Abstract
Hydroxamate-based histone deacetylase inhibitors (HDACIs) have been approved as therapeutic agents by the US Food and Drug Administration for use in oncology applications. While the potential utility of such HDACIs in other areas of medicinal chemistry is tremendous, there are significant concerns that “pan-HDAC inhibitors” may be too broadly acting and/or toxic for clinical use beyond oncology. In addition to the isozyme selectivity challenge, the potential mutagenicity of hydroxamate-containing HDAC inhibitors represents a major hindrance in their application to other therapeutic areas. Herein we report on the mutagenicity of known hydroxamates, discuss the mechanisms responsible for their genotoxicity, and review some of the current alternatives to hydroxamates. We conclude that the hydroxamate group, while providing high-potency HDACIs, is not necessarily the best zinc-binding group for HDACI drug discovery.
Keywords: histone deacetylase inhibitors, hydroxamate, Lossen rearrangement, mutagenicity
Introduction
Hydroxamates are a class of organic compounds containing the functional group C(O)-N(R)-OH. Their carbonyl and N-hydroxy groups are capable of complexing metals in a bidentate fashion, making them excellent ligands for a number of metals of biological importance including zinc, iron, and nickel. Among those metals, zinc is one of the most frequently occurring metals in metalloenzymes (>300 enzymes).[1] In particular, zinc-dependent endopeptidases, matrix metalloproteases (MMPs), and histone deacetylases (HDACs), which play significant roles in the growth, promotion, and spread of tumors, have been recognized as potential targets for cancer therapy in past decades. Hundreds of inhibitors against MMPs and HDACs have been or are being developed, and many of them contain a hydroxamate group as the zinc-binding group (ZBG). More than 50 MMP inhibitors have been investigated in clinical trials; however, all of them failed for reasons that include inadequate trial design, poor ADMET properties (metabolic instability, low oral bioavailability, and toxicity), and limited knowledge of the complexity of MMP function.[2]
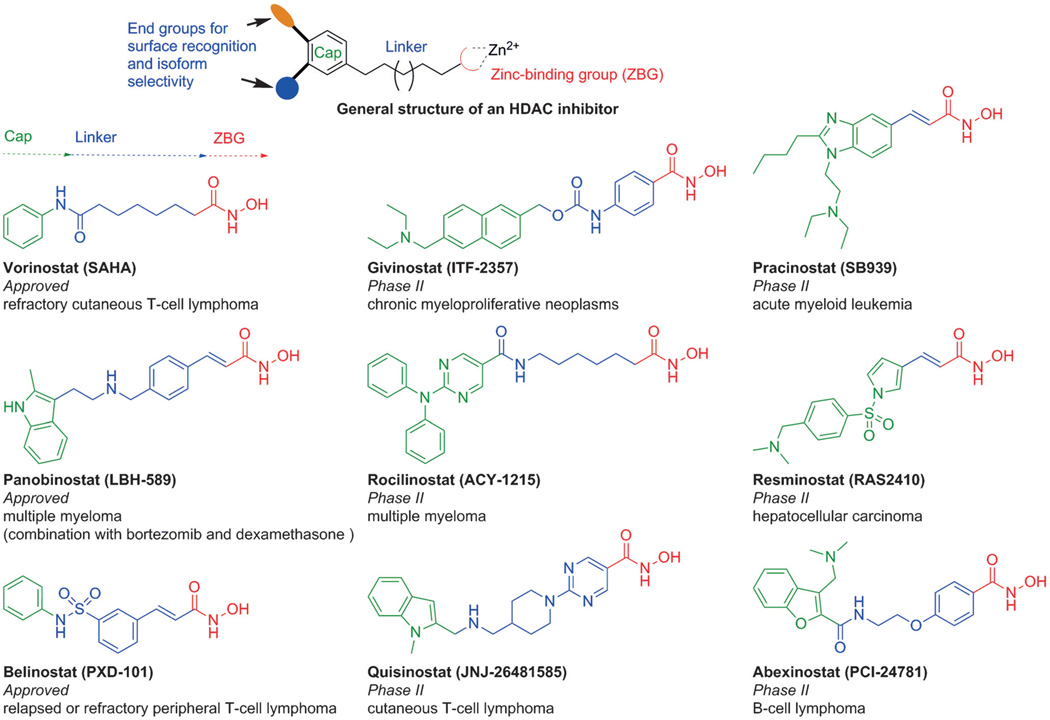
Unlike the story of MMP inhibitor discovery, three broad-spectrum HDAC inhibitors (HDACIs) bearing a hydroxamic acid function (vorinostat, panobinostat, and belinostat) have been approved as drugs, even though they still exhibit poor pharmacokinetics (rapid clearance) and selectivity profiles (multiple off-target interactions, including the hERG cardiac ion channel), and give rise to dose-limiting toxicity concerns.[3] In the meantime, more than ten hydroxamate-based pan-HDACIs are under clinical investigation for the treatment of various cancers (Figure 1).[4] Outside oncology, the targeting of specific HDAC isoforms is being pursued as a promising therapeutic strategy for neurological disorders,[5] based on the finding that several selective HDACIs bearing hydroxamate groups exhibit neuroprotective properties and improve cognition and motor activity, as well as other behaviors in animal models of depression,[6] Alzheimer’s disease (AD),[7] Huntington’s disease (HD),[8] Charcot– Marie–Tooth disease (CMT), etc.[9] The potential utility of HDAC inhibitors appears to be tremendous, but further translation of these ideas to the clinic will ultimately require the design of potent, isozyme-selective, and drug-like molecules that have minimal side effects. In general, HDACIs comprise three main motifs: a cap group that interacts with the surface of the enzyme, a linker that occupies a hydrophobic channel, and a metal chelator that coordinates with the zinc ion at the bottom of the catalytic pocket (Figure 1). A properly optimized cap group can improve both potency and selectivity, presumably through its ability to engage in appropriate contacts with residues on the enzyme surface. There is growing interest in developing HDAC-specific blockers for clinical use beyond oncology; nevertheless, significant concern is now emerging that the potential mutagenicity of the hydroxamate group would present an obstacle.
Figure 1.
General structure of HDACIs and structures of therapeutically relevant HDACIs.
Mutagenicity
Mutagenic chemicals may exert their effect through substitutions of nucleotides in DNA as well as insertion or deletion of nucleotides, chromosomal breakages, and the rearrangement of chromosomes. Because of these mechanisms of action, mutagens are also potential carcinogens, and this concern has driven most of the mutagenicity testing programs. The hydroxamate group and its derivatives were found to be mutagenic as early as three decades ago.[10] Ames tests indicated that some alkyl and aryl hydroxamic acids lead to obvious mutation in strains of the bacterium Salmonella typhimurium.[11] O-Acetyl-benzohydroxamate, a mutagen, was observed to form adducts with DNA and polyguanylic acid in vitro.[12] Moreover, some hydroxamic acid–metal complexes show effective capacity for DNA cleavage.[13] These interactions with DNA may cause DNA damage, mutation, and cancer development. There is substantial evidence to demonstrate that the HDAC tool compound trichostatin A (TSA), which shares structural similarity with vorinostat, shows mutagenicity in vitro. In 2006, it was reported that DNA damage is induced in leukemic cells in response to treatment with TSA.[14] In the same year, both clastogenic and aneugenic properties of TSA in human TK6 lymphoblastoid cells were reported, which indicated that exposure to TSA causes chromosomal aberrations, DNA damage, and aneuploidy in this cell line.[15] In 2013, a positive genotoxicity result for TSA was obtained within the acceptable toxic dose range in the TK6 cell-line-hosted GADD45a–GFP assay, which has high sensitivity and specificity in the detection of genotoxic carcinogens and in vivo genotoxicants. This result also suggested that the genotoxicity of TSA may contribute to its antitumor activity.[16]
In many cases it is difficult to locate information on the mutagenic properties of HDAC drug candidates containing a hydroxamate group that are being advanced to clinical trials. However, detailed genotoxic assessments for approved HDACIs have been described in the pharmacology review materials submitted to the US Food and Drug Administration (FDA) (Table 1).[17] Vorinostat is mutagenic in the Ames test; is clastogenic in rodents (i.e., Chinese hamster ovary (CHO)) cells, but not in normal human lymphocytes; and is positive in an in vivo mouse micronucleus assay.[18] Both belinostat and panobinostat are mutagenic in the Ames test and L5178Y mouse lymphoma cells. Both panobinostat and vorinostat are negative for chromosomal aberrations in cultured purified human blood lymphocytes; however, in the case of panobinostat, an increased frequency of endoreduplication was observed, indicating an increase in the number of chromosomes. All hydroxamate HDACIs on the market are Ames positive and cause chromosomal aberrations in rodent cells. These drugs are being used only for cancer, as this undesired side effect can, to a certain extent, be tolerated in a disease considered to be life threatening. Certainly, for use in diseases that would require chronic, longer-term use of an HDACI, it would be preferable to have inhibitors available that are not genotoxic.
Table 1.
Mutagenicity assay results of approved hydroxamate-based HDAC inhibitors.[a]
| Mutagenicity Assays | Vorinostat | Belinostat | Panobinostat |
|---|---|---|---|
| In vitro reverse mutation assay in bacteria (Ames test) | Positive (S. typhimurium TA97a, TA100, TA989, TA1535, TA1537, TA1538) | Positive (TA98, TA100, TA102, TA1535, TA1537) | Positive (TA97a) |
| In vitro assay in mammalian cells (chromosomal aberrations) |
Mutagenic in Chinese hamster ovary (CHO) cells Non-clastogenic in cultured purified human blood lymphocytes Marked suppression of DNA synthesis in CHO cells |
Mutagenic in L5178Y mouse lymphoma cells |
Negative for chromosomal aberrations in cultured purified human blood lymphocytes, but an increased frequency of endoreduplication was observed, indicating an increase in the number of chromosomes Positive for DNA damage in L5178Y mouse lymphoma cells |
| In vivo clastogenicity assay in rodents (micronucleus assay) | Clastogenic, 2–3-fold increase in micronucleated polychromatic erythrocytes (PCEs) in mouse bone marrow | Clastogenic, induces a significant increase in micronucleated PCEs at nontoxic dose of 260 mg kg−1 day −1 | Not performed |
All data were obtained from the pharmacology review materials for the Center for Drug Evaluation and Research (CDER), US Food and Drug Administration (FDA).
Proposed mutagenicity mechanism
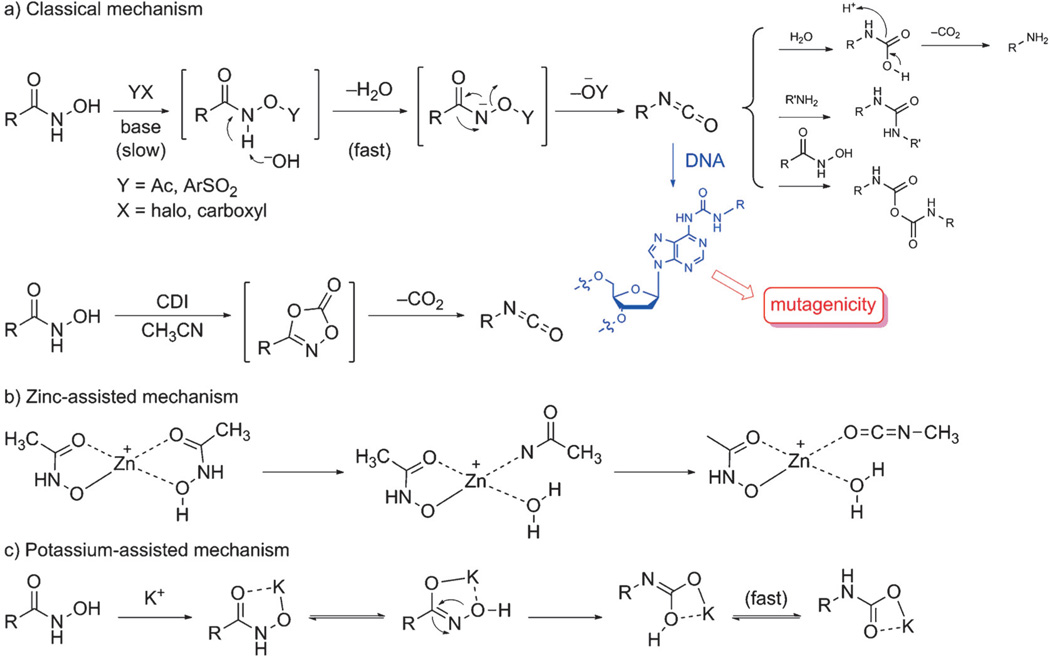
According to the current hypothesis, hydroxamate mutagenicity is caused by a Lossen rearrangement, a reaction which transforms an activated hydroxamate into the corresponding isocyanate.[19] In the laboratory, the Lossen rearrangement is carried out by using preformed O-activated hydroxamic acid derivatives or by in situ activation of free hydroxamic acids with various activating agents such as carbonyldiimidazole (CDI) (Scheme 1a).[20] The O-activated hydroxamates undergo deprotonation of the amide nitrogen atom under basic conditions, and then the anionic intermediates rearrange rapidly to the isocyanates. The isocyanates are unstable, and they easily undergo hydrolysis or react with available nucleophiles. Moreover, aliphatic and aromatic isocyanates are known to react readily with imidazole groups within enzymes to form ureatype adducts which are susceptible to hydrolysis or can be attacked by nucleophilic species other than water, for example, nucleophilic groups residing on DNA (Scheme 1a).[21]
Scheme 1.
Proposed Lossen rearrangement mechanisms for hydroxamate anions.
The occurrence of the Lossen rearrangement under physiological conditions has been reported.[19] Thus it has been shown that 2-naphthohydroxamic acid can be converted into its O-acetyl derivative by acetyl-CoA present in bacteria. This process is a key step in priming a hydroxamic acid for the Lossen rearrangement. The presence of the isocyanate intermediate derived from the acetylated hydroxamate has been confirmed by trapping with aniline present in the reaction buffer, resulting in the formation of urea. Moreover, it has been shown that incubation of 2-naphthohydroxamic acid as well as its O-acetyl derivative with TA98 cell suspensions results in the formation of adducts with bacterial DNA. Furthermore, it was reported that 2-aminonaphthalene formed by way of the Lossen rearrangement with subsequent hydrolysis and decarboxylation was the major metabolite in the presence of the TA98 tester strain. The amide metabolite, naphthalene-2-carboxamide, was also detected, which was formed by reduction of the O-acetylated hydroxamate under catalysis by NADH/NADPH. Only small amounts of the corresponding carboxylic acid were detected in the presence of bacteria TA98 at each time point. Therefore, the amount of the concomitantly released hydroxylamine, which is also a mutagen, is likely insufficient to cause the observed mutagenicity.[10a]
Of further note is the observation that 2-aminonaphthalene can be generated from its precursor, O-acetyl-2-naphthohydroxamic acid, in the absence of bacteria, indicating that the Lossen rearrangement proceeds in the absence of any enzyme catalysis. These results taken in total would strongly support the idea that the reactive isocyanates stemming from the Lossen rearrangement are the cause of the mutagenicity of hydroxamates. The rate-limiting step in this variant of the Lossen rearrangement is the formation of an activated hydroxamate, and the rate of the reaction is directly proportional to the relative acidity of the conjugate acid of the anionic leaving group.[22]
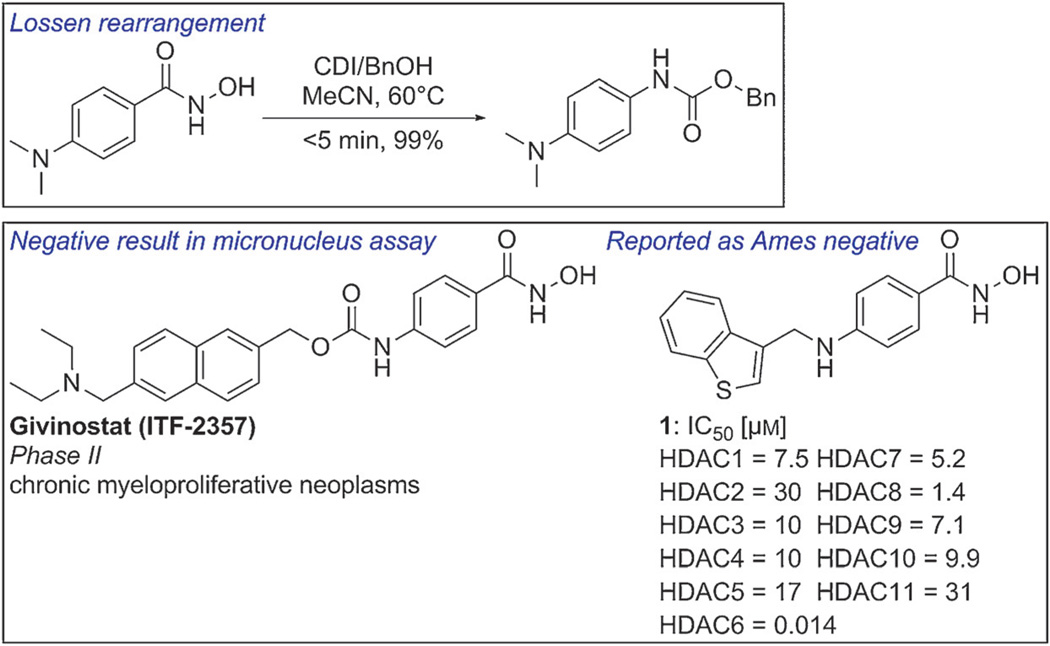
Most reported Lossen rearrangements take place in an alkaline medium, in the presence of a catalyst, or under neutral conditions. However, these observations are made under standard reaction conditions used in synthetic chemistry, and Lossen rearrangements in a biological environment do not necessarily proceed by the same mechanism, and may in fact exhibit reactivity patterns different from those observed in a synthesis context. For instance, 4-(dimethylamino)benzohydroxamic acid is a very active substrate for the Lossen rearrangement. The reaction is complete within 5 min in 99% yield under conditions using CDI/CH3CN.[20b] However, the Phase II compound givinostat, bearing a structurally related moiety, has been reported to have no genotoxic effects in the mouse lymphoma assay and the chromosomal aberration assay in vitro, and in the micronucleus test and unscheduled DNA synthesis test in vivo.[23] Moreover, another 4-aminobenzohydroxamic acid derivative incorporating a benzothiophene ring was found to be Ames negative when tested using four strains of S. typhimurium (TA98, TA100, TA1535, and TA1537; Figure 2, compound 1).[24] However, as cytotoxicity was observed at concentrations of >50 µm in all the tester strains, this result may obscure the genotoxicity of the compound, and further evaluation of the mutagenicity of compound 1 is certainly warranted, and in particular would require study in the in vitro micronucleus test for chromosomal damage. It is clear that our present knowledge does not provide a definitive answer as to whether a Lossen rearrangement will or will not occur in a biological system. The exception to this statement would be the case in which the nitrogen atom bears an additional substituent. It is also possible that simple hydrolysis of the hydroxamate with formation of hydroxylamine, a mutagen, may compete with the Lossen reaction.
Figure 2.
Sample Lossen rearrangement and structures for HDACIs containing a 4-amino-N-hydroxybenzamide moiety.
Hydroxamates exhibit much more efficient binding to zinc-containing enzymes than compounds containing other zinc-complexing groups such as carboxylic or phosphinic acids. The low acidity of hydroxamates (pKa values ~8.5) suggests that they are not appreciably deprotonated at physiological pH. They therefore approach the active sites as neutral molecules,[25] and deprotonation to form a strongly chelating hydroxamate anion takes place only after coordination to the zinc cation.[26] Metal-assisted mechanisms for the Lossen rearrangement, which are subject to only a very small energy barrier, should therefore be taken into consideration. In an example of the zinc-triggered Lossen rearrangement, two acetohydroxamic acid molecules bond with zinc as neutral or anionic ligands. Hydrogen migration from the nitrogen atom to the hydroxy group leads to the formation of an unsaturated complex in which zinc bears anionic acetohydroxamate, water, and acetylnitrene as ligands. The nitrene can undergo rearrangement to yield the more stable methylisocyanate (Scheme 1b).[27] Recently, it has been observed that deprotonation occurred at the oxygen atom for the zinc complex, instead of at the nitrogen atom as in the potassium complex. The potassium-mediated Lossen rearrangement is initiated by hydrogen migration from the nitrogen atom to the oxygen atom to form an N-deprotonated hydroxamate–metal complex, which is of similar energy as the original complex.[28] Next, the metal hydroxamate undergoes Lossen rearrangement, followed by facile hydrogen migration from oxygen to nitrogen, to yield a metal carbamate (Scheme 1c). Although these metal-assisted Lossen rearrangements may provide another route for the transformation of hydroxamates in biological systems in addition to the acetylase-initiated pathway, further studies to assess their actual contribution in biological systems may be warranted.
Alternatives to Hydroxamates for HDACIs
The best strategy to circumvent mutagenicity issues related to the hydroxamate function in HDACIs is to develop alternatives to hydroxamate as ZBGs. To date, alternative ZBGs present in the most advanced HDACIs that are either in clinical use or under investigation are carboxylic acid (i.e., valproic acid), ortho-aminoanilide (i.e., chidamide, entinostat, mocetinostat, tacedinaline, etc.), and thiol (i.e., romidepsin).
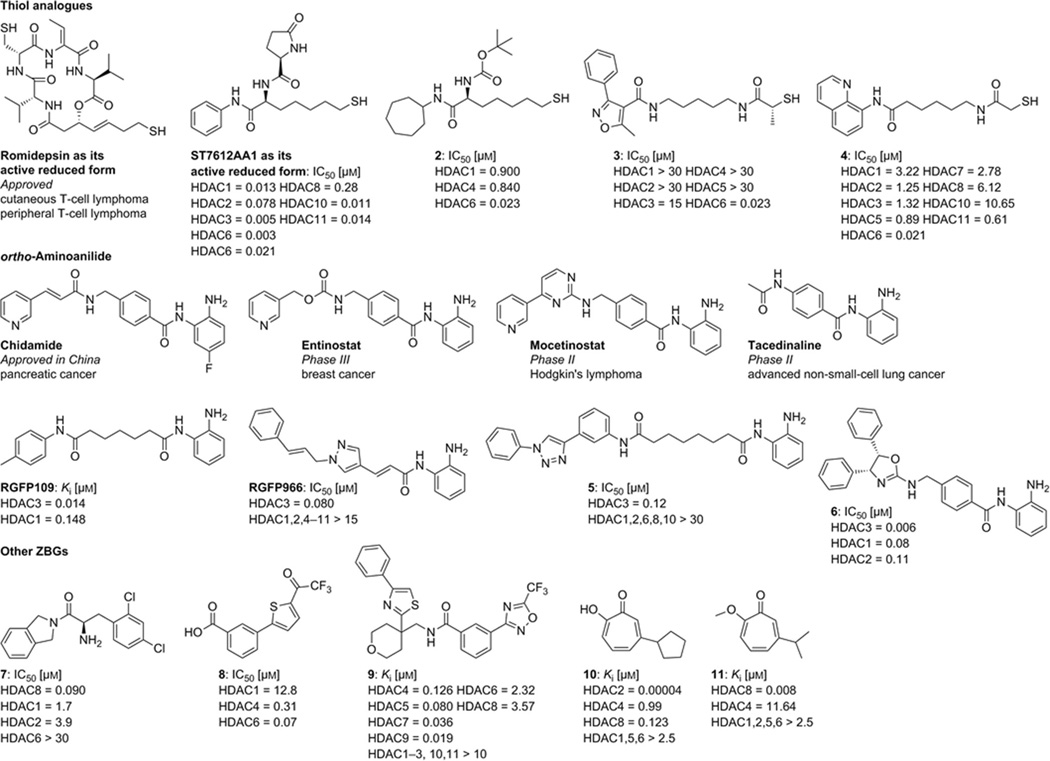
The prototypical example for a thiol ZBG is the natural product romidepsin, a cyclic peptide that has been approved for the treatment of cutaneous and peripheral T-cell lymphoma. Romidepsin is a prodrug whose disulfide bond can be reduced within the cellular environment to release the free thiol.[29] It has been shown to be non-mutagenic in the Ames test as well as in the mouse lymphoma assay in vitro. Moreover, romidepsin is non-clastogenic in an in vivo rat bone marrow micronucleus assay when tested at the maximum tolerated dose.[30] Recently, a preclinical study of ST7612AA1, a thioacetate prodrug, confirmed that the prodrug is rapidly absorbed and converted into the active thiol form after administration of a single oral dose to mice.[31] The thiol group thus represents a relatively potent zinc-chelating group, and various thiol-based HDACIs and their thioate prodrugs have been widely investigated over the last decade.[32] Notably, one thiol-based compound bearing a cycloheptyl cap group (Figure 3, 2)[33] and several mercaptoacetamides (Figure 3, 3 and 4)[34] were found to display potent HDAC6-selective inhibition in enzyme-and cell-based assays. Moreover, the neuroprotective capacity of mercaptoacetamide-based HDAC6Is (Figure 3, 3 and 4) has been demonstrated in primary cortical neurons and in a mouse model of AD.[5f, 34a, 35] However, it is important to note that some mutagenic potential has been found for thiol-containing compounds such as glutathione (Ames positive even at concentrations found in mammalian tissues), cysteine ethyl ester, and l- and d-penicillamine.[36] Accordingly, the mutagenic safety of compounds bearing a thiol group as an alternative ZBG should be investigated.
Figure 3.
HDACIs with thiol, aminoanilide, and atypical zinc-binding groups.
As noted above, the ortho-aminoanilides are an important class of HDACIs in clinical trials and include compounds such as chidamide (approved in China for the treatment of pancreatic cancer), entinostat (Phase III), mocetinostat (Phase II), and tacedinaline (Phase II). Most of the ortho-aminoanilide-based HDACIs display selective inhibition of the class I HDAC isoforms in addition to HDAC8. Several ortho-aminoanilides possessing alkyl or alkenyl linkers (Figure 3, RGFP109, RGFP966, and compound 5) or a chiral oxazoline cap (Figure 3, compound 6) were found to be HDAC3 selective.[37]
Over the years, novel zinc-binding motifs have emerged and could provide complementary selectivity profiles. Compound 7 containing an amino acid amide as the ZBG showed preferential inhibition of HDAC8.[38] Compound 8, processing a trifluoroacetylthiophene ZBG, shows selectivity against HDAC4/6.[39] A class IIa-selective HDACI bearing a trifluoromethyloxadiazole moiety as a unique chelating ZBG was identified by screening approximately two million individual compounds from GlaxoSmithKline’s diversity collection, and it exhibited good potency for the class IIa HDACs 4, 5, 7, and 9 (Figure 3, compound 9).[40] The β-thujaplicin analogue 10, containing a tropolone as the ZBG, is the first discovered highly potent and selective HDAC2 inhibitor. Interestingly, β-thujaplicin methyl ether (11) has been shown to be a highly potent and selective HDAC8 inhibitor.[41]
Perspectives
Despite the numerous clinical trials in progress using hydroxamate-bearing HDACI, it is difficult to gain ready access to the mutagenicity data that are likely present for many of these compounds. However, the available data reviewed in this paper suggest that a number of HDACIs bearing hydroxamates do indeed exert mutagenic effects. Nonetheless, hydroxamates still attract much attention in HDACI drug discovery because of their remarkable zinc chelating capability, isoform selectivity, and efficacy in vivo. The tremendous potential of HDACIs for the treatment of neurological disorders can be expected to drive further investigations into the mechanism of hydroxamate-induced mutagenicity and of Lossen rearrangements in biological environments, as there may be opportunities to identify non-mutagenic hydroxamate-based HDACIs. Moreover, non-hydroxamate zinc-binding groups represent a solid alternative for developing non-mutagenic HDACIs, and we anticipate that screening methods like those used in the discovery of compound 9 will lead to further breakthroughs in this area.
Acknowledgments
The authors gratefully acknowledge Dr. Werner Tueckmantel for reviewing the article and providing comments. We are indebted to the US National Institutes of Health (NS079183) for their support of our HDAC program.
References
- 1.McCall KA, Huang C, Fierke CA. J. Nutr. 2000;130:1437S–1446S. doi: 10.1093/jn/130.5.1437S. [DOI] [PubMed] [Google Scholar]
- 2.Vandenbroucke RE, Libert C. Nat. Rev. Drug Discovery. 2014;13:904–927. doi: 10.1038/nrd4390. [DOI] [PubMed] [Google Scholar]
- 3.a) Xu S, De Veirman K, Vanderkerken K, Van Riet I. Bone. 2013;57:384–385. doi: 10.1016/j.bone.2013.08.024. [DOI] [PubMed] [Google Scholar]; b) Giavini E. Birth Defects Res. Part B. 2007;80:417–418. doi: 10.1002/bdrb.20128. [DOI] [PubMed] [Google Scholar]; c) Wise LD, Turner KJ, Kerr JS. Birth Defects Res. Part B. 2007;80:57–68. doi: 10.1002/bdrb.20104. [DOI] [PubMed] [Google Scholar]; d) Doi T, Hamaguchi T, Shirao K, Chin K, Hatake K, Noguchi K, Otsuki T, Mehta A, Ohtsu A. Int. J. Clin. Oncol. 2013;18:87–95. doi: 10.1007/s10147-011-0348-6. [DOI] [PubMed] [Google Scholar]; e) Hamberg P, Woo MM, Chen LC, Verweij J, Porro MG, Zhao L, Li W, van der Biessen D, Sharma S, Hengelage T, de Jonge M. Cancer Chemother. Pharmacol. 2011;68:805–813. doi: 10.1007/s00280-011-1693-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Savelieva M, Woo MM, Schran H, Mu S, Nedelman J, Capdeville R. Eur. J. Clin. Pharmacol. 2015;71:663–672. doi: 10.1007/s00228-015-1846-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Warren KE, McCully C, Dvinge H, Tjornelund J, Sehested M, Lichenstein HS, Balis FM. Cancer Chemother. Pharmacol. 2008;62:433–437. doi: 10.1007/s00280-007-0622-5. [DOI] [PubMed] [Google Scholar]; h) Goey AK, Sissung TM, Peer CJ, Trepel JB, Lee MJ, Tomita Y, Ehrlich S, Bryla C, Balasubramaniam S, Piekarz R, Steinberg SM, Bates SE, Figg WD. J. Clin. Pharmacol. 2015 doi: 10.1002/jcph.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Finazzi G, Vannucchi AM, Martinelli V, Ruggeri M, Nobile F, Specchia G, Pogliani EM, Olimpieri OM, Fioritoni G, Musolino C, Cilloni D, Sivera P, Barosi G, Finazzi MC, Di Tollo S, Demuth T, Barbui T, Rambaldi A. Br. J. Haematol. 2013;161:688–694. doi: 10.1111/bjh.12332. [DOI] [PubMed] [Google Scholar]; b) Santo L, Hideshima T, Kung AL, Tseng JC, Tamang D, Yang M, Jarpe M, van Duzer JH, Mazitschek R, Ogier WC, Cirstea D, Rodig S, Eda H, Scullen T, Canavese M, Bradner J, Anderson KC, Jones SS, Raje N. Blood. 2012;119:2579–2589. doi: 10.1182/blood-2011-10-387365. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Venugopal B, Baird R, Kristeleit RS, Plummer R, Cowan R, Stewart A, Fourneau N, Hellemans P, Elsayed Y, McClue S, Smit JW, Forslund A, Phelps C, Camm J, Evans TR, de Bono JS, Banerji U. Clin. Cancer Res. 2013;19:4262–4272. doi: 10.1158/1078-0432.CCR-13-0312. [DOI] [PubMed] [Google Scholar]; d) Zorzi AP, Bernstein M, Samson Y, Wall DA, Desai S, Nicksy D, Wainman N, Eisenhauer E, Baruchel S. Pediatr. Blood Cancer. 2013;60:1868–1874. doi: 10.1002/pbc.24694. [DOI] [PubMed] [Google Scholar]; e) Kitazono S, Fujiwara Y, Nakamichi S, Mizugaki H, Nokihara H, Yamamoto N, Yamada Y, Inukai E, Nakamura O, Tamura T. Cancer Chemother. Pharmacol. 2015;75:1155–1161. doi: 10.1007/s00280-015-2741-8. [DOI] [PubMed] [Google Scholar]; f) Choy E, Flamand Y, Balasubramanian S, Butrynski JE, Harmon DC, George S, Cote GM, Wagner AJ, Morgan JA, Sirisawad M, Mani C, Hornicek FJ, Duan Z, Demetri GD. Cancer. 2015;121:1223–1230. doi: 10.1002/cncr.29175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Rumbaugh G, Sillivan SE, Ozkan ED, Rojas CS, Hubbs CR, Aceti M, Kilgore M, Kudugunti S, Puthanveettil SV, Sweatt JD, Rusche J, Miller CA. Neuropsychopharmacology. 2015;40:2307–2316. doi: 10.1038/npp.2015.93. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Govindarajan N, Rao P, Burkhardt S, Sananbenesi F, Schluter OM, Bradke F, Lu J, Fischer A. EMBO Mol. Med. 2013;5:52–63. doi: 10.1002/emmm.201201923. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Fischer A, Sananbenesi F, Mungenast A, Tsai LH. Trends Pharmacol. Sci. 2010;31:605–617. doi: 10.1016/j.tips.2010.09.003. [DOI] [PubMed] [Google Scholar]; d) Bang SR, Ambavade SD, Jagdale PG, Adkar PP, Waghmare AB, Ambavade PD. Pharmacol. Biochem. Behav. 2015;134:65–69. doi: 10.1016/j.pbb.2015.04.011. [DOI] [PubMed] [Google Scholar]; e) Sen A, Nelson TJ, Alkon DL. J. Neurosci. 2015;35:7538–7551. doi: 10.1523/JNEUROSCI.0260-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Sung YM, Lee T, Yoon H, DiBattista AM, Song JM, Sohn Y, Moffat EI, Turner RS, Jung M, Kim J, Hoe HS. Exp. Neurol. 2013;239:192–201. doi: 10.1016/j.expneurol.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jochems J, Boulden J, Lee BG, Blendy JA, Jarpe M, Mazitschek R, Van Duzer JH, Jones S, Berton O. Neuropsychopharmacology. 2014;39:389–400. doi: 10.1038/npp.2013.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Selenica ML, Benner L, Housley SB, Manchec B, Lee DC, Nash KR, Kalin J, Bergman JA, Kozikowski A, Gordon MN, Morgan D. Alzheimer’s Res. Ther. 2014;6:12. doi: 10.1186/alzrt241. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang L, Liu C, Wu J, Tao JJ, Sui XL, Yao ZG, Xu YF, Huang L, Zhu H, Sheng SL, Qin C. J. Alzheimer’s Dis. 2014;41:1193–1205. doi: 10.3233/JAD-140066. [DOI] [PubMed] [Google Scholar]; c) Kim C, Choi H, Jung ES, Lee W, Oh S, Jeon NL, Mook-Jung I. PloS one. 2012;7:e42983. doi: 10.1371/journal.pone.0042983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guedes-Dias P, de Proenca J, Soares TR, Leitao-Rocha A, Pinho BR, Duchen MR, Oliveira JM. Biochim. Biophys. 2015;1852:2484–2493. doi: 10.1016/j.bbadis.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 9.d’Ydewalle C, Krishnan J, Chiheb DM, Van Damme P, Irobi J, Kozikowski AP, Van den Berghe P, Timmerman V, Robberecht W, Van Den Bosch L. Nat. Med. 2011;17:968–974. doi: 10.1038/nm.2396. [DOI] [PubMed] [Google Scholar]
- 10.a) Wang CY. Mutat. Res. 1977;56:7–12. doi: 10.1016/0027-5107(77)90235-4. [DOI] [PubMed] [Google Scholar]; b) Munakata K, Mochida H, Kondo S, Suzuki Y. J. Pharmacobio-dyn. 1980;3:557–561. doi: 10.1248/bpb1978.3.557. [DOI] [PubMed] [Google Scholar]
- 11.a) Wang CY, Lee LH. Antimicrob. Agents Chemother. 1977;11:753–755. doi: 10.1128/aac.11.4.753. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wei CI, Cohen MD, Swartz DD, Fernando SY, Corbett MD. Toxicol. Lett. 1985;24:111–116. doi: 10.1016/0378-4274(85)90148-1. [DOI] [PubMed] [Google Scholar]; c) Lipczynska-Kochany E, Iwamura H, Takahashi K, Hakura A, Kawazoe Y. Mutat. Res. 1984;135:139–148. doi: 10.1016/0165-1218(84)90114-9. [DOI] [PubMed] [Google Scholar]
- 12.Skipper PL, Tannenbaum SR, Thilly WG, Furth EE, Bishop WW. Cancer Res. 1980;40:4704–4708. [PubMed] [Google Scholar]
- 13.a) Canova-Davis E, Eng M, Mukku V, Reifsnyder DH, Olson CV, Ling VT. Biochem. J. 1992;285:207–213. doi: 10.1042/bj2850207. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hashimoto S, Itai K, Takeuchi Y, Nakamura Y. Nucleic Acids Symp. Ser. 1997;37:7–8. doi: 10.1515/hc.1997.3.4.307. [DOI] [PubMed] [Google Scholar]
- 14.Gaymes TJ, Padua RA, Pla M, Orr S, Omidvar N, Chomienne C, Mufti GJ, Rassool FV. Mol. Cancer Res. 2006;4:563–573. doi: 10.1158/1541-7786.MCR-06-0111. [DOI] [PubMed] [Google Scholar]
- 15.Olaharski AJ, Ji Z, Woo JY, Lim S, Hubbard AE, Zhang L, Smith MT. Toxicol. Sci. 2006;93:341–347. doi: 10.1093/toxsci/kfl068. [DOI] [PubMed] [Google Scholar]
- 16.Johnson D, Walmsley R. Mutat. Res. 2013;751:96–100. doi: 10.1016/j.mrgentox.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 17.a) Pharmacology review for vorinostat. http://www.accessdata.fda.gov/drugsatfda_docs/nda/2006/021991s000_Zolinza_PharmR.pdf.; b) Pharmacology review for belinostat. http://www.accessdata.fda.gov/drugsatfda_docs/nda/2014/206256Orig1s000PharmR.pdf.; c) Pharmacology review for panobinotat. http://www.accessdata.fda.gov/drugsatfda_docs/nda/2015/205353Orig1s000PharmR.pdf.
- 18.Kerr JS, Galloway S, Lagrutta A, Armstrong M, Miller T, Richon VM, Andrews PA. Int. J. Toxicol. 2010;29:3–19. doi: 10.1177/1091581809352111. [DOI] [PubMed] [Google Scholar]
- 19.Lee MS, Isobe M. Cancer Res. 1990;50:4300–4307. [PubMed] [Google Scholar]
- 20.a) Hoshino Y, Okuno M, Kawamura E, Honda K, Inoue S. Chem. Commun. 2009:2281–2283. doi: 10.1039/b822806j. [DOI] [PubMed] [Google Scholar]; b) Dubé P, Nathel NF, Vetelino M, Couturier M, Aboussafy CL, Pichette S, Jorgensen ML, Hardink M. Org. Lett. 2009;11:5622–5625. doi: 10.1021/ol9023387. [DOI] [PubMed] [Google Scholar]
- 21.Groutas WC, Giri PK, Crowley JP, Castrisos JC, Brubaker MJ. Biochem. Biophys. Res. Commun. 1986;141:741–748. doi: 10.1016/s0006-291x(86)80235-2. [DOI] [PubMed] [Google Scholar]
- 22.Pereira MMA, Santos PP. In: The Chemistry of Hydroxylamines, Oximes, and Hydroxamic Acids. Rappoport Z, Liebman JF, editors. New York: Wiley; 2008. pp. 343–499. [Google Scholar]
- 23.Saccone V, Consalvi S, Puri PL, Mascagni P. Italfarmaco SPA, editor. WO2013114413A1. 2013 [Google Scholar]
- 24.De Vreese R, Van Steen N, Verhaeghe T, Desmet T, Bougarne N, De Bosscher K, Benoy V, Haeck W, Van Den Bosch L, D’Hooghe M. Chem. Commun. 2015;51:9868–9871. doi: 10.1039/c5cc03295d. [DOI] [PubMed] [Google Scholar]
- 25.a) Somoza JR, Skene RJ, Katz BA, Mol C, Ho JD, Jennings AJ, Luong C, Arvai A, Buggy JJ, Chi E, Tang J, Sang BC, Verner E, Wynands R, Leahy EM, Dougan DR, Snell G, Navre M, Knuth MW, Swanson RV, McRee DE, Tari LW. Structure. 2004;12:1325–1334. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]; b) Dowling DP, Gantt SL, Gattis SG, Fierke CA, Christianson DW. Biochemistry. 2008;47:13554–13563. doi: 10.1021/bi801610c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cross JB, Duca JS, Kaminski JJ, Madison VS. J. Am. Chem. Soc. 2002;124:11004–11007. doi: 10.1021/ja0201810. [DOI] [PubMed] [Google Scholar]
- 27.Ducháčková L, Roithová J. Chemistry. 2009;15:13399–13405. doi: 10.1002/chem.200901645. [DOI] [PubMed] [Google Scholar]
- 28.Jašikova L, Hanikỳřová E, Škríba A, Jašik J, Roithová J. J. Org. Chem. 2012;77:2829–2836. doi: 10.1021/jo300031f. [DOI] [PubMed] [Google Scholar]
- 29.Furumai R, Matsuyama A, Kobashi N, Lee KH, Nishiyama M, Nakajima H, Tanaka A, Komatsu Y, Nishino N, Yoshida M, Horinouchi S. Cancer Res. 2002;62:4916–4921. [PubMed] [Google Scholar]
- 30.Product monograph for romidepsin. http://www.celgenecanada.net/pdfs/istodax_product_monograph_english_version.pdf. [Google Scholar]
- 31.Vesci L, Bernasconi E, Milazzo FM, De Santis R, Gaudio E, Kwee I, Rinaldi A, Pace S, Carollo V, Giannini G, Bertoni F. Oncotarget. 2015;6:5735–5748. doi: 10.18632/oncotarget.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ying Y, Taori K, Kim H, Hong J, Luesch H. J. Am. Chem. Soc. 2008;130:8455–8459. doi: 10.1021/ja8013727. [DOI] [PubMed] [Google Scholar]
- 33.Itoh Y, Suzuki T, Kouketsu A, Suzuki N, Maeda S, Yoshida M, Nakagawa H, Miyata N. J. Med. Chem. 2007;50:5425–5438. doi: 10.1021/jm7009217. [DOI] [PubMed] [Google Scholar]
- 34.a) Kalin JH, Zhang H, Gaudrel-Grosay S, Vistoli G, Kozikowski AP. ChemMedChem. 2012;7:425–439. doi: 10.1002/cmdc.201100522. [DOI] [PubMed] [Google Scholar]; b) Kozikowski AP, Chen Y, Gaysin AM, Savoy DN, Billadeau DD, Kim KH. ChemMedChem. 2008;3:487–501. doi: 10.1002/cmdc.200700314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozikowski AP, Chen Y, Gaysin A, Chen B, D’Annibale MA, Suto CM, Langley BC. J. Med. Chem. 2007;50:3054–3061. doi: 10.1021/jm070178x. [DOI] [PubMed] [Google Scholar]
- 36.a) Glatt H, Protic-Sablji CM, Oesch F. Science. 1983;220:961–963. doi: 10.1126/science.6342137. [DOI] [PubMed] [Google Scholar]; b) Dreessen B, Westphal G, Bunger J, Hallier E, Muller M. Mutat. Res. 2003;539:157–166. doi: 10.1016/s1383-5718(03)00160-8. [DOI] [PubMed] [Google Scholar]; c) Stark AA, Arad A, Siskindovich S, Pagano DA, Zeiger E. Mutat. Res. 1989;224:89–94. doi: 10.1016/0165-1218(89)90007-4. [DOI] [PubMed] [Google Scholar]
- 37.a) Chen Y, He R, Chen Y, D’Annibale MA, Langley B, Kozikowski AP. ChemMedChem. 2009;4:842–852. doi: 10.1002/cmdc.200800461. [DOI] [PubMed] [Google Scholar]; b) Rai M, Soragni E, Chou CJ, Barnes G, Jones S, Rusche JR, Gottesfeld JM, Pandolfo M. PloS one. 2010;5:e8825. doi: 10.1371/journal.pone.0008825. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Malvaez M, McQuown SC, Rogge GA, Astarabadi M, Jacques V, Carreiro S, Rusche JR, Wood MA. Proc. Natl. Acad. Sci. USA. 2013;110:2647–2652. doi: 10.1073/pnas.1213364110. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Marson CM, Matthews CJ, Atkinson SJ, Lamadema N, Thomas NS. J. Med. Chem. 2015;58:6803–6818. doi: 10.1021/acs.jmedchem.5b00545. [DOI] [PubMed] [Google Scholar]
- 38.Whitehead L, Dobler MR, Radetich B, Zhu Y, Atadja PW, Claiborne T, Grob JE, McRiner A, Pancost MR, Patnaik A, Shao W, Shultz M, Tichkule R, Tommasi RA, Vash B, Wang P, Stams T. Bioorg. Med. Chem. 2011;19:4626–4634. doi: 10.1016/j.bmc.2011.06.030. [DOI] [PubMed] [Google Scholar]
- 39.Ontoria JM, Altamura S, Di Marco A, Ferrigno F, Laufer R, Muraglia E, Palumbi MC, Rowley M, Scarpelli R, Schultz-Fademrecht C, Serafini S, Steinkuhler C, Jones P. J. Med. Chem. 2009;52:6782–6789. doi: 10.1021/jm900555u. [DOI] [PubMed] [Google Scholar]
- 40.Lobera M, Madauss KP, Pohlhaus DT, Wright QG, Trocha M, Schmidt DR, Baloglu E, Trump RP, Head MS, Hofmann GA, Murray-Thompson M, Schwartz B, Chakravorty S, Wu Z, Mander PK, Kruidenier L, Reid RA, Burkhart W, Turunen BJ, Rong JX, Wagner C, Moyer MB, Wells C, Hong X, Moore JT, Williams JD, Soler D, Ghosh S, Nolan MA. Nat. Chem. Biol. 2013;9:319–325. doi: 10.1038/nchembio.1223. [DOI] [PubMed] [Google Scholar]
- 41.Ononye SN, VanHeyst MD, Oblak EZ, Zhou W, Ammar M, Anderson AC, Wright DL. ACS Med. Chem. Lett. 2013;4:757–761. doi: 10.1021/ml400158k. [DOI] [PMC free article] [PubMed] [Google Scholar]