis B. Vernalization-induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene. Proc Natl Acad Sci USA. 2009;106:8386–91. doi: 10.1073/pnas.0903566106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefers N, Dang KK, Kumimoto RW, Bynum WE, Tayrose G, Holt BF. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009;149:625–641. doi: 10.1104/pp.108.130591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson TJ, McIntyre CL, Collet C, Xue GP. Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol Biol. 2007;65:77–92. doi: 10.1007/s11103-007-9200-9. [DOI] [PubMed] [Google Scholar]
- Stephenson TJ, McIntyre CL, Collet C, Xue GP. TaNF-YC11, one of the light-upregulated NF-YC members in Triticum aestivum, is co-regulated with photosynthesis-related genes. Funct Integr Genomics. 2010;10:265–276. doi: 10.1007/s10142-010-0158-3. [DOI] [PubMed] [Google Scholar]
- Stephenson TJ, McIntyre CL, Collet C, Xue GP. TaNF-YB3 is involved in the regulation of photosynthesis genes in Triticum aestivum. Funct Integr Genomics. 2011 doi: 10.1007/s10142-011-0212-9. [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Shen Y, Chang HC, Hou YL, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C, Belachew A, Repetti PP, Reuber TL, Ratcliffe OJ. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010;187:57–66. doi: 10.1111/j.1469-8137.2010.03251.x. [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Dennis ES, Peacock WJ. The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 2007;12:352–357. doi: 10.1016/j.tplants.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310:1031–1034. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- Wei X, Xu J, Guo H, Jiang L, Chen S, Yu C, Zhou Z, Hu P, Zhai H, Wan J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010;153:1747–1758. doi: 10.1104/pp.110.156943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. The Plant Cell. 2006;18:2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue WY, Xing YZ, Weng XY, Zhao Y, Tang WJ, Wang L, Zhou HJ, Yu SB, Xu CG, Li XH, Zhang QF. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Kagaya Y, Toyoshima R, Kagaya M, Takeda S, Hattori T. Arabidopsis NF-YB subunits LEC1 and LEC1-like activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 2009;58:843–856. doi: 10.1111/j.1365-313X.2009.03817.x. [DOI] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of wheat vernalization gene VRN1. Proc Natl Acad Sci USA. 2003;100:6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004;303:1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Dubcovsky J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA. 2006;103:19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Xie ZY, Glover BJ. Asymmetric evolution of duplicate genes encoding the CCAAT-binding factor NF-Y in plant genomes. New Phytol. 2005;165:623–631. doi: 10.1111/j.1469-8137.2004.01260.x. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Nishida H, Zhu J, Nitcher R, Distelfeld A, Akashi Y, Kato K, Dubcovsky J. Vrn-D4 is a vernalization gene located on the centromeric region of chromosome 5D in hexaploid wheat. Theor Appl Genet. 2010;120:543–552. doi: 10.1007/s00122-009-1174-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1. Generation of a monoclonal antibody for ZCCT1 protein
S2. Generation of a ZCCT2 protein carrying the wild type CCT domain by site-directed mutagenesis
S3. Validation of interactions between ZCCT1 and NF-Y proteins by in vitro Co-Immunoprecipitation (Co-IP)
Table S1. PCR Primers used for yeast two-hybrid constructs (restriction sites are underlined).
Table S2. Primers used to generate GST and MAP-tagged fusion proteins (restriction sites are underlined).
Table S3. Primers used in yeast three-hybrid constructs (restriction sites are underlined)
Table S4. Primers used in the generation of gateway constructs
Figure S1. Schematic diagrams of wheat ZCCT1, CO1 and CO2 protein regions used in the yeast two-hybrid constructs.
Figure S2. Yeast two-hybrid analysis of interactions between ZCCT1-CCT truncation and wheat NF-Y proteins.
Figure S3. Yeast two-hybrid analysis of interactions between yeast HAP (NF-Y) and wheat CCT-domain proteins.
Figure S4. Effect of mutations in Arginine (R) amino acids within the CCT domain on the strength of the interactions between ZCCT and CO.