Abstract
The DNA mismatch repair (MMR) machinery in mammals plays critical roles in both mutation avoidance and spermatogenesis. Meiotic analysis of knockout mice of two different MMR genes, Mlh1 and Mlh3, revealed both male and female infertility associated with a defect in meiotic crossing over. In contrast, another MMR gene knockout, Pms2 (Pms2ko/ko), which contained a deletion of a portion of the ATPase domain, produced animals that were male sterile but female fertile. However, the meiotic phenotype of Pms2ko/ko males was less clear-cut than for Mlh1- or Mlh3-deficient meiosis. More recently, we generated a different Pms2 mutant allele (Pms2cre), which results in deletion of the same portion of the ATPase domain. Surprisingly, Pms2cre/cre male mice were completely fertile, suggesting that the ATPase domain of Pms2 is not required for male fertility. To explore the difference in male fertility, we examined the Pms2 RNA and found that alternative splicing of the Pms2cre allele results in a predicted Pms2 containing the C-terminus, which contains the Mlh1-interaction domain, a possible candidate for stabilizing Mlh1 levels. To study further the basis of male fertility, we examined Mlh1 levels in testes and found that whereas Pms2 loss in Pms2ko/ko mice results in severely reduced levels of Mlh1 expression in the testes, Mlh1 levels in Pms2cre/cre testes were reduced to a lesser extent. Thus, we propose that a primary function of Pms2 during spermatogenesis is to stabilize Mlh1 levels prior to its critical crossing over function with Mlh3.
Keywords: Pms2, Mlh1 stability, male infertility
1. Introduction
The DNA mismatch repair (MMR) machinery in mammals carries out critical roles in both mutation avoidance and meiosis. Evidence for these dual roles came from early studies of various MMR knockout mice, which showed, in addition to the expected phenotypes of increased spontaneous mutation and cancer [1–3], the unanticipated male and/or female infertility [4–10]. Analyses of Mlh1 and Mlh3 gene knockout mice revealed both male and female infertility associated with a defect consistent with the two proteins having a critical role in meiotic crossing over [5–7]. Other studies in mice and yeast indicate that Mlh1 and Mlh3 form a heterodimer critical for normal meiotic crossing over [11]. In addition, the Mlh3 protein possesses an endonuclease function that is critical for normal levels of reciprocal recombination between homologs [12], which in turn is absolutely essential for normal meiosis [13].
In contrast to the male and female infertility exhibited by both Mlh1 and Mlh3 knockout mice, Pms2 knockout (Pms2ko) mice were male sterile, but female fertile. Analysis of Pms2 null spermatogenesis showed a greatly reduced number of sperm of abnormal morphology [4]. However, the phenotype of Pms2-deficient male meiosis was less interpretable than was the case for Mlh1-deficient meiosis, which clearly showed a greatly increased number of univalent chromosomes in anaphase, characteristic of a failure of crossing over [5]. In contrast, the phenotype of Pms2ko male meiosis was relatively subtle, suggesting a possible defect in chromosome pairing during prophase of male meiosis [4]. Here, based on unexpected findings with a mouse model intended to facilitate stochastic alteration of target genes, we show that the Pms2 ATPase domain is not necessary for normal meiosis. Based on initial results, we suggest that Pms2 has an important role in stabilizing Mlh1 during spermatogenesis.
2. Materials and Methods
2.1 Mice and DNA
Mice were housed in a specific pathogen free HEPA filtered room and were fed a diet of LabDiet PicoLab Rodent Diet 20. All experiments were approved by the IACUC committee at OHSU. Pms2ko [4] and Pms2cre [14] mice were generated and genotyped as previously described. Mlh1 [5], Mlh3 [7] and Pms1 [2] knockout mice were generated and genotyped as previously described. Frozen sections of testes tissue were prepared as previously described [15]. Testis volume was calculated by estimating the radius determined with a dissecting scope and using the formula, volume = 4/3xπxr3. Sanger sequencing was performed at the OHSU DNA sequencing core using either PCR products or Topo TA (Invitrogen) cloned PCR products.
2.2 RNA analyses
RNA was extracted from fresh or frozen testes using the miRNeasy Mini Kit (Qiagen). cDNA was made from RNA using random hexamers and the SuperScript III First Strand Synthesis Kit (Invitrogen). Quantitative PCR for Pms2 levels was performed using 0.5X Sybr Green (Invitrogen) in a Rotor-Gene Q (Qiagen). Expression level of genes neighboring Pms2 was calculated from band intensity with Adobe Photoshop. Primers used are listed in Supplemental Table 1.
2.3 Protein analyses
Each sample tested by Western represented a single testis or thymus extracted from individual mice of the relevant genotypes. Total protein was extracted from fresh or frozen tissue with Brij 150 Lysis buffer (0.01M Tris, 2mM EDTA, 0.15M NaCl, 1% Brij 97, 0.1% NP40, 25 nM Leupeptin, 25 nM Aprotinin, 25 nM AEBSF). Samples were then treated as previously described [16]. Briefly, samples were heated in SDS sample buffer at 95 °C for 5 min, and 20–50 μg total protein run on an 8% SDS-PAGE gel. The protein was transferred to a PVDF membrane and probed simultaneously with mouse anti-human MLH1 (Cat #: 51–1327GR), PMS2 (Cat #: 556415), and MSH6 (Cat #: 610918) antibodies (BD Biosciences) at dilutions of 1:1000, 1:500, and 1:2500, respectively. Membranes were then probed with goat anti-mouse HRP secondary antibody (Jackson ImmunoResearch) at a dilution of 1:1000. The secondary antibody was visualized using enhanced chemiluminescense (Perkin Elmer). Mlh1 signal within each lane was normalized to the Msh6 signal using Adobe Photoshop. Multiple, independent Westerns were performed with each blot containing at least one wild-type (WT) sample in addition to experimental samples for normalization across the different genotypes and blots. Student t-test was used to determine significance between Mlh1 levels in the Pms2ko and Pms2cre testes.
3. Results
3.1 Male fertility despite lack of an intact Pms2 ATPase motif
As reported previously, we developed a mouse model designed to facilitate stochastic activation of Cre recombinase by targeting an out-of-reading frame cre sequence to the Pms2 locus, thus creating the Pms2cre allele [14]. The targeting strategy placed Cre expression under the control of the Pms2 promoter and as for the original Pms2ko allele [4], resulted in deletion of exon 2, which encodes a portion of a ATPase domain critical for mutation avoidance (Figure 1A). As expected from our analyses of Pms2ko/ko mice [4], Pms2cre/cre mice are strong mutators [15, 17]. To our great surprise, we found that Pms2cre/cre male mice, in contrast to Pms2ko/ko males, were fully fertile, producing normal size litters (Table 1) and exhibiting normal sperm morphology (Figure 1B). The fertility of Pms2cre/cre male mice which lack an intact ATPase domain indicates that the ATPase of Pms2 is dispensable for normal spermatogenesis.
Figure 1.
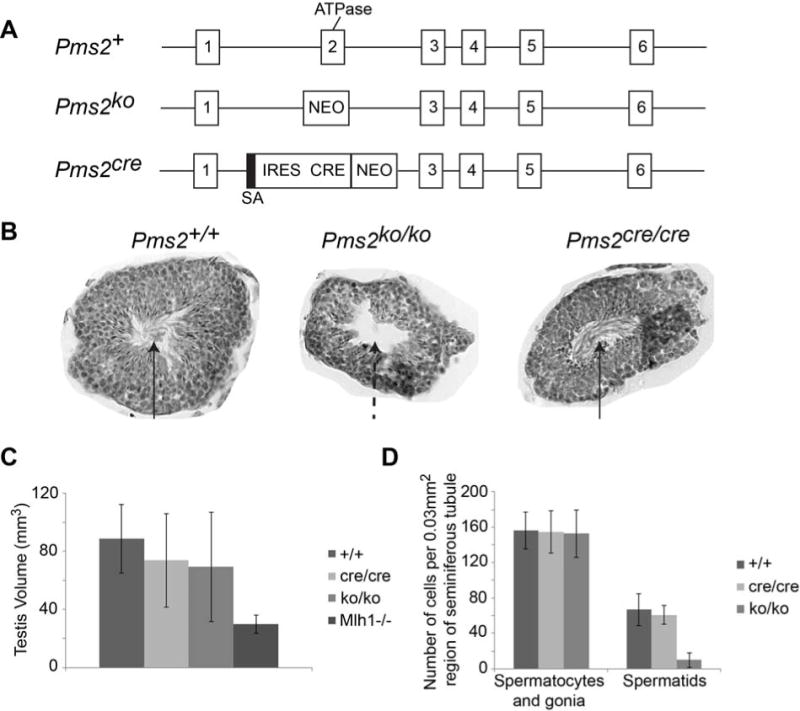
Pms2 ATPase activity is not necessary for male fertility. A) Diagram of Pms2 gene targeting. Exon 2, which contains the ATPase domain, was removed after targeting Pms2 allele in both the Pms2ko and Pms2cre mice. Thus homozygous mice (Pms2ko/ko or Pms2cre/cre) do not have fully functional Pms2. Pms2ko targeted allele has insertion of a neomycin cassette (NEO). Pms2cre targeted allele has insertion of a splice acceptor (SA), internal ribosome entry site (IRES), Cre gene, and neomycin cassette. B) Images of seminiferous tubules. Pms2ko/ko testes do not have fully functional spermatids, as evidenced by the lack of flagella in the lumen (arrows). Pms2cre/cre testes look similar to Pms2+/+ testes. C) The total testis volume is only minimally changed in Pms2cre/cre (n=4 testes) or Pms2ko/ko (n=3 testes) mice compared to Pms2+/+ (n=5 testes). Mlh1−/− testis were significantly smaller (n=3) (ANOVA p=0.05). D) The numbers of spermatocytes and gonia were similar between Pms2cre/cre (n=3 seminiferous tubule sections) or Pms2ko/ko (n=4 seminiferous tubule sections) testes compared to Pms2+/+ (n=3 seminiferous tubule sections), but the numbers of spermatids were decreased in Pms2ko/ko testes (ANOVA p=0.001).
Table 1.
Fecundity of Pms2-mutant male mice.
|
Pms2 Genotype |
# Males |
Total # of pups | Avg. # of Pups/Litter |
|---|---|---|---|
| +/+ | 17 | 106 | 6.2 |
| ko/+ | 10 | 61 | 6.1 |
| cre/+ | 8 | 49 | 6.1 |
| cre/cre | 5 | 31 | 6.2 |
3.2 Alternative splicing in Pms2cre testis
We first examined RNA expression levels of Pms2 and closely linked genes to determine if transgene insertion of the targeting cassette resulted in aberrant regulation. We found no differences between Pms2+/+, Pms2cre/cre, and Pms2ko/ko testes in the expression levels of Pms2 (Figure 2A) or six neighboring genes (Cyth3, Usp42, Eif2ak1, Ankrd61, Aimp2, and Rsph10b2) (Figure 2B), suggesting that the differences in fertility between genotypes was not caused by changes in RNA expression due to transgene insertion. In addition, because loss of Usp42 function results in male infertility [18], we sequenced the exons from Usp42 in Pms2ko mice and did not find any truncating or nonsense mutations.
Figure 2.

Pms2cre testes have alternative RNA splicing. A) Pms2 RNA levels are no different between the different genotypes (n=2 qPCRs for each genotype). B) RNA levels of neighboring genes are not affected by transgene insertion (n=6 genes for each genotype). C) Gel showing alternative splicing in Pms2cre testes compared to Pms2ko testes. Pms2ko RNA is spliced from exon 1 to 3 (loss of exon 2). Pms2cre RNA has 2 different species, either spliced from exon 1 to 4 (loss of exon 2 and 3) or spliced from exon 1 to 5 (loss of exon 2, 3 and 4). D) Diagram of alternative splicing in RNA from Pms2cre testes, which can result in an in frame RNA when spliced from exon 1 to 5 (loss of 330 basepairs). E) Alternative splicing from exon 1 to exon 5 in Pms2cre results in a predicted functional protein (missing 110 amino acids in the N terminus).
Because Pms2 exon 2 was deleted from the DNA, any changes in Pms2 expression levels would nonetheless result in an out-of-frame Pms2 protein. However, alternative splicing of the Pms2 RNA for the Pms2cre allele might result in a Pms2 polypeptide different from the Pms2ko allele. Therefore, we compared Pms2 RNA species from the different alleles by amplifying Pms2 cDNA from exon 1 to exon 5 and confirmed by sequencing (Figure 3). We obtained results consistent with Pms2cre RNA being spliced differently than Pms2ko RNA in the testes. As predicted, for both Pms2cre and Pms2ko, exon 2 containing a portion of the ATPase domain [4] was absent in both cases from the cDNA. As predicted, Pms2ko RNA was spliced from exon 1 to exon 3, which results in an out-of-frame protein (Figure 2C). However, Pms2cre RNA is spliced in two different ways: 1) From exon 1 to exon 4, which is out of frame, and 2) from exon 1 to exon 5, which places the remainder of the RNA in frame (Figure 2D). Therefore, a fraction of Pms2cre RNA from testes should generate a Pms2 protein that contains an intact C-terminus. We analyzed further Pms2cre testis tissue via Western analysis, using a C-terminal specific monoclonal antibody, but this failed to provide direct evidence of Pms2 C-terminus expression. However, Western analysis is not a particularly sensitive technique. The Pms2 protein has three known functional domains: ATPase, endonuclease and Mlh1 interaction domain, which based on our RNA splicing analysis is predicted to be expressed by Pms2cre but not the Pms2ko allele (Figure 2E). These results, in combination with previous results showing that male mice mutated in the Pms2-endonuclease motif were fertile [19], suggested to us that the MutL interaction domain is the critical domain for male fertility. Therefore, we examined the stability of Mlh1 following Pms2 loss in somatic (thymus) and germline (testis) tissues.
Figure 3.

Images of ABI Trace files for sequencing of splice variants in Pms2 RNA. Sequencing was performed on cDNA PCR products, which were cloned into pCR 2.1-TOPO vectors. Pms2+ allele from testes has normal splicing from exon 1 to exon 2. Pms2ko allele from testes splices from exon 1 to exon 3 (exon 2 is deleted from the DNA). Pms2cre allele from testes has alternative splice forms with a splice from exon 1 to exon 4 and from exon 1 to exon 5.
3.3 Loss of both Pms2 and Pms1 destabilizes Mlh1 in mouse thymus
Previous studies had shown that Pms2 and Mlh1 form the MutLα heterodimer via interaction between their C-termini [20–23]. Furthermore, loss of this interaction due to Mlh1 mutation resulted in greatly reduced levels of Pms2 [20] strongly suggesting that formation of the heterodimer was necessary for Pms2 protein stability. However, cultured cells [1, 16] or thymus from Pms2ko/ko mice had no detectable effect on the level of Mlh1 (Figure 4A). Mlh1 also forms heterodimers with Pms1 (MutLβ) or Mlh3 (MutLγ) [24, 25]. To determine which Mlh1 partners are important for Mlh1 stability, we first examined thymus tissue from mice with Pms1 or Mlh3 gene knockouts and different gene knockout combinations with Pms2. In the thymus, only gene knockouts of both Pms2 and Pms1 resulted in destabilization of Mlh1, whereas Mlh3 gene knockout had minimal effect on Mlh1 stability (Figure 4A). These results indicate that Pms2 in conjunction with Pms1 is critical for Mlh1 stabilization in the mouse thymus.
Figure 4.
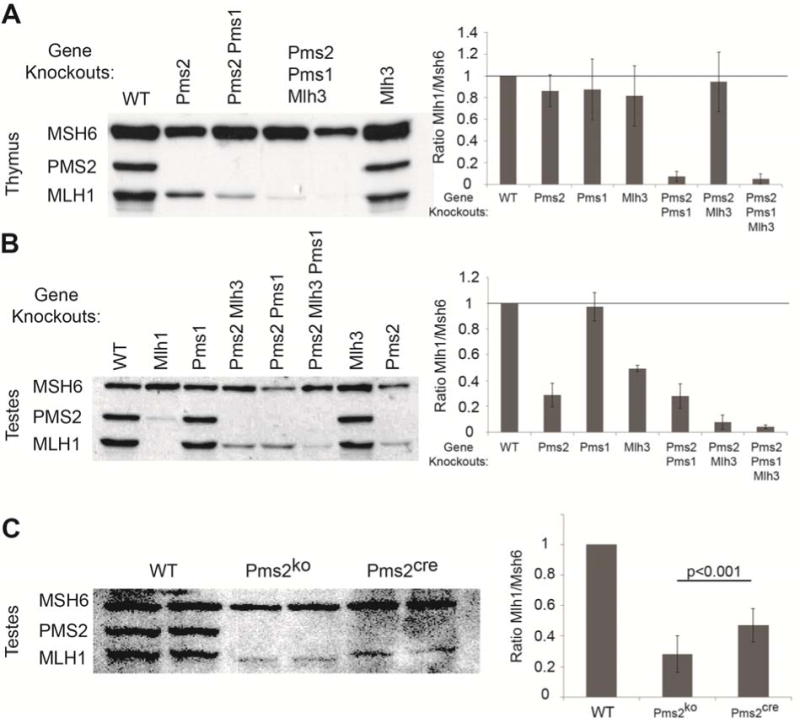
Pms2 is important for Mlh1 stability in mouse testis, but not thymus. A) Western analyses on total proteins from mouse thymi. Mlh1 levels are not affected by a single gene knockout of Pms2ko (n=6 thymi), Pms1 (n=7 thymi) or Mlh3 (n=3 thymi) compared to wild-type (WT). The double gene knockout of Pms2ko and Pms1 (n=6 thymi) reduced Mlh1 stability, while Pms2ko and Mlh3 (n=2 thymi) had minimal effect of Mlh1 stability. The triple gene knockout (n=5 thymi) reduced Mlh1 levels similar to the Pms2ko and Pms1 double gene knockout. B and C) Western analyses on total proteins from mouse testes. B) Mlh1 levels are not affected by Pms1 (n=4 testes) gene knockout alone compared to WT. Mlh1 levels in the testes were reduced by a single gene knockout of either Mlh3 (n=2 testes) or, surprisingly, Pms2ko (n=11 testes) alone. The double gene knockout of Pms2ko and Pms1 (n=3 testes) was not different from Pms2ko alone on Mlh1 stability, while Pms2ko and Mlh3 (n=5 testes) further reduced Mlh1 stability compared to the single gene knockouts. The triple gene knockout (n=2 testes) reduced Mlh1 levels similar to the Pms2ko and Mlh3 double knockout. C) Interestingly, Pms2ko/ko testes (n=11 testes) have a greater reduction in Mlh1 levels compared to Pms2cre/cre testes (n=15 testes) (t-test p<0.001).
3.4 Loss of Pms2 alone is sufficient to destabilize Mlh1 in mouse testis
Because Mlh1 is critical for meiotic crossing over, we asked whether loss of Pms2 in the testes of Pms2ko/ko mice would destabilize Mlh1. Western analysis of whole protein isolates revealed that Mlh1 levels were in fact decreased by ~70% in the testes of Pms2ko/ko mice (Figure 4B), indicating a role for Pms2 in stabilizing Mlh1 in the mouse testis. Gene knockout of Mlh3, the key heterodimeric partner of Mlh1 for meiotic crossover, resulted in Mlh1 instability, but not to the same degree as the Pms2 gene knockout (Figure 4B). Gene knockouts of both Pms2 and Mlh3 resulted in the lowest level of Mlh1. In contrast, Pms1 gene knockout had a minimal effect on Mlh1 stability (Figure 4B). These results show the importance of Pms2 in maintaining normal Mlh1 levels in the mouse testis and implicate the destabilization of Mlh1 as the basis for Pms2ko/ko male infertility.
Because Pms2cre/cre male mice are fertile, we next examined Mlh1 levels in the testes of Pms2cre/cre mice and found that Mlh1 was reduced only 40–50% (Figure 4C), equating to a ~2-fold higher expression level than for Pms2ko/ko mice, supporting increased stabilization of Mlh1. Comparing protein expression levels in whole testes between Pms2cre/cre and Pms2ko/ko mice is valid because, in contrast to Mlh1−/− mice [6], Pms2ko/ko testes, although exhibiting reduced spermatid numbers, were similar in overall size and contained similar numbers of spermatocytes and spermatogonia when compared to wild-type and Pms2cre/cre mice (Figure 1C–D) [4]. Thus, we suggest that the higher Mlh1 levels in Pms2cre/cre testes relative to Pms2ko/ko accounts for the observed fertility. The apparent increased stabilization of Mlh1 levels in Pms2cre/cre mice and alternative splicing of the Pms2cre allele resulting in expression of the Mlh1-interacting domain, in contrast to the Pms2ko allele [4], suggest that Pms2 is important for male fertility due to its role in the stabilization of Mlh1 during spermatogenesis. Further studies, for example the analysis of staged male germ cells are required to substantiate this hypothesis.
4. Discussion
Implications for the function of Pms2 in male meiosis can be made based on previous studies and the findings presented here. There are three known functional domains/motifs within mammalian Pms2, which are important for mutation avoidance: 1) The ATPase motif located in the N-terminal region of the protein [26–28], 2) the endonuclease motif located in the C-terminal region [29–31] and, 3) a domain in the C-terminus that is necessary for heterodimerization with Mlh1 [20–23]. Despite the absence of critical residues within the ATPase domain in Pms2cre/cre male mice, these mice are fully fertile, indicating that the ATPase activity of Pms2 is not necessary for normal spermatogenesis. Edelmann and colleagues derived mice with a missense mutation predicted to disrupt function of the endonuclease motif located in the C-terminal region of Pms2 [19]. They reported that males homozygous for the mutation were strong mutators, but fertile, strongly suggesting that the endonuclease activity of Pms2, although necessary for mutation avoidance, was dispensable for spermatogenesis [19].
Our findings presented here suggest that the fertility of Pms2cre/cre males is due to the production of a Pms2 polypeptide that contains the C-terminal Mlh1-interacting domain, which is not expressed in Pms2ko/ko mice. Although modest, we propose that the resultant increase in Mlh1 protein in Pms2cre testes compared to Pms2ko testes, and presumably in spermatocytes, fosters adequate formation of the Mlh1/Mlh3 heterodimer, which based on studies in yeast [32], mice [33], and humans [34] is necessary for normal levels of crossing over and the resultant normal disjunction of homologs during first division meiosis [10]. These studies could be furthered by examining spermatocyte chromosome spreads from Pms2cre/cre mice for Mlh1 foci, adding the alternative splice variant back to the Pms2ko mice to rescue the male infertility, or by generating Mlh1 hypomorphic mice to determine if there is a threshold level of Mlh1 needed for spermatogenesis and gametogenesis. In addition, based on the fertility of Pms2ko/ko females [4], and the findings presented here, we further suggest that stabilization of Mlh1 protein levels by the presence Pms2 is not critical during oogenesis. In summary, our results indicate that the ATPase of Pms2 is dispensable for proper spermatogenesis and presumably meiosis. Further, based on our results and those of others mentioned above, we propose that the primary function of Pms2 protein in spermatogenesis and meiosis is to stabilize Mlh1 protein levels, thus ensuring adequate levels prior to formation of the cross-over and meiosis-critical Mlh1/Mlh3 heterodimer.
Supplementary Material
Highlights.
A fully intact Pms2 ATPase domain is not necessary for male fertility
Pms2 and Pms1 are important for Mlh1 stability in mouse thymus
Pms2 is primarily important for Mlh1 stability in mouse testis
Acknowledgments
We would like to thank Dr. Paula Cohen for critical comments on the manuscript. RML was funded by NIH grant 2R01GM032741-28.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement:
The authors declare that there are no conflicts of interest.
References
- 1.Yao X, Buermeyer AB, Narayanan L, Tran D, Baker SM, Prolla TA, Glazer PM, Liskay RM, Arnheim N. Different mutator phenotypes in Mlh1- versus Pms2-deficient mice. Proc Natl Acad Sci U S A. 1999;96:6850–6855. doi: 10.1073/pnas.96.12.6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prolla TA, Baker SM, Harris AC, Tsao JL, Yao X, Bronner CE, Zheng B, Gordon M, Reneker J, Arnheim N, Shibata D, Bradley A, Liskay RM. Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nat Genet. 1998;18:276–279. doi: 10.1038/ng0398-276. [DOI] [PubMed] [Google Scholar]
- 3.Narayanan L, Fritzell JA, Baker SM, Liskay RM, Glazer PM. Elevated levels of mutation in multiple tissues of mice deficient in the DNA mismatch repair gene Pms2. Proc Natl Acad Sci U S A. 1997;94:3122–3127. doi: 10.1073/pnas.94.7.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker SM, Bronner CE, Zhang L, Plug AW, Robatzek M, Warren G, Elliott EA, Yu J, Ashley T, Arnheim N, Flavell RA, Liskay RM. Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell. 1995;82:309–319. doi: 10.1016/0092-8674(95)90318-6. [DOI] [PubMed] [Google Scholar]
- 5.Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie DM, Monell C, Arnheim N, Bradley A, Ashley T, Liskay RM. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 6.Edelmann W, Cohen PE, Kane M, Lau K, Morrow B, Bennett S, Umar A, Kunkel T, Cattoretti G, Chaganti R, Pollard JW, Kolodner RD, Kucherlapati R. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85:1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- 7.Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, Thomas J, Cheng J, Touchman JW, Green ED, Schwartzberg P, Collins FS, Cohen PE. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet. 2002;31:385–390. doi: 10.1038/ng931. [DOI] [PubMed] [Google Scholar]
- 8.Kolas NK, Cohen PE. Novel and diverse functions of the DNA mismatch repair family in mammalian meiosis and recombination. Cytogenetic and genome research. 2004;107:216–231. doi: 10.1159/000080600. [DOI] [PubMed] [Google Scholar]
- 9.Svetlanov A, Cohen PE. Mismatch repair proteins, meiosis, and mice: understanding the complexities of mammalian meiosis. Experimental cell research. 2004;296:71–79. doi: 10.1016/j.yexcr.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 10.Kan R, Sun X, Kolas NK, Avdievich E, Kneitz B, Edelmann W, Cohen PE. Comparative analysis of meiotic progression in female mice bearing mutations in genes of the DNA mismatch repair pathway. Biology of reproduction. 2008;78:462–471. doi: 10.1095/biolreprod.107.065771. [DOI] [PubMed] [Google Scholar]
- 11.Kolas NK, Svetlanov A, Lenzi ML, Macaluso FP, Lipkin SM, Liskay RM, Greally J, Edelmann W, Cohen PE. Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J Cell Biol. 2005;171:447–458. doi: 10.1083/jcb.200506170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishant KT, Plys AJ, Alani E. A mutation in the putative MLH3 endonuclease domain confers a defect in both mismatch repair and meiosis in Saccharomyces cerevisiae. Genetics. 2008;179:747–755. doi: 10.1534/genetics.108.086645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Svetlanov A, Baudat F, Cohen PE, de Massy B. Distinct functions of MLH3 at recombination hot spots in the mouse. Genetics. 2008;178:1937–1945. doi: 10.1534/genetics.107.084798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller AJ, Dudley SD, Tsao JL, Shibata D, Liskay RM. Tractable Cre-lox system for stochastic alteration of genes in mice. Nat Methods. 2008;5:227–229. doi: 10.1038/NMETH.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer JM, Miller AJ, Shibata D, Liskay RM. Different phenotypic consequences of simultaneous versus stepwise Apc loss. Oncogene. 2012;31:2028–2038. doi: 10.1038/onc.2011.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson JR, Erdeniz N, Nguyen M, Dudley S, Liskay RM. Conservation of functional asymmetry in the mammalian MutLalpha ATPase. DNA Repair (Amst) 2010;9:1209–1213. doi: 10.1016/j.dnarep.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer JM, Schepers AG, Clevers H, Shibata D, Liskay RM. Occult progression by Apc-deficient intestinal crypts as a target for chemoprevention. Carcinogenesis. 2014;35:237–246. doi: 10.1093/carcin/bgt296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adissu HA, Estabel J, Sunter D, Tuck E, Hooks Y, Carragher DM, Clarke K, Karp NA, Newbigging S, Jones N, Morikawa L, White JK, McKerlie C. Histopathology reveals correlative and unique phenotypes in a high-throughput mouse phenotyping screen. Disease models & mechanisms. 2014;7:515–524. doi: 10.1242/dmm.015263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Oers JM, Roa S, Werling U, Liu Y, Genschel J, Hou H, Jr, Sellers RS, Modrich P, Scharff MD, Edelmann W. PMS2 endonuclease activity has distinct biological functions and is essential for genome maintenance. Proc Natl Acad Sci U S A. 2010;107:13384–13389. doi: 10.1073/pnas.1008589107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohd AB, Palama B, Nelson SE, Tomer G, Nguyen M, Huo X, Buermeyer AB. Truncation of the C-terminus of human MLH1 blocks intracellular stabilization of PMS2 and disrupts DNA mismatch repair. DNA Repair (Amst) 2006;5:347–361. doi: 10.1016/j.dnarep.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Brieger A, Plotz G, Hinrichsen I, Passmann S, Adam R, Zeuzem S. C-terminal fluorescent labeling impairs functionality of DNA mismatch repair proteins. PLoS ONE. 2012;7:e31863. doi: 10.1371/journal.pone.0031863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X, Platt JL, Cascalho M. Dimerization of MLH1 and PMS2 limits nuclear localization of MutLalpha. Mol Cell Biol. 2003;23:3320–3328. doi: 10.1128/MCB.23.9.3320-3328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerrette S, Acharya S, Fishel R. The interaction of the human MutL homologues in hereditary nonpolyposis colon cancer. J Biol Chem. 1999;274:6336–6341. doi: 10.1074/jbc.274.10.6336. [DOI] [PubMed] [Google Scholar]
- 24.Leung WK, Kim JJ, Wu L, Sepulveda JL, Sepulveda AR. Identification of a second MutL DNA mismatch repair complex (hPMS1 and hMLH1) in human epithelial cells. J Biol Chem. 2000;275:15728–15732. doi: 10.1074/jbc.M908768199. [DOI] [PubMed] [Google Scholar]
- 25.Flores-Rozas H, Kolodner RD. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc Natl Acad Sci U S A. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ban C, Yang W. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell. 1998;95:541–552. doi: 10.1016/s0092-8674(00)81621-9. [DOI] [PubMed] [Google Scholar]
- 27.Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends in biochemical sciences. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 28.Tomer G, Buermeyer AB, Nguyen MM, Liskay RM. Contribution of human mlh1 and pms2 ATPase activities to DNA mismatch repair. J Biol Chem. 2002;277:21801–21809. doi: 10.1074/jbc.M111342200. [DOI] [PubMed] [Google Scholar]
- 29.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 30.Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 31.Erdeniz N, Nguyen M, Deschenes SM, Liskay RM. Mutations affecting a putative MutLalpha endonuclease motif impact multiple mismatch repair functions. DNA Repair (Amst) 2007;6:1463–1470. doi: 10.1016/j.dnarep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang TF, Kleckner N, Hunter N. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc Natl Acad Sci U S A. 1999;96:13914–13919. doi: 10.1073/pnas.96.24.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcon E, Moens P. MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics. 2003;165:2283–2287. doi: 10.1093/genetics/165.4.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenzi ML, Smith J, Snowden T, Kim M, Fishel R, Poulos BK, Cohen PE. Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis i in human oocytes. Am J Hum Genet. 2005;76:112–127. doi: 10.1086/427268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


