Abstract
Cells must duplicate their mass in order to proliferate. Glucose and glutamine are the major nutrients consumed by proliferating mammalian cells, but the extent to which these and other nutrients contribute to cell mass is unknown. We quantified the fraction of cell mass derived from different nutrients and find that the majority of carbon mass in cells is derived from other amino acids, which are consumed at much lower rates than glucose and glutamine. While glucose carbon has diverse fates, glutamine contributes most to protein, and this suggests that glutamine’s ability to replenish TCA cycle intermediates (anaplerosis) is primarily used for amino acid biosynthesis. These findings demonstrate that rates of nutrient consumption are indirectly associated with mass accumulation and suggest that high rates of glucose and glutamine consumption support rapid cell proliferation beyond providing carbon for biosynthesis.
Introduction
Rapidly proliferating cells have different metabolic needs from non-proliferating cells. During each cell cycle, proliferating cells must synthesize all of the components needed to duplicate cell mass (Lunt and Vander Heiden, 2011). One metabolic feature common to many proliferating cells is high glycolytic flux to lactate in the presence of oxygen, a phenomenon referred to as aerobic glycolysis or the Warburg effect. Why proliferating cells, including cancer cells, consume large quantities of glucose only to excrete the majority of this carbon as lactate is a subject of debate (Brand, 1985; Brand et al., 1986; DeBerardinis et al., 2008; Gatenby and Gillies, 2004; Hsu and Sabatini, 2008; Hume et al., 1978; Jiang and Deberardinis, 2012; Koppenol et al., 2011; Lunt and Vander Heiden, 2011; Newsholme et al., 1985; Vander Heiden et al., 2009; Vazquez et al., 2010). One widely held hypothesis is that high glycolytic flux allows intermediates to be shunted into anabolic pathways to support biomass accumulation (Brand, 1985; Chaneton et al., 2012; Faubert et al., 2013; Hsu and Sabatini, 2008; Hume et al., 1978; Jiang and Deberardinis, 2012; Jiang et al., 2011; Lunt and Vander Heiden, 2011; Newsholme et al., 1985; Shestov et al., 2014; Vander Heiden et al., 2009; Wang and Green, 2012). Many proliferating mammalian cells also consume substantial quantities of glutamine, and glutamine is also hypothesized to provide material for biosynthesis (Brand, 1985; Brand et al., 1986; Daye and Wellen, 2012; DeBerardinis et al., 2007; Hsu and Sabatini, 2008; Wang and Green, 2012). Glutamine or other carbon sources can add new carbon to the tri-carboxylic acid (TCA) cycle (anaplerosis) in order for TCA cycle intermediates to be removed from the cycle and used for the production of new macromolecules in cells (Daye and Wellen, 2012; DeBerardinis and Cheng, 2010; DeBerardinis et al., 2007; Lunt and Vander Heiden, 2011; Newsholme et al., 1985; Wang and Green, 2012), although the extent to which glutamine or other nutrients contribute to biomass has not been determined.
Implicit in these hypotheses is the notion that the most consumed nutrients are also the major contributors to biosynthesis and, therefore, to cell mass. This hypothesis has not been rigorously tested, yet it forms the basis for modeling efforts to understand cancer metabolism (Cascante et al., 2002; Shestov et al., 2014; Shlomi et al., 2011). In Escherichia coli, the sources of cell mass and their fates have been carefully quantified (Roberts et al., 1955), and for prototrophic strains grown in defined minimal media, medium composition constrains the nutrients available to produce new biomass. This is not the case for mammalian cells; in both tissues and in cell culture mammalian cells are exposed to diverse metabolic substrates. While the relative uptake of various nutrients has been examined in several systems (Jain et al., 2012; Paredes et al., 1998), the extent to which any contributes to biomass is unknown.
Various qualitative fates of glucose and glutamine in proliferating cells have been extensively studied, and recent work has suggested that metabolism of other nutrients can be important for proliferation, although net contribution of these alternate fuels to cell mass has not been quantified (Comerford et al., 2014; Kamphorst et al., 2013; Labuschagne et al., 2014; Maddocks et al., 2013; Schoors et al., 2015; Schug et al., 2015). In this study we quantitatively determined the sources of cell mass in rapidly proliferating mammalian cells. Surprisingly, we found that glucose and glutamine, the two nutrients with the highest consumption rates, are not the major contributors of carbon to cell mass. Instead, other amino acids, which are consumed at much lower rates, together account for the majority of carbon in cells. Examining the biosynthetic fates of these nutrients for mass acquisition provides a framework for considering how metabolism supports cell proliferation.
Results
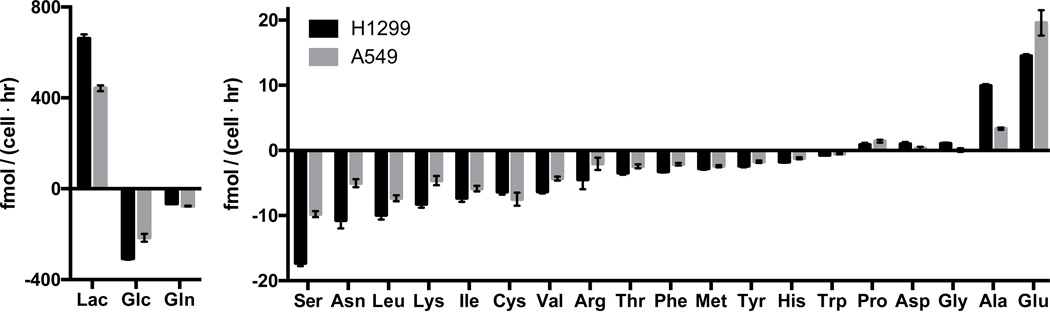
To begin examining the contribution of nutrients to mammalian cell mass, we measured the consumption and excretion rates of glucose, lactate, and amino acids in H1299 and A549 non-small cell lung cancer cell lines, cells that have been shown previously to rely on aerobic glycolysis for proliferation (Christofk et al., 2008) (Figure 1). Similar to other mammalian cell lines (Figure S1A) (Jain et al., 2012), glucose, followed by glutamine, was the most consumed nutrient, and consumption of both exceeded that of serine, the third most consumed metabolite by several fold. Consistent with high aerobic glycolysis in these cells, glucose was consumed at approximately half the rate that lactate was excreted. Most lactate is derived from glucose (Figure S1B) (Brand, 1985; Hume et al., 1978), and two lactate molecules can be derived from one glucose molecule. Since the difference between glucose uptake and lactate excretion equals the maximum possible rate of glucose contribution to cell mass, the rate of new mass addition from glucose must be small relative to the rate of glycolysis.
Figure 1. Rapidly proliferating mammalian cells in culture consume glucose and glutamine in excess of other nutrients.
Consumption and excretion rates of glucose, lactate, and amino acids by H1299 and A549 cells. Nutrients are ranked in descending order by absolute magnitude of their fluxes. Each bar represents the slope from a linear fit of N=3 replicate, ± standard error. Standard three-letter abbreviations are used for amino acids; Glc, glucose; Lac, lactate.
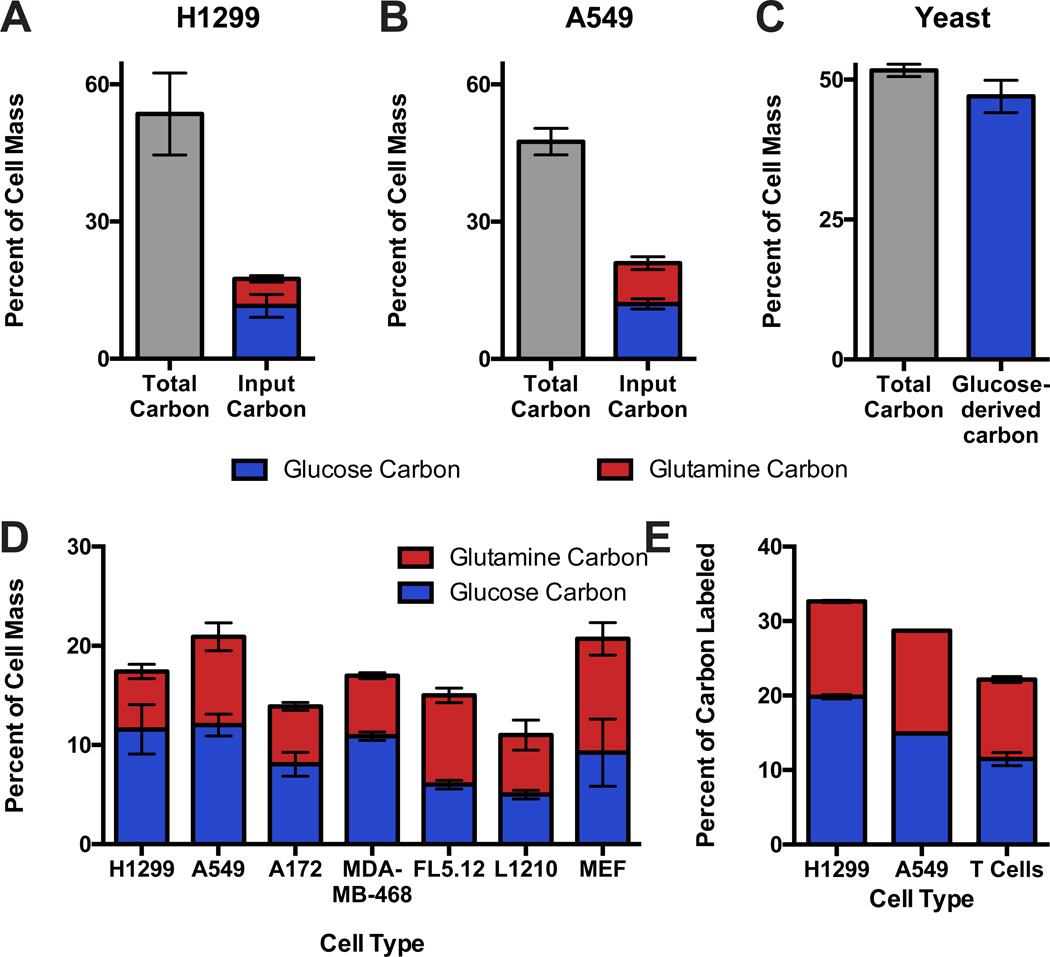
To determine how different nutrients contribute carbon to cell mass, H1299 and A549 cells were grown in the presence of glucose uniformly labeled with carbon-14 ([U-14C]-glucose, i.e. all carbon atoms substituted with carbon-14) until >95% of cell mass had turned over, allowing the proportion of cell dry mass derived from glucose carbon to be determined (Figure S2, see methods). Surprisingly, glucose carbon only contributed to around 10% of cell mass even though cells are approximately 50% carbon by dry weight (Figure 2A,B, S2B). To confirm that this method is capable of accounting for the contribution of glucose to cell mass, the approach was repeated in prototrophic Saccharomyces cerevisiae strain SK1. The dry mass of this strain is also approximately 50% carbon, and when SK1 yeast were cultured in minimal medium containing glucose as the sole carbon source, this carbon could be accounted for using carbon-14 incorporation from [U-14C]-glucose (Figure 2C).
Figure 2. Neither glucose nor glutamine contributes the majority of carbon present in proliferating mammalian cells.
The fraction of cell dry mass consisting of carbon in (A) H1299 and (B) A549 cancer cells exceeds the fraction of cell mass labeled by glucose or glutamine. (C) In SK1 prototrophic yeast, the fraction of cell mass labeled by glucose as the sole carbon source is equal to the fraction of cell mass composed of carbon. (D) The contributions of glucose and glutamine to cell mass are similar across mammalian cells. (E) The fraction of cellular carbon derived from glucose or glutamine in activated primary mouse T cells. Each bar represents the average of N=3 replicates, ±S.D.
Glutamine is the most abundant amino acid in plasma (McMenamy et al., 1957), and, like glucose, can be rapidly consumed by proliferating cells. To measure the contribution of glutamine to cell mass, we assessed carbon-14 labeling from [U-14C]-glutamine (Figure 2A,B). Like glucose, glutamine carbon did not contribute to more than 10% of cell mass, suggesting that the majority of cellular carbon is not derived from glucose or glutamine. To ensure that low incorporation of glucose and glutamine carbon was not influenced by base medium choice, carbon-14 labeling of cells cultured in RPMI 1640 and DMEM were compared, with no differences observed (Figure S2C). We also confirmed that labeling was saturated during these experiments; as subsequent passaging in medium with labeled glucose or glutamine did not increase the fraction of cell mass labeled by these nutrients (Figure S2D,E).
To confirm these findings using an orthogonal approach, cells were grown in medium containing [U-13C]-glucose or glutamine, such that none of the carbon in either nutrient was unlabeled. After culturing cells in this media until >95% of cell mass was produced de novo, the fraction of cellular carbon labeled by carbon-13 was measured by isotope ratio mass spectrometry (IRMS), and the fractional labeling observed was consistent with values derived from carbon-14 incorporation (Figure S2F).
Other cell lines representing different tissues of origin, oncogenic drivers, and species also incorporated glucose and glutamine carbon to a similar extent as H1299 and A549 cells (Table S1, Figure 2D). This finding suggests that many immortalized cells derive substantial cell mass from a source other than glucose and glutamine. To determine if cell mass is acquired similarly by normal proliferating mammalian cells, we examined both primary embryonic fibroblasts (MEFs) and primary T cells derived from mice. Both MEFS and activated proliferating T cells exhibited comparable levels of carbon incorporation from glucose and glutamine, suggesting that a large proportion of cell mass is also derived from other nutrients in these cells (Figure 2D,E).
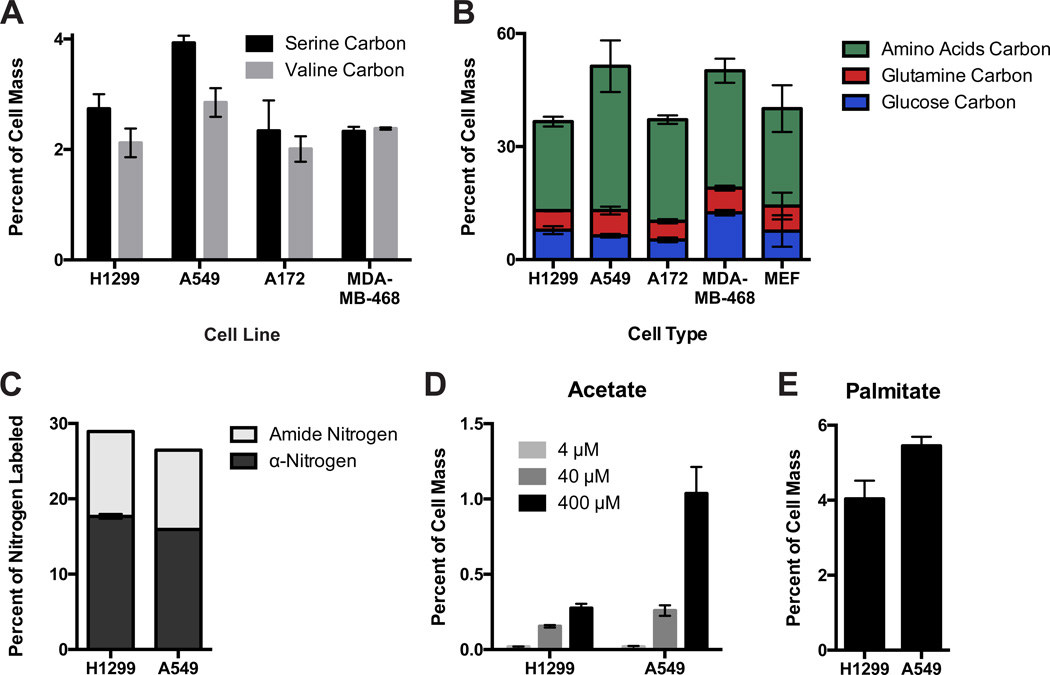
Mammalian cells are thought to consist largely of protein (Alberts et al., 2008; Bonarius et al., 1996; Mourant et al., 2005), so we hypothesized that amino acids could be a major contributor to cell mass. Mammalian cells cannot synthesize many amino acids, and often have access to both essential and non-essential amino acids in their environment. For example, valine, an essential amino acid, must be taken up from the environment, but serine, a non-essential amino acid, may be synthesized de novo. Carbon from these amino acids, which have different fates in central carbon metabolism and a different ability to contribute to non-protein biomass, each labeled 2–4% of cell mass (Figure 3A). To examine how amino acids in general contribute to cell mass, several mammalian cell lines and primary MEFs were grown in medium containing a pool of fifteen [U-14C]-amino acids (which did not include glutamine). This medium was modified from RPMI 1640 in a manner that supported normal cell proliferation in order to facilitate quantitative determination of cell labeling by these amino acids (see methods, Figure S3A,B). Together, amino acids were found to label the majority of carbon in cells and, together with the carbon derived from glucose and glutamine, can account for most of the carbon mass in mammalian cells (Figure 3B).
Figure 3. Amino acids contribute the majority of cell mass for proliferating mammalian cells.
(A) Serine and valine carbon each contribute 2–4% of cell dry mass in mammalian cells. (B) A pooled mixture of fifteen amino acids can label the majority of cellular carbon in proliferating mammalian cells. Amino acid mass contribution was determined by culturing cells in modified RPMI (Table S2, Figure S3A) with [U-14C]-amino acids. Mass contribution of glucose and glutamine as determined in D are also presented for comparison. (C) The fraction of cellular nitrogen derived from glutamine α- and amide-nitrogen atoms. (D) Acetate carbon is a minor contributor to cell mass, and the net contribution is dependent on acetate concentration. (E) Contribution of serum palmitate to cell mass as determined by incorporation of [U-14C]-palmitate. Each bar represents the average of N=3 replicates, ±S.D.
In addition to providing carbon, glutamine contains two nitrogen atoms and can be a nitrogen source for cultured cells (DeBerardinis and Cheng, 2010). The amide nitrogen can be transferred to asparagine and nucleotides, and the α- (amino) nitrogen can be transaminated to non-essential amino acids. Both nitrogen atoms can also be incorporated into protein as glutamine, and hydrolysis reactions allow either to be excreted as ammonia. To determine the extent to which glutamine contributes nitrogen to biomass of H1299 and A549 cells, cells were cultured in the presence of either [amide-15N]- or [α-15N]-glutamine, and the contribution of each to cellular nitrogen was measured by IRMS after labeling had reached steady state (Figures 3C, S3C,D). The amide- and α-nitrogen atoms, respectively, accounted for approximately 11% and 17% of cellular nitrogen. Interestingly, glutamine nitrogen altogether contributed less than half of the nitrogen in each of the cell lines analyzed, suggesting that the rest is derived from other sources such as amino acids.
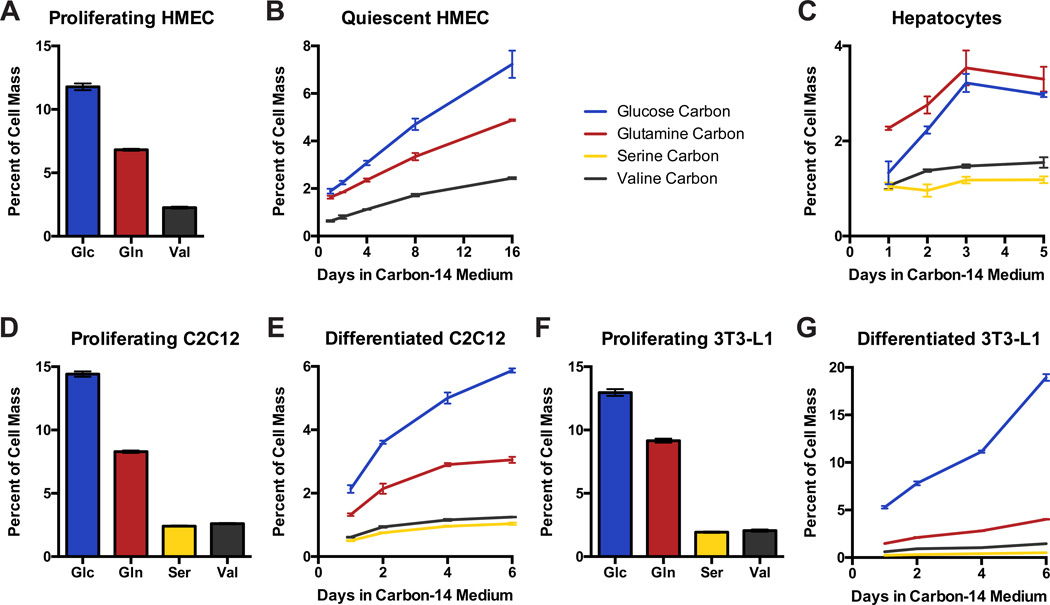
We next sought to determine the extent to which glucose and amino acids contribute carbon mass to non-proliferating cells. These cells do not have the biosynthetic demand to duplicate cell mass, but are not thought to be metabolically quiescent (Lemons et al., 2010) and likely turnover some proportion of cell mass. To determine the mass contribution to quiescent cells, we examined non-transformed human mammary epithelial cells (HMECs) that reversibly arrest when epidermal growth factor (EGF) is withdrawn (Figure S4A) (Stampfer et al., 1993). Proliferating HMECs incorporate carbon from glucose, glutamine, serine, and valine to an extent comparable to other proliferating cells, and interestingly, labeling of non-proliferating HMECs approached similar steady state values, suggesting that most of their cell mass turns over after several weeks (Figure 4A,B). We also determined the contributions of these nutrients to carbon cell mass of post-mitotic cells: primary hepatocytes cultured ex vivo, as well as myocytes and adipocytes differentiated in vitro from C2C12 and 3T3-L1 cells, respectively. Similar to HMECs, hepatocytes, myocytes, and adipocytes were labeled by each of the tracers used, indicating some cell mass turnover (Figures 4C–G, S4B–E). However, in most cases labeling approached steady state values lower than those observed for proliferating cells. This suggests that, in contrast to the HMECs, only a fraction of cell mass in these differentiated cells is turned over. Importantly, undifferentiated C2C12 myoblasts and undifferentiated 3T3-L1 pre-adipocytes incorporated glucose and amino acids to an extent similar to other proliferating cells (Figure 4D,F). Like hepatocytes and myocytes, adipocytes incorporated amino acids at levels lower than proliferating cells (Figure 4G). Glucose, however, labeled a substantial portion of adipocyte cell mass, consistent with their role in lipid storage (Figure S4E). The non-proliferating cells analyzed illustrate that different cell types use different metabolic programs. Hepatocytes and myocytes partially turn over cell mass, using glucose and amino acids to similar extents. Adipocytes are metabolically specialized, incorporating large amounts of glucose carbon, and HMECs abundantly incorporate amino acids into cell mass regardless of their proliferation status.
Figure 4. Carbon contribution to non-proliferating cell mass.
Using carbon-14 tracers, carbon from glucose (Glc), glutamine (Gln), serine (Ser), and valine (Val) was traced into cell mass of: (A) proliferating human mammary epithelial cells (HMEC), (B) arrested HMEC, (C) primary hepatocytes, (D) proliferating C2C12 myoblasts, (E) differentiated C2C12 myocytes, (F) proliferating 3T3-L1 fibroblasts, and (G) 3T3-L1 differentiated into adipocytes. Carbon incorporation into proliferating cells is shown at steady state, whereas incorporation over time is shown for non-proliferating cells. Each bar represents the average of N=3 replicates, ±S.D.
The large contribution of amino acids to cell mass despite relatively low uptake suggests that other nutrients consumed at low rates might contribute to cell mass. For instance, acetate can play a role in fueling cell proliferation (Comerford et al., 2014; Schug et al., 2015). Acetate is absent from tissue culture base media, so we measured acetate levels in fetal bovine serum (FBS) and in the dialyzed FBS used in our labeling studies (Figure S3E). In bovine serum, acetate was approximately 400 µM, and in tissue culture medium containing 10% FBS or 10% dialyzed FBS, acetate levels were approximately 40 µM or 4 µM, respectively. Carbon from acetate did not label cells appreciably when cultured in media with 10% serum, but began to approach 1% of cell dry mass when 400 µM acetate was present in the medium (Figure 3D), a concentration higher than what is typically observed in human serum (Richards et al., 1976; Tollinger et al., 1979).
Cells can also scavenge serum fatty acids as another potential source of mass (Bailey et al., 1972; Kamphorst et al., 2013; Schoors et al., 2015). Quantitative assessment of total fatty acid contribution is complicated by the fact that the fatty acid composition of serum is complex and cannot be recapitulated using isotope labeled tracers. However, to assess the magnitude of individual fatty acid contribution, we examined the incorporation of palmitate (C16:0) into cell mass. Palmitate is the most abundant fatty acid in both serum and in cell membranes (Kilsdonk et al., 1992; Raatz et al., 2001; Vajreswari et al., 1990). Palmitate is present at around 260 µM in FBS (Figure S3F), however this value includes both free fatty acid and esterified palmitate in lipids. [U-14C]-palmitate provided as a free fatty acid labeled approximately 5% of H1299 and A549 cell mass when considered in relation to total palmitate (free fatty acid and esterified) in media (Figure 3E). These data indicate that exogenous lipids can also be a contributor to mass; however this assessment may overestimate the contribution because most serum fatty acids are esterified in lipids such as triglycerides (Raatz et al., 2001). Esterified fatty acids taken up by cells are first hydrolyzed by lipases to generate free fatty acids that mix with free fatty acid pools in cells (Walther and Farese, 2012). However, the amount of carbon incorporated from [1-14C]-palmitate was one sixteenth of that incorporated from [U-14C]-palmitate (Figure S3G). Because the labeled carbon atom in [1-14C]-palmitate is the first carbon lost during fatty acid oxidation, these data suggest that each molecule of palmitate acquired by cells is either completely catabolized or completely incorporated into cell mass. Based on similar assumptions, carbon from [1-14C]-oleate (C18:1), the second most abundant serum fatty acid (Figure S3F) (Raatz et al., 2001), also could account for approximately 5% of cell mass (Figure S3G).
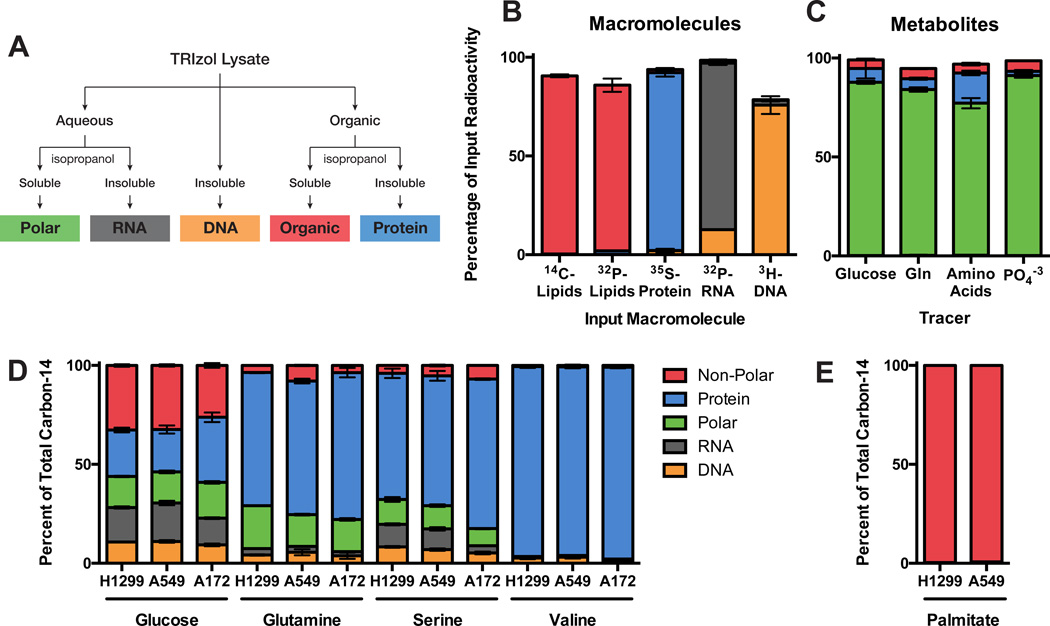
Lipids can be scavenged from the environment or synthesized de novo, with glucose and glutamine serving as potential substrates for de novo biosynthesis (Kamphorst et al., 2013; Metallo et al., 2012; Mullen et al., 2012). To understand the contributions of glucose and glutamine specifically to lipids, we adapted a bi-phasic extraction protocol to separate and recover lipids (and other non-polar compounds) as well as the other major macromolecule classes: RNA, DNA, protein, and polar metabolites (Figure 5A). This approach can quantitatively recover these macromolecules from a cell lysate with high purity, as validated using radioactive standards that were isolated by orthogonal means (Figure 5B,C). Fractionation of radiolabeled cells using this approach also yields full recovery of input radioactivity (Figure S5A). Consistent with previous studies (Kamphorst et al., 2013; Metallo et al., 2012; Mullen et al., 2012), carbon-14 from both glucose and glutamine was recovered in the lipid fraction of H1299, A549, and A172 cells, with greater glucose contribution to lipids than glutamine (Figure 5D). Substantial amounts of glucose carbon were recovered in each of the macromolecule fractions investigated, but the largest proportion of glucose was diverted to the non-polar fraction suggesting that glucose is a major source of material for de novo lipogenesis.
Figure 5. Glucose, glutamine, and other amino acids have diverse biosynthetic fates.
(A) Scheme used to fractionate cells into different macromolecular classes based on differential solubility is shown. Material not precipitated from the aqueous phase is referred to as the polar fraction, and material not precipitated from the organic phase is referred to as the non-polar fraction. (B) Radioactive macromolecules were independently synthesized and purified from HEK293 cells and then used to assess yield and purity of the fractions resulting from the scheme in (A). (C) Radioactive small molecules derived from different nutrients were extracted from HEK293 cells and used to assess yield and purity of the polar fraction resulting from the scheme in (A). (D and E) The relative contributions of (D) glucose, glutamine, serine, and valine, and (E) exogenous palmitate to different macromolecule fractions were determined for H1299, A549, and A172 cells. Each bar represents the average of N=3 replicates, ±S.D.
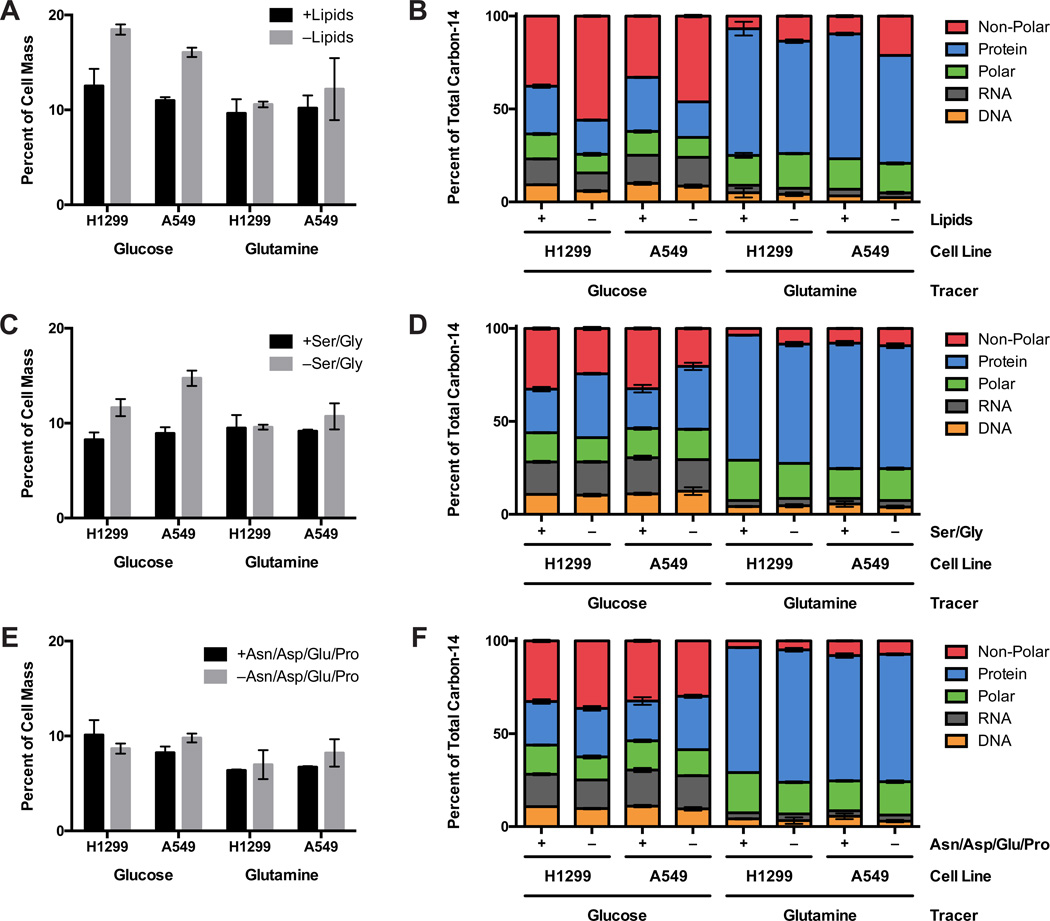
To assess the contribution of lipid biosynthesis from glucose relative to lipid scavenging from the environment, we measured the increase in glucose incorporation into the lipid fraction when proliferating cells were deprived of exogenous lipids. To do this, cells were cultured in serum stripped of lipids by non-denaturing organic extraction (Figure S3F). Any increase in glucose contribution to fatty acids under these conditions should reflect the contribution of exogenous lipids to mass when cells are cultured in lipid-replete conditions. Importantly, cell mass was not significantly changed when cells were cultured in medium containing fatty-acid-stripped serum (Figure S6A). Substantially more glucose carbon was incorporated into cell mass when cells were cultured without lipids, such that approximately half of carbon from glucose was diverted to the non-polar fraction under these conditions (Figure 6A,B). These data are consistent with cells obtaining substantial lipid mass from their environment when available (Figures 5E, S5B). These data are also consistent with the observed equal contributions of [U-14C]-palmitate and [1-14C]-palmitate to cell mass (Figure S3G). No detectable change in net glutamine carbon contribution to cell mass was observed when cells were cultured in the absence of lipids; however, the relative incorporation of glutamine carbon into non-polar material approximately doubled in this condition. Finally, the difference in the proportion of cell mass derived from glucose when cells are grown with or without serum fatty acids corresponds well with the measured contribution of palmitate and oleate carbon to cell mass when these fatty acids are present (Figures 3E, S3G). This corroborates the notion that these two fatty acids are the predominant mass contributors from exogenous lipids and argues that lipid mass in these cells is obtained from a combination of scavenging and de novo synthesis from glucose. Assuming that cellular lipids are derived from only glucose, glutamine, and extracellular lipids, the above data suggest that 60–70% of lipid carbon is derived from exogenous sources, while 20–30% is derived from glucose and ~5% from glutamine.
Figure 6. Glucose carbon incorporation into cell mass is increased when non-essential nutrients are absent.
Glucose and glutamine incorporation and utilization in H1299 and A549 cells grown in RPMI 1640 (A) containing normal serum or serum stripped of lipids; (B) with or without serine and glycine; or (C) with or without asparagine, aspartate, proline, and glutamate. Each bar represents the average of N=3 replicates, ±S.D.
Glucose carbon can enter many biosynthetic pathways (Hume et al., 1978; Lunt and Vander Heiden, 2011), and glucose contributes to all macromolecular fractions (Figure 5D). One non-redundant function of glucose is to provide ribose for nucleotide biosynthesis and a quarter of glucose carbon was traced to RNA and DNA. Some amino acids might also contribute to pathways beyond protein synthesis. To understand how cells utilize amino acids, we traced glutamine, serine, and valine carbon into macromolecules (Figure 5D). Both glutamine and serine can be metabolized to other biosynthetic precursors. Although glutamine anapleurosis for the TCA cycle is hypothesized to provide material that can satisfy biosynthetic demands beyond protein biosynthesis (Daye and Wellen, 2012; DeBerardinis and Cheng, 2010; DeBerardinis et al., 2007; Lunt and Vander Heiden, 2011; Metallo et al., 2012; Mullen et al., 2012), protein was by far the major fate of glutamine carbon. Apart from a small contribution to the non-polar fraction, minor amounts of glutamine carbon were also recovered in nucleic acids. Glutamine carbon incorporation was 3–4 times greater than serine or valine carbon incorporation (Figures 2,3), but nonetheless most glutamine carbon was recovered with the protein fraction. Because glutamine is not over-represented in cell protein (Bonarius et al., 1996), this finding suggests that glutamine anapleurosis provides carbon for other TCA cycle-derived amino acids. Serine carbon exhibited similar relative incorporation into cell mass as glutamine carbon. While the majority of serine carbon was found in protein, serine carbon was also recovered in nucleic acids in agreement with the known role of serine in providing both one-carbon units and glycine for nucleotide biosynthesis (Labuschagne et al., 2014; Lunt and Vander Heiden, 2011). Valine carbon was traced exclusively into protein. If glucose, glutamine, and serine are assumed to be the sole carbon sources for nucleic acids (glycine is likely a minor contributor as it is excreted by these cells, see Figure 1), these data suggest that glucose supplies 60–80% of nucleotide carbon, and glutamine and serine supply 10–20% and ~15%, respectively.
It is possible that under some conditions glucose and/or glutamine carbon contribute more to cell mass. Several amino acids are non-essential for cells, and cells cultured without those amino acids may become more dependent on glucose, glutamine, or other nutrients. To identify which amino acids or combinations or amino acids are non-essential for cells in culture, H1299 and A549 cells were cultivated in media lacking combinations of non-essential amino acids (Figure S6B,C). Two amino acids that can be derived from glycolysis, serine and glycine, and four amino acids that can be derived from the TCA cycle, asparagine, aspartate, glutamate, and proline can be removed from medium without stopping proliferation of these cells. Removal of glycine or the TCA-cycle amino acids had minimal effect on doubling times of the cells tested (Figure S6B,C), and removal of either the TCA-cycle amino acids or both serine and glycine did not significantly alter cell mass (Figure S6A). In the absence of serine and glycine cells incorporate more glucose carbon (Figure 6C), and the increase in incorporation is approximately two times greater than the amount of carbon incorporated from serine. More glucose carbon also contributes to protein in this condition (Figure 6D), but the relative fate of glucose carbon does not differ much from serine/glycine-replete conditions, most likely because serine and glycine also contribute to nucleotides and lipids (Lunt and Vander Heiden, 2011). Since serine and glycine are synthesized from glycolytic intermediates, glutamine incorporation and utilization are not changed when cells are cultured in the absence of these amino acids (Figure 6D).
Without the TCA-cycle amino acids asparagine, aspartate, glutamate, and proline, neither glucose nor glutamine carbon incorporation into cell mass changes (Figure 6E), and their relative biosynthetic fates are not altered (Figure 6F). This suggests that independent of the presence or absence of these four TCA-derived amino acids, glutamine provides much of the carbon for these amino acids even when they are available in the media. Because glutamine carbon mostly contributes to protein and because its incorporation into cell mass is 3–4 times greater than that of serine or valine, these data are consistent with TCA cycle anapleurosis from glutamine producing these amino acids in greater quantities than other macromolecular precursors. Consistent with this hypothesis, based on known pathways, the contribution of glutamine carbon to RNA and DNA only occurs via glutamine-generated aspartate.
Glucose and glutamine utilization is altered in hypoxia: glucose contributes less carbon and glutamine contributes more carbon to lipogenic acetyl-CoA, and cells become increasingly dependent on scavenged lipids (Kamphorst et al., 2013; Metallo et al., 2012). In H1299 cells, glucose and glutamine incorporation was not altered when cells were cultured in 1% oxygen, whereas incorporation of both was decreased in A549 cells while serine and valine incorporation were relatively unchanged in hypoxia (Figure S6D,E). Importantly, HIF1α was stabilized in cells cultured in 1% oxygen confirming a normal response to low oxygen (Figure S6F), These results suggest that, although metabolic specific pathways are regulated by hypoxia, the incorporation of glucose and glutamine carbon into cell mass is not grossly altered.
Discussion
The finding that mammalian cells derive the majority of their mass from exogenously supplied amino acids provides a model for efficient cell growth. By utilizing biosynthetic end products available in the environment rather than synthesizing them de novo, cells will require less ATP, redox equivalents, and carbon to support cell proliferation. Consistent with this hypothesis, carbon from available fatty acids is used exclusively to generate non-polar material, and any fatty acid oxidation does not appear to provide carbon to non-lipid biomass. In some cell types, fatty acid carbon has been traced into dNTPs, but the net contribution of this carbon source has not been quantified (Schoors et al., 2015). In this study, valine carbon was only found in protein, with no evidence that catabolism of this amino acid provides carbon for non-protein biomass. Whole protein can also be scavenged as an alternative source of amino acids for cells (Commisso et al., 2013). Because we can account for protein mass in the cells from the uptake of free amino acids, protein scavenging does not appear to be a major contributor to cell carbon under the conditions tested. Nevertheless, in conditions where free amino acids are limiting, mobilizing amino acids from bulk protein may be important (Commisso et al., 2013), particularly if minimizing de novo amino acid biosynthesis is favored in proliferating cells.
The finding that amino acids together contribute the majority of cell mass is in agreement with the notion that cells are composed primarily of protein (Alberts et al., 2008; Dolfi et al., 2013; Mourant et al., 2005) and is consistent with genomic models of mammalian metabolism (Thiele et al., 2013). The nutrients used to support the growth of other organisms also argues that direct use of amino acids nutrients is a common means to support cell proliferation. Both fertilized bird eggs and germinating plant seeds are examples of closed systems comprised primarily of protein, amino acid, and lipid stores that support rapid gains in organismal mass in the absence of other exogenous nutrients (Boulter and Barber, 1963; Moran, 2007; Willems et al., 2014). Rapid cell proliferation in these systems is also consistent with the notion that proliferating cells avoid de novo biosynthesis where possible. Glycolytic flux is also crucial to development, but it may be more important for ATP and redox balance than for direct production of biosynthetic carbon (Kučera et al., 1984), and the sugar reserves found in seeds contribute primarily to polysaccharides in seedlings (Abdul-Baki, 1969).
The observation that glucose incorporation can be increased to compensate for the absence of some nutrients implies that there is a degree of modularity in macromolecular biosynthesis. Cells are known to have increased dependence on glucose-derived serine when serine is limiting (Chaneton et al., 2012; Labuschagne et al., 2014), and when cells are grown without serine and glycine, the increase in glucose incorporation is roughly twice the amount of carbon derived from serine. Similarly, the increase in glucose incorporation in the absence of lipids corresponded closely to the contributions of palmitate and oleate to cell mass. These findings argue that cell composition and the biosynthetic components required for proliferation do not change across these conditions.
Modeling efforts have sought to explain rapid glucose and glutamine consumption by maximizing the efficiency of consumed carbon incorporation into biomass (Cascante et al., 2002; Shestov et al., 2014; Shlomi et al., 2011). However, rapid consumption of a particular nutrient need not correlate with cell mass incorporation and it appears that the large fluxes through glycolysis and glutaminolysis support proliferation in ways other than supplying carbon. Although the contribution of glucose carbon to cell mass is small, both glucose carbon and glucose metabolism are still important for proliferation. For instance, duplication of nucleic acids is needed for cell proliferation and this requires ribose from glucose or another bioavailable carbohydrate. Glucose and glutamine metabolism also have important roles in contributing non-carbon material to biosynthesis, including energy and reducing equivalents, as well as nitrogen. In many cell lines and in some tumors, glutamine is a major source of nitrogen and can provide anapleurotic TCA cycle carbon. Both glutamine nitrogen atoms contributed abundantly to cellular nitrogen, with the α-nitrogen contributing slightly more. Because the α-nitrogen is used for de novo amino acid synthesis that contribute to both protein and nucleic acids, while the amide-nitrogen contributes primarily to nucleic acids, the increased abundance of proteins in cells likely accounts for this difference.
The primary contribution of glutamine to biomass production in proliferating cells is to the protein fraction. Although non-glutamine amino acids are the dominant source of protein carbon mass, the contribution of glutamine is in excess of that expected of glutamine salvage alone because glutamine atoms are incorporated into other amino acids and TCA cycle intermediates. Indeed, while glutamine may not be utilized as a TCA cycle substrate in all cells in vivo (Davidson et al., 2016; Marin-Valencia et al., 2012; Tardito et al., 2015; Yuneva et al., 2012), our data suggests important incorporation of glutamine-derived carbon in TCA cycle intermediates regardless of whether they are freely available in the media. Aspartate, for example, is poorly transported by cells outside of the prostate and the nervous system (Lao et al., 1993; Storck et al., 1992), and therefore must be synthesized. Glutamine is by far the most abundant amino acid in tissue culture media; facile transport of this amino acid and high glutaminase activity observed in cultured cells (DeBerardinis and Cheng, 2010) yield a rapid influx of glutamine carbon to generate glutamate and proline, which are both excreted by the cells studied. High concentrations of glutamate can enable cell growth in the absence of glutamine, indicating that, relative to glutamine, glutamate import is limited (Eagle et al., 1956). Alternatively, de novo synthesis of TCA-cycle amino acids may provide some benefit to proliferating mammalian cells that exceeds the cost of their synthesis. Collectively, our data argue that the importance of rapid glycolysis and glutaminolysis for proliferating cells lies in an ability to generate metabolic products beyond biomass carbon.
Experimental Procedures
Cell Culture
Cells were maintained in RPMI 1640 or DMEM (without pyruvate) supplemented with 10% heat-inactivated fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (100 µg/mL) using standard tissue culture techniques. When cells were grown in the absence of specific amino acids, RPMI 1640 lacking these amino acids was supplemented with 10% heat-inactivated dialyzed FBS and all other medium components did not differ relative to normal RPMI 1640. RPMI containing modified amino acids was prepared by dissolving glucose and amino acids into RPMI 1640 base. The composition of this medium is detailed in Table S2 and Figure S3A. Medium lacking lipids was RPMI 1640 supplemented with 10% lipid-stripped heat-inactivated FBS (see Supplementary Experimental Procedures). Prototrophic SK1 S. cerevisiae were grown in liquid cultures of synthetic dextrose minimal medium (0.67% yeast nitrogen base without amino acids, 2% glucose) containing glucose as the sole carbon source.
Measurement of nutrient consumption and excretion
Cells number was monitored and medium collected from cultures of exponentially proliferating cells. Glucose, glutamine, and lactate concentrations were measured on a YSI-7100 MBS (Yellow Springs Instruments). Medium amino acids were quantified by gas chromatography-mass spectrometry (GC-MS). Consumption was normalized to the area under the curve of a growth curve.
Mass Spectrometry Analysis
Samples were analyzed by gas-chromatography coupled to mass spectrometry (GC-MS) as described previously (Lewis et al., 2014). Samples were derivitized with MOX reagent (Thermo Scientific) and N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide with 1% tert-butyldimethylchlorosilane (Sigma Aldrich). Fatty acids were derivitized to fatty acid methyl esters in methanol with 2% sulphuric acid. After derivitization, samples were analyzed by GC-MS, using a DB-35MS column (Agilent Technologies) in an Agilent 7890A gas chromatograph coupled to an Agilent 5975C mass spectrometer. Data were analyzed using in-house software described previously (Lewis et al., 2014).
Carbon-14 labeling studies
Unless otherwise indicated, mammalian cells were grown in RPMI 1640 containing 10% dialyzed heat-inactivated FBS supplemented with carbon-14 tracer. Media were supplemented tracers purchased from American Radiolabeled Chemicals or Perkin Elmer (see Supplementary Experimental Procedures). To ensure labeling steady state was reached, cells were grown in the presence of carbon-14 until >95% of cellular material had turned over. Cells were washed three times with phosphate-buffered saline (PBS) to remove unincorporated carbon-14, collected by trypsinization, and incorporated carbon-14 was quantified by liquid scintillation counting. To determine the fraction of cell dry mass derived from a specific nutrient, carbon-14 incorporated into cells was adjusted by the molar ratio of the carbon-14 tracer to the corresponding carbon-12 nutrient in the media. The mass of incorporated carbon was normalized to the culture mass (cell number multiplied by single cell mass, see Supplementary Experimental Procedures).
Stable isotope labeling studies
Cells were cultured in RPMI 1640 supplemented with 10% heat-inactivated dialyzed FBS and containing either [U-13C]-glucose or [U-13C]-glutamine (Cambridge Isotopes Laboratories) in place of the corresponding unlabeled nutrient. Contribution of glucose or glutamine to total cellular carbon was measured after multiple passages in medium containing carbon-13. Contribution of glutamine to total cellular nitrogen was measured for cells grown in RPMI 1640 supplemented as above but containing [amide-15N]- or [α-15N]-glutamine (supplemented at 4% of total glutamine). Carbon-13 and nitrogen-15 enrichment (i.e. 13C:12C and 15N:14N) were measured by isotope ratio mass spectrometry (IRMS) (see Supplementary Experimental Procedures).
Cell fractionation
Cells were lysed using the TRIzol reagent (Life Technologies) and RNA, DNA, and protein were extracted and purified by bi-phasic extraction according to the manufacturer’s instructions (Chomczynski, 1993). Briefly, following initial lysis, insoluble material was considered to be DNA; RNA was precipitated from the aqueous phase, and the remaining soluble material was termed the “polar fraction”; protein was precipitated from the organic phase, and the remaining soluble material was termed the “non-polar fraction”. Radioactivity in each fraction was quantified by liquid scintillation counting. To evaluate the efficacy of this purification scheme, radioactive molecules of a defined macromolecular class were spiked into a TRIzol lysate of unlabeled HEK293 cells prior to fractionation (see Supplementary Experimental Procedures), and radioactivity in the resulting fractions measured. Yield from fractionation was assessed by comparing the sums of the quantities of radioactivity in each fraction to the amount of input radioactive tracer (Figure 2B,C).
Supplementary Material
Highlights.
Glucose and glutamine are not the sources of the majority of mammalian cell mass
Non-glutamine amino acids provide abundant carbon and nitrogen to proliferating cells
Non-proliferating mammalian cells exhibit variable degrees of cell mass turnover
Nutrient fates are determined, showing glutamine contributes primarily to protein
Acknowledgments
This work was supported by NIH grants U54CA121852, R01CA168653, P30CA14051, R01HL108006 (J.C.R.), U54CA143874 (S.R.M.), and K08DK090147 (M.L.S.); the Burroughs Wellcome Fund; and the Damon Runyon Cancer Research Foundation. A.M.H. is a Howard Hughes Medical Institute International Student Research fellow and was a Vertex Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: A.M.H. planned and carried out all experiments. V.C.H. and S.R.M. contributed measurements of cell mass. L.V.D. assisted with primary cell and post-mitotic cell culture. M.O.J. and J.C.R. analyzed primary T cells. M.L.S. performed nitrogen-15 IRMS. A.M.H. and M.G.V.H. designed the study, analyzed the data, and wrote the manuscript.
References
- Abdul-Baki AA. Metabolism of barley seed during early hours of germination. Plant physiology. 1969;44:733–738. doi: 10.1104/pp.44.5.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 5. New York, NY: Garland Science; 2008. [Google Scholar]
- Bailey JM, Howard BV, Dunbar LM, Tillman SF. Control of lipid metabolism in cultured cells. Lipids. 1972;7:125–134. doi: 10.1007/BF02532600. [DOI] [PubMed] [Google Scholar]
- Bonarius HP, Hatzimanikatis V, Meesters KP, de Gooijer CD, Schmid G, Tramper J. Metabolic flux analysis of hybridoma cells in different culture media using mass balances. Biotechnology and bioengineering. 1996;50:299–318. doi: 10.1002/(SICI)1097-0290(19960505)50:3<299::AID-BIT9>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Boulter D, Barber JT. Amino-acid metabolism in germinating seeds of Vicia faba L. in relation to their biology. New Phytologist. 1963;62:301–316. [Google Scholar]
- Brand K. Glutamine and glucose metabolism during thymocyte proliferation. Pathways of glutamine and glutamate metabolism. The Biochemical journal. 1985;228:353–361. doi: 10.1042/bj2280353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand K, Leibold W, Luppa P, Schoerner C, Schulz A. Metabolic alterations associated with proliferation of mitogen-activated lymphocytes and of lymphoblastoid cell lines: evaluation of glucose and glutamine metabolism. Immunobiology. 1986;173:23–34. doi: 10.1016/S0171-2985(86)80086-9. [DOI] [PubMed] [Google Scholar]
- Cascante M, Boros LG, Comin-Anduix B, de Atauri P, Centelles JJ, Lee PW. Metabolic control analysis in drug discovery and disease. Nature biotechnology. 2002;20:243–249. doi: 10.1038/nbt0302-243. [DOI] [PubMed] [Google Scholar]
- Chaneton B, Hillmann P, Zheng L, Martin AC, Maddocks OD, Chokkathukalam A, Coyle JE, Jankevics A, Holding FP, Vousden KH, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques. 1993;15:532–534. 536–537. [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Comerford SA, Huang Z, Du X, Wang Y, Cai L, Witkiewicz AK, Walters H, Tantawy MN, Fu A, Manning HC, et al. Acetate dependence of tumors. Cell. 2014;159:1591–1602. doi: 10.1016/j.cell.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, Bauer MR, Jha AK, O’Brien JP, Pierce KA, et al. Environment impacts the metabolic dependencies of Ras-driven non-small cell lung cancer. Cell metabolism. 2016 doi: 10.1016/j.cmet.2016.01.007. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daye D, Wellen KE. Metabolic reprogramming in cancer: unraveling the role of glutamine in tumorigenesis. Seminars in cell & developmental biology. 2012;23:362–369. doi: 10.1016/j.semcdb.2012.02.002. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Cheng T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell metabolism. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfi SC, Chan LL, Qiu J, Tedeschi PM, Bertino JR, Hirshfield KM, Oltvai ZN, Vazquez A. The metabolic demands of cancer cells are coupled to their size and protein synthesis rates. Cancer & metabolism. 2013;1:20. doi: 10.1186/2049-3002-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagle H, Oyama VI, Levy M, Horton CL, Fleischman R. The growth response of mammalian cells in tissue culture to L-glutamine and L-glutamic acid. J Biol Chem. 1956;218:607–616. [PubMed] [Google Scholar]
- Faubert B, Boily G, Izreig S, Griss T, Samborska B, Dong Z, Dupuy F, Chambers C, Fuerth BJ, Viollet B, et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell metabolism. 2013;17:113–124. doi: 10.1016/j.cmet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nature reviews Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Hume DA, Radik JL, Ferber E, Weidemann MJ. Aerobic glycolysis and lymphocyte transformation. The Biochemical journal. 1978;174:703–709. doi: 10.1042/bj1740703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Deberardinis RJ. Cancer metabolism: When more is less. Nature. 2012;489:511–512. doi: 10.1038/489511a. [DOI] [PubMed] [Google Scholar]
- Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, Yang X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nature cell biology. 2011;13:310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst JJ, Cross JR, Fan J, de Stanchina E, Mathew R, White EP, Thompson CB, Rabinowitz JD. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8882–8887. doi: 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilsdonk EP, Dorsman AN, van Gent T, van Tol A. Effect of phospholipid fatty acid composition of endothelial cells on cholesterol efflux rates. Journal of lipid research. 1992;33:1373–1382. [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nature reviews Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- Kučera P, Raddatz E, Baroffio A. Oxygen and glucose uptakes in the early chick embryo. In: Seymour R, editor. Respiration and metabolism of embryonic vertebrates. Netherlands: Springer; 1984. pp. 299–309. [Google Scholar]
- Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, Maddocks OD. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell reports. 2014;7:1248–1258. doi: 10.1016/j.celrep.2014.04.045. [DOI] [PubMed] [Google Scholar]
- Lao L, Franklin RB, Costello LC. High-affinity L-aspartate transporter in prostate epithelial cells that is regulated by testosterone. Prostate. 1993;22:53–63. doi: 10.1002/pros.2990220108. [DOI] [PubMed] [Google Scholar]
- Lemons JM, Feng XJ, Bennett BD, Legesse-Miller A, Johnson EL, Raitman I, Pollina EA, Rabitz HA, Rabinowitz JD, Coller HA. Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol. 2010;8:e1000514. doi: 10.1371/journal.pbio.1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo CM. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Molecular cell. 2014;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annual review of cell and developmental biology. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang XL, Rajagopalan KN, Maddie M, Vemireddy V, Zhao Z, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell metabolism. 2012;15:827–837. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMenamy RH, Lund CC, Oncley JL. Unbound amino acid concentrations in human blood plasmas. The Journal of clinical investigation. 1957;36:1672–1679. doi: 10.1172/JCI103568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran ET. Nutrition of the Developing Embryo and Hatchling. Poultry Science. 2007;86:1043–1049. doi: 10.1093/ps/86.5.1043. [DOI] [PubMed] [Google Scholar]
- Mourant JR, Short KW, Carpenter S, Kunapareddy N, Coburn L, Powers TM, Freyer JP. Biochemical differences in tumorigenic and nontumorigenic cells measured by Raman and infrared spectroscopy. Journal of biomedical optics. 2005;10:031106. doi: 10.1117/1.1928050. [DOI] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsholme EA, Crabtree B, Ardawi MS. The role of high rates of glycolysis and glutamine utilization in rapidly dividing cells. Bioscience reports. 1985;5:393–400. doi: 10.1007/BF01116556. [DOI] [PubMed] [Google Scholar]
- Paredes C, Sanfeliu A, Cardenas F, Cairo JJ, Godia F. Estimation of the intracellular fluxes for a hybridoma cell line by material balances. Enzyme Microb Tech. 1998;23:187–198. [Google Scholar]
- Raatz SK, Bibus D, Thomas W, Kris-Etherton P. Total fat intake modifies plasma fatty acid composition in humans. The Journal of nutrition. 2001;131:231–234. doi: 10.1093/jn/131.2.231. [DOI] [PubMed] [Google Scholar]
- Richards RH, Dowling JA, Vreman HJ, Feldman C, Weiner MW. Acetate levels in human plasma. Proceedings of the Clinical Dialysis and Transplant Forum. 1976;6:73–79. [PubMed] [Google Scholar]
- Roberts RB, Cowie DB, Abelson PH, Bolton ET, Britten RJ. Studies of biosynthesis in Escherichia coli. Washington, D. C.: Carnegie Institution of Washington; 1955. Composition of the cells: Fractionation and hydrolysis. [Google Scholar]
- Schoors S, Bruning U, Missiaen R, Queiroz KC, Borgers G, Elia I, Zecchin A, Cantelmo AR, Christen S, Goveia J, et al. Fatty acid carbon is essential for dNTP synthesis in endothelial cells. Nature. 2015;520:192–197. doi: 10.1038/nature14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, Alam IS, Goodwin LM, Smethurst E, Mason S, Blyth K, et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer cell. 2015;27:57–71. doi: 10.1016/j.ccell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestov AA, Liu X, Ser Z, Cluntun AA, Hung YP, Huang L, Kim D, Le A, Yellen G, Albeck JG, et al. Quantitative determinants of aerobic glycolysis identify flux through the enzyme GAPDH as a limiting step. eLife. 2014;3 doi: 10.7554/eLife.03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T, Benyamini T, Gottlieb E, Sharan R, Ruppin E. Genome-scale metabolic modeling elucidates the role of proliferative adaptation in causing the Warburg effect. PLoS computational biology. 2011;7:e1002018. doi: 10.1371/journal.pcbi.1002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stampfer MR, Pan CH, Hosoda J, Bartholomew J, Mendelsohn J, Yaswen P. Blockage of EGF receptor signal transduction causes reversible arrest of normal and immortal human mammary epithelial cells with synchronous reentry into the cell cycle. Exp Cell Res. 1993;208:175–188. doi: 10.1006/excr.1993.1236. [DOI] [PubMed] [Google Scholar]
- Storck T, Schulte S, Hofmann K, Stoffel W. Structure, expression, and functional analysis of a Na(+)-dependent glutamate/aspartate transporter from rat brain. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:10955–10959. doi: 10.1073/pnas.89.22.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardito S, Oudin A, Ahmed SU, Fack F, Keunen O, Zheng L, Miletic H, Sakariassen PO, Weinstock A, Wagner A, et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nature cell biology. 2015;17:1556–1568. doi: 10.1038/ncb3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele I, Swainston N, Fleming RM, Hoppe A, Sahoo S, Aurich MK, Haraldsdottir H, Mo ML, Rolfsson O, Stobbe MD, et al. A community-driven global reconstruction of human metabolism. Nature biotechnology. 2013;31:419–425. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollinger CD, Vreman HJ, Weiner MW. Measurement of acetate in human blood by gas chromatography: effects of sample preparation, feeding, and various diseases. Clinical chemistry. 1979;25:1787–1790. [PubMed] [Google Scholar]
- Vajreswari A, Narayanareddy K, Rao PS. Fatty acid composition of erythrocyte membrane lipid obtained from children suffering from kwashiorkor and marasmus. Metabolism: clinical and experimental. 1990;39:779–782. doi: 10.1016/0026-0495(90)90118-v. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez A, Liu J, Zhou Y, Oltvai ZN. Catabolic efficiency of aerobic glycolysis: the Warburg effect revisited. BMC systems biology. 2010;4:58. doi: 10.1186/1752-0509-4-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Farese RV., Jr Lipid droplets and cellular lipid metabolism. Annual review of biochemistry. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Green DR. Metabolic reprogramming and metabolic dependency in T cells. Immunological reviews. 2012;249:14–26. doi: 10.1111/j.1600-065X.2012.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems E, Decuypere E, Buyse J, Everaert N. Importance of albumen during embryonic development in avian species, with emphasis on domestic chicken. Worlds Poultry Science Journal. 2014;70:503–517. [Google Scholar]
- Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Mates JM, Alonso FJ, Wang C, Seo Y, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell metabolism. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.