Abstract
Both silent information regulator 1 (SIRT1) and hypoxia inducible factor 1 (HIF-1) have been found to play important roles in the pathophysiology of Parkinson’s disease (PD). However, their mechanisms and their relationship still require further study. In the present study, we focused on the change and relationship of SIRT1 and HIF-1α in PD. PD cell models were established by using methyl-4-phenylpyridinium (MPP+), which induced inhibition of cell proliferation, cell cycle arrest and apoptosis. We found that the expression of HIF-1α and its target genes VEGFA and LDHA increased and that SIRT1 expression was inhibited in MPP+ treated cells. With further analysis, we found that the acetylation of H3K14 combined with the HIF-1α promoter was dramatically increased in cells treated with MPP+, which resulted in the transcriptional activation of HIF-1α. Moreover, the acetylation of H3K14 and the expression of HIF-1α increased when SIRT1 was knocked down, suggesting that SIRT1 was involved in the epigenetic regulation of HIF-1α. At last, phenformin, another mitochondrial complex1 inhibitor, was used to testify that the increased HIF-1a was not due to off target effects of MPP+. Therefore, our results support a link between PD and SIRT1/HIF-1α signaling, which may serve as a clue for understanding PD.
Keywords: Parkinson’s disease, Hypoxia inducible factor-1, Silent information regulator 1, Acetylation, H3K14, Epigenetics
1. Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disease, and the underlying mechanism implicated in the disease is still unknown [1,2]. To investigate the pathogenesis of the disease, a variety of PD models have been established by researchers [3]. Among them, 1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) is commonly used, which can induce cell cycle arrest, cell death and inhibit cell proliferation by binding to complex I of the electron transport chain or affecting cell cycle-related protein expression [3–7]. However, little attention has been shown to the protective adaptation of the dopaminergic cells themselves when faced with neurotoxin.
Silent information regulator 1 (SIRT1), a nuclear protein, is highly expressed in both embryo and adult brain. Its neuroprotective effect is widely studied in neurodegenerative diseases, including PD. The level of SIRT1 in dopaminergic neurons is significantly down-regulated in toxic models of PD [8–10]. Caloric restriction, glucose analogue or resveratrol, which activate SIRT1, could alleviate the loss and injury of dopaminergic neurons in PD models [11,12]. Overexpression of SIRT1 has been shown to suppress the formation of a-synuclein aggregates by activating molecular chaperones in animal and cell models [13,14]. The protective effects of SIRT1 are mediated by deacetylating histones and many non-histones, including p53, FOXO, HIF-1α, PPARs, NF-kB, Nrf-2, etc [14].
Histone acetylation plays a significant role in regulating gene transcription [15,16]. Acetylation neutralizes the positive charge of the histone tails, making for a flabby nucleosome structure, which is beneficial for transcription factors to bind. Two main enzymes, histone acetyltransferases (HAT) and histone deacetylases (HDAC), participate in the process of acetylation [17]. Our previous studies found that SIRT1 could inhibit the transcription of p53 by decreasing H3K9 acetylation and increasing H3K9 tri-methylation [18]. However, the regulation of SIRT1 on other resides of histones remains to be elucidated in PD.
Hypoxia inducible factor-1 (HIF-1) is a transcription factor that is regulated by oxygen concentration [19]. Its downstream target genes, pathways and proteins can regulate the cell response due to a hypoxia-like stimulation [20]. HIF-1 is composed of the HIF-1α and HIF-1 β subunits; the latter is constitutively expressed in the nucleus, whereas HIF-1α is regulated by the environment [21–23]. Besides hypoxia, HIF-1α can be activated by environmental stimuli under normoxic conditions [23,24]. Previous studies have found that the stability and function of HIF-1 is primarily regulated by posttranslational modifications [24]. However, little attention has been given to HIF-1’s transcriptional levels.
In the present study, we investigated the changes of SIRT1 and HIF-1α expression in MPP+ treated cells. Additionally, we analyzed the regulation of the HIF-1α gene by SIRT1 at the epigenetic level.
2. Materials and methods
2.1. Cell culture and treatments
SH-SY5Y cells were routinely grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (GIBCO, Gaithersburg, MD, USA) and cultured at 37 °C under humidified 5% CO2 atmosphere. MPP+ (Sigma–Aldrich, St. Louis, MO, USA) and phenformin (Selleckchem, Houston, USA) were freshly dissolved in phosphate buffered saline (PBS) at a stock concentration at 125 mM and 50 mM which was stored at −20 °C. MPP+ and phenformin were further diluted in serum free DMEM to achieve the final concentrations.
2.2. Assessment of cell viability
The number of inhibited cells was measured by using a CCK-8 assay according to the manufacturer’s instructions (Cell Counting Kit-8; Beyotime, Shanghai, China), as previously described. Briefly, SH-SY5Y cells were seeded into 96-well plates with 5000 cells in each well. On the second day, cells were treated with MPP+ at different concentrations and times, and cells treated with vehicle only were used as control. After a specific time interval, one-tenth volume of CCK-8 solution was added to each well to incubate for 2 h at 37 °C. The well containing only the culture medium was regarded as blanks. Absorption was measured using a spectrophotometer (Bio Tek, VT, USA) at 450 nm. The cell inhibition rate was calculated as 1 − [(mean OD of one group-blank)/(mean OD of the control-blank)]. All experiments were independently repeated at least three times.
2.3. RNA extraction, RT-PCR, and real-time PCR
Total RNA was extracted using Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. All RNA samples were quantified and reverse-transcribed into cDNA using the ReverTra Ace-α first strand cDNA synthesis kit (Toyobo Co., Ltd., Osaka, Japan). qRT-PCR was conducted using a RealPlex4 real-time PCR detection system from Eppendorf Co Ltd (Hamburg, Germany), with SYBR-green real-time PCR Master Mix (Toyobo Co., Ltd., Osaka, Japan) used as the detection dye. A comparative threshold cycle (Ct) was used to determine the relative gene expression normalized to 18s RNA. For each sample, the Ct values of the genes were normalized using the formula δ Ct = Ct_ genes − Ct_18s RNA. To determine relative expression levels, the following formula was used δδ Ct = δ Ct _all groups −δ Ct _blank control group. The values used to plot relative expression of markers were calculated using the expression 2−δδCt. The cDNA of each gene was amplified with primers as previously described. The following primers were used: HIF1α-F GCGCGAACGACAAGAAA; HIF1α-R:GAAGTGGCAACTGATGAGCA; VEGFA-F: TCGGGCCTCCGAAACCATGA; VEGFA-R: CCTGGTGAGAGATCTGGTTC; LDHA-F: ATGGCCTGTGCCATCAGTAT; LDHA-R: TTCTAAGGAAAAGGCTGCCA; 18s rRNA-F: CAGCCACCCGAGATTGAGCA; 18s RNA-R:TAGTAGCGACGGGCGGTGTG. Data are presented as mean ± standard error of three independent experiments in three real-time PCR replicates.
2.4. Immunoblotting assay
Total proteins were isolated with a mammalian cell lysis/extraction kit (Sigma–Aldrich, St. Louis, MO, USA) according to the manufacturer’s protocol and equal amount of the protein were separated on SDS-polyacrylamide gel electrophoresis gels and transferred to polyvinylidene fluoride (PVDF) membranes. After blocking in 5% non-fat milk prepared in Tris-buffered saline containing 0.05% Tween-20 (TBST) for 45 min at room temperature, the PVDF membranes were then incubated with specific primary antibodies: anti-SIRT1, anti-CDK4 (Cell Signaling Technology, Inc. Danvers, MA, USA), anti-HIF-1α antibody (Abcam, San Francisco, USA) respectively and anti-VEGFA, LDHA (Protein Tech Group, Chicago, USA). An immunoblot for β-Actin (1:1000; Cell Signaling Technology, Inc, Danvers, MA, USA) was performed to demonstrate equal protein loading. Then the membrane was washed with TBST for 3 times for 15 min each. After incubation with the secondary antibody for 45 min at 37 °C and washed with TBST, the immunoreactivity was visualized by an NIR scanning device (Odyssey scanner, I-COR Bioscience). Specific proteins were quantified using Image J software.
2.5. Annexin V-FITC/PI assays
Apoptosis was evaluated using an AnnexinV-FITC/PI kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. After treatment under various conditions, approximately 105 cells from each group were harvested and washed twice with PBS. Then, 195 μL Annexin V-FITC binding buffer was added to the resuspended cells, followed by 5 μL Annexin V-FITC. After vortexing gently, all the cells were incubated for 30 min at 37 °C without light. Cells were then centrifuged at 1000 × g for 5 min, after removing the supernatant, and 190 μL binding buffer with 10 μL PI was added to the cells. Flow cytometry were used to count the number of cells that underwent apoptosis.
2.6. RNA interference (RNAi) and transfection
siRNA oligomers targeting SIRT1 (si-SIRT1) and a scrambled oligomer (si-control) were purchased from Genepharma Co. Ltd. (Shanghai, China) and the transfection method was performed according to the manufacturer’s instructions. Briefly, SH-SY5Y cells were transfected with 0.3 μg siRNA-SIRT1 or a siRNA-control, respectively, by electroporation (or Lipofectamine 2000 reagent (Invitrogen Life Technologies, Carlsbad, CA, USA). Cells were treated with MPP+ after siRNA transfection. The sequence of siRNA is as followed: SIRT1 siRNA1 F: UACAAAUCAGGCAAGAUGCUGUUGC R: GCAACAGCAUCUUGCCUGAUUUGUA
SIRT1 siRNA2: F: GCAAUAGGCCUCUUAAUUA R: UAAUUA AGAGGCCUAUUGC
-
SIRT1 siRNA3
F: UUCAACAUUCCUAGAAGUUUGUACUUC
R: GAAGUACAAACUUCUAGGAAUGUUGAA;
-
SIRT1 siRNA4
F: ACAGUUUCAUAGAGCCAUGAAGUAU R: AUACUUCAUGG CUCUAUGAAACUGU
2.7. ChIP assay
The ChIP assay was performed using a ChIP chromatin immunoprecipitation kit (Beyotime, Shanghai, China). SH-SY5Y cells (1 × 107) were fixed in 10 ml of DMEM with 1% formaldehyde at 37 °C for 10 min and decross-linked with glycine. Cells were resuspended in 1 ml of SDS Lysis Buffer containing 1 × Protease Inhibitor Cocktail II. The ChIP procedure was performed as described in Ref. [25]. The chromatin solution was incubated with primary antibodies against H3K14-ac or control immunoglobulin G and protein A/G plus-agarose immunoprecipitation reagent overnight at 4 °C with rotation. Human IgG was used as the negative control and DNA (Input) as positive control. Immunoprecipitates were eluted from the protein A/G plus-agarose reagent and precipitated. Then the purified DNA was subjected to PCR amplification with the specific PCR primers to human HIF-1α promoter: 5′-GCGCGAACGACAAGAAA-3′ and 5′-GAAGTGGCAACTGATGAGCA-3′.
2.8. Statistical analysis
Each experiment was performed at least three times. Data were presented as mean ± standard error, and the differences were evaluated using t-tests. P values less than 0.05 were considered statistically significant.
3. Results
3.1. MPP+ caused a decrease in cell viability, cell cycle arrest and apoptosis
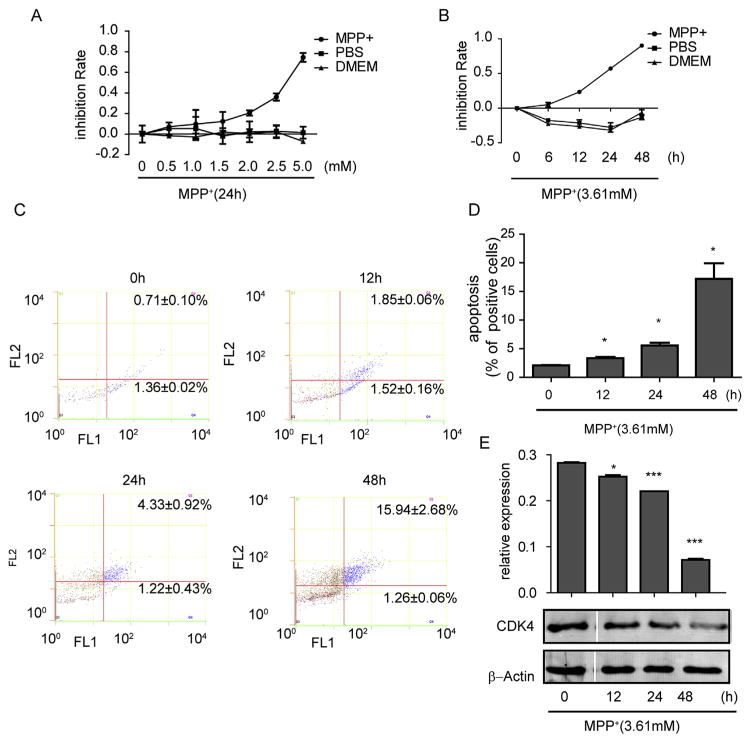
To evaluate the toxicity of MPP+ to SH-SY5Y cells, we first examined the cell inhibition rate by the CCK-8 method. Our results showed that MPP+ dose- and time-dependently suppressed the growth of SH-SY5Y cells (Fig. 1A–B). By using IC 50 software, we selected the concentration of 3.61 mM for further experiments.
Fig. 1.
MPP+ caused inhibition of cell viability, cell cycle arrest and apoptosis. (A) SH-SY5Y cells were treated with MPP+ for 24 h at various concentrations or (B) at 3.61 mM (IC50) for different time periods. (C) Cell apoptosis was measured by flow cytometry and quantified (D) under different treatment times. The cell cycle-related CDK4 protein was examined by western blot (E). Data are expressed as mean ± standard error. *P < 0.05, **P < 0.01, ***P < 0.001, compared to 0 h.
We further studied the effect of MPP+ on apoptosis by using flow cytometry. SH-SY5Y cells were treated with MPP+ at 3.61 mM for different lengths of time, and we found that the number of apoptotic cells significantly increased (Fig. 1C–D) Meantime, the level of CDK4 decreased with the extended time of MPP+ treatment, further supporting the data above (Fig. 1E). Our results suggest that as time increases MPP+ can significantly inhibit the cell cycle and promote apoptosis.
3.2. MPP+ significantly changed SIRT1 and HIF-1α in vitro
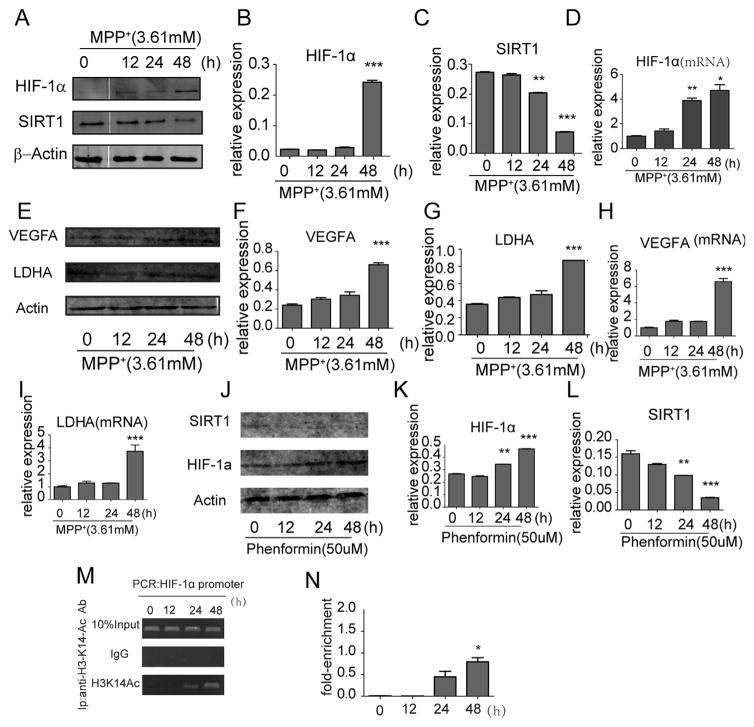
To determine whether SIRT1 and HIF-1α were altered in MPP+ treated cells, the protein levels of SIRT1 and HIF-1α were detected by western blot following MPP+ treatment. Similar to our previous report [26], SIRT1 was significantly decreased at 24hr in our current study whereas HIF-1α levels were up-regulated at 48 h (Fig. 2A–C). Next, we examined whether there was an enhancement of HIF-1α at the transcriptional levels by qRT-PCR. Indeed, HIF-1α mRNA levels increased after MPP+ treatment for 24 h (Fig. 2D). In order to know whether the changes affected the functionality of HIF-1, we tested the variations of VEGFA and LDHA which were HIF-1 dependent. We found the level of mRNA and protein level of these two both increased at 48 h (Fig. 2E–I), illustrating the functional change of HIF-1α. Meanwhile, to prove the increase HIF-1α was not due to off target effects of MPP+, we chose phenformin, another mitochondrial complex 1 inhibitor, to treat cells, results showed that SIRT1 was decreased and HIF-1α was increased (Fig. 2J–L), which were just as what MPP+ did.
Fig. 2.
The change of SIRT1 and HIF-1 and its target genes in MPP+ and phenformine treated SH-SY5Y cells and the mechanisms behind it. Western blot assay (A) and quantification of SIRT1 (B) and HIF-1α (C) after MPP+ treatment. (D) qRT-PCR analysis of HIF-1α expression in SH-SY5Y cells after MPP+ treatment at different time points. (E) Western blot assay and quantification of VEGFA (F) and LDHA (G) after MPP+ treatment. qRT-PCR analysis of expression of VEGFA (H) and LDHA (I) in MPP+ treated cells. (J) Western blot assay and quantification of SIRT1 (K) and HIF-1α (L) in phenformin treated cells. (M) A specific H3K14-ac antibody was applied to detect the H3K14-ac level at different times (N). *P < 0.05, ** P < 0.01, ***P < 0.001, compared to 0 h. Data were expressed as mean ± standard error.
3.3. Acetylated H3K14 was increased in MPP+ treated cells
To further investigate the mechanism underlying the regulation of HIF-1α by MPP+, we utilized the ChIP assay to analyze the presence of H3K14-ac in the HIF-1α promoter region. After the various treatments described above, the cross-linked DNA fragments were immunoprecipitated using an anti-H3K14-ac antibody and subjected to PCR. The level of PCR product could be used as an indicator of the amount of H3K14-ac that bound to HIF-1α promoter. We found the levels of H3K14-ac in the HIF-1α promoter region is increased after treatment with MPP+ for 24 h, especially at the 48 h time-point which was accord with the increased level of HIF-1α mRNA, indicating that MPP+ promotes the transcription of HIF-1α (Fig. 2M–N).
3.4. Increased levels of HIF-1α was dependent on SIRT1
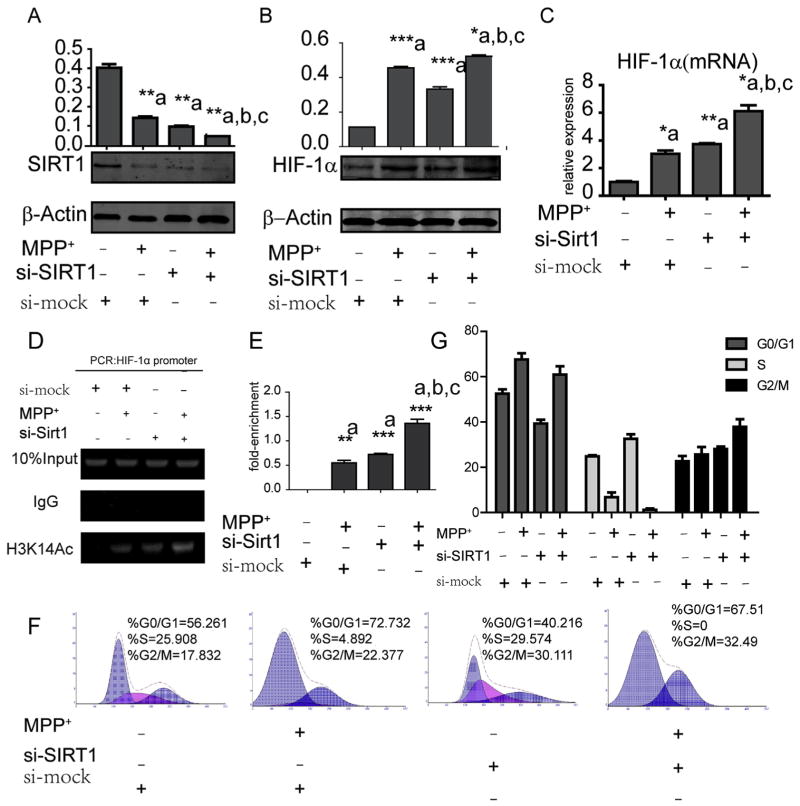
To determine whether SIRT1 was involved in MPP+-induced HIF-1α, we transfected SH-SY5Y cells with SIRT1 small interfering RNA (siRNA) and observed a dramatic decrease in SIRT1 protein levels (Fig. 3A). We found that HIF-1α mRNA and protein levels significantly increased upon SIRT1 downregulation. Next, we explored the effect of siRNA together with MPP+ treatment on the expression of SIRT1 and HIF-1α. The results indicated that the synergistic effects of siRNA and MPP+ further decreased the level of SIRT1 compared with siRNA or MPP+ alone. Accordingly, the mRNA and protein levels of HIF-1α increased more than with the single treatments (Fig. 3B–C). Moreover, SH-SY5Y cells transfected with SIRT1 siRNA exhibited a significant increase in H3K14-ac levels compared with control cells (Fig. 3D–E). When cells were treated with MPP+ and siRNA-SIRT1 combined, the H3K14-ac levels was the highest among all groups (Fig. 3E). Furthermore, we tested cell cycle alterations by flow cytometry (Fig. 3F–G) and found that cells transfected with SIRT1 siRNA arrested in G2/M phase. Correspondingly, the combined treatment of MPP+ and si-SIRT1 locked all cells in G0-G1 or G2-M phase.
Fig. 3.
Alterations of SIRT1, HIF-1α, ac-H3K14 and the cell cycle after si-SIRT1 treatment. Western blot and quantification of SIRT1 (A) and HIF-1α (B) protein after MPP+ treatment, siRNA transfection or both. (C) qRT-PCR assay used to test HIF-1α changes at the mRNA level. (D) A specific H3K14-ac antibody was applied to detect the H3K14-ac level after different treatments (E) *P < 0.05, **P < 0.01, ***P < 0.001, compared to controls (a), MPP+ treatment only (b) or si-SIRT1 only (c). Flow cytometry (F) and quantification (G) of the stage of cell cycle after different treatment was analyzed. Data are expressed as mean ± standard error.
4. Discussion
In this study, we provide the evidence that HIF-1α and its downstream target genes is upregulated when SH-SY5Y cells encounter MPP+, and the change of the gene is mediated through the up-regulation of histone acetylation.
MPP+, a inhibitor of complex I of the respiratory chain, is assumed to cause neuron death by energy failure. However, by using Omics technologies, several researchers have found the upstream network of responses took place in MPP+ treated cells preceding the final decision on cell death [27]. In the present experiments, an increase in HIF-1α and its downstream genes expression was shown when SH-SY5Y cells were exposed to MPP+, which may be related with generation of reactive oxygen species (ROS) induced by the altered the electron flow toward oxygen at a subunit of complex I [27]. In normoxic state, the 402 and 564 proline residues of HIF-1α are hydroxylated by prolyl hydroxylase enzymes (PHDs); hydroxylated HIF-1α can be identified by von Hippel-Lindau (VHL), which is then recognized by a ubiquitin E3 ligase complex. Next, HIF-1α is degraded in the proteasome pathway [21]. ROS can stabilize HIF-1α by inhibiting PHDs [28]. While, in this study, This change of HIF-1α mainly occurred at the transcription level because both the mRNA and protein levels of HIF-1α were upregulated, and the increase of protein was delayed compared with the mRNA, which may be related to the proteasome degradation of HIF-1α under normal oxygen conditions. Meanwhile, phenformin, another mitochondrial complex 1 inhibitor, could also increase the expression of HIF-1α, which further proved that the increased HIF-1α in MPP+ treated cells was not due to its off target effects.
In addition to posttranslational modification, a growing number of studies have shown that HIF-1α can be regulated at the transcriptional level. Herein, we explored the effect of histone acetylation to the expression of HIF-1α. The lysine on the N-terminal tail of histones can be acetylated by histone acetyltransferases (HATs), which diminishes the electrostatic affinity between histone proteins and DNA, loosening the nucleosome and enhancing transcription [29–31]. Many studies have confirmed the neuroprotective role of histone acetylation in PD. Chen P et al. found that valproate acid (VPA), which is a histone deacetylase inhibitor, protects midbrain DA neurons from LPS or MPP+ induced neurotoxicity [32]. Trichostatin A (TSA), a commonly used histone deacetylase (HDAC) inhibitor, exerts an early neuroprotection in PD models induced by MPP+ [33]. Herein, to verify the epigenetic regulation of HIF-1α in MPP+ cells, we used the ChIP assay; the acetylation level of H3K14 was enhanced by time, suggesting that the increased level of HIF-1α expression caused by MPP+ was related to an increase in H3K14-ac in the promoter region. Meanwhile, consistent with our former study, we confirmed that MPP+ decreased the level of SIRT1 in a time-dependent manner.
To further elucidate the relationship between SIRT1 and HIF-1α, we transfected cells with SIRT1 siRNA. The mRNA and protein levels of HIF-1α increased when SIRT1 was knocked down. To further confirm the role of SIRT1, we carried out a ChIP assay after SIRT1 interference. The results showed that knockdown of SIRT1 could promote the expression of H3K14-ac; moreover, the level of H3K14-ac was higher than in MPP+ treated alone cells, which may be related to cell injury by the neurotoxicant. Therefore, the increased expression of HIF-1α is related to enhanced H3K14-ac levels, which resulted from SIRT1 knockdown. Likewise, our results from the cell cycle analysis measured by flow cytometer showed similar results by demonstrating that either MPP+ or si-SIRT1 could arrest the cell cycle.
Our study demonstrated that MPP+ could reduce SIRT1 in cells, which induced alterations of histone epigenetic modifications, further activating HIF-1α transcription. This process might be an important mechanism for the interaction between HIF-1α and SIRT1.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (NSFC) [grant numbers 81371410, 81171205], the National Basic Research Program of China (973 Program) [grant number 2011CB707506] and the Biomedical Multidisciplinary Program of Shanghai Jiao Tong University [grant number YG2014MS31 to Yun-Cheng Wu], the National Natural Science Foundation of China [grant number 81202811], the Project of China Postdoctoral Science Foundation [grant number 2014M550250], and the Shanghai Municipal Health Bureau Fund [grant number 2012432 to Te Liu].
Abbreviations
- PD
Parkinson’s disease
- SIRT1
silent information regulator 1
- HIF-1
hypoxia inducible factor 1
- MPTP
1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- MPP+
methyl-4-phenylpyridinium
- CCK8
cell counting kit-8
- ChIP
chromatin immunoprecipitation
- DMEM
Dulbecco’s Modified Eagle’s medium
- siRNA
small interfering RNA
- PHD
prolyl hydroxylase
- HAT
histone acetyltransferase
- ROS
reactive oxygen species
Footnotes
Conflict of interest statement
The authors report no conflicts of interest with this study.
Contributor Information
Te Liu, Email: liute1979@126.com.
Yun-Cheng Wu, Email: yunchw@medmail.com.cn.
References
- 1.Feng Y, Liu T, Li XQ, Liu Y, Zhu XY, Jankovic J, Pan TH, Wu YC. Neuro-protection by Orexin-A via HIF-1alpha induction in a cellular model of Parkinson’s disease. Neurosci Lett. 2014;579C:35–40. doi: 10.1016/j.neulet.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 2.Kai Y. The content of intracellular mitochondrial DNA is decreased by 1-methyl-4-phenylpyridinium ion (MPP+) J Biol Chem. 1997;272:9605–9608. doi: 10.1074/jbc.272.15.9605. [DOI] [PubMed] [Google Scholar]
- 3.Pizarro JG, Junyent F, Verdaguer E, Jordan J, Beas-Zarate C, Pallas M, Camins A, Folch J. Effects of MPP+ on the molecular pathways involved in cell cycle control in B65 neuroblastoma cells. Pharmacol Res. 2010;61:391–399. doi: 10.1016/j.phrs.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Hartley A, Stone JM, Heron C, Cooper JM, Schapira AH. Complex I inhibitors induce dose-dependent apoptosis in PC12 cells: relevance to Parkinson’s disease. J Neurochem. 1994;63:1987–1990. doi: 10.1046/j.1471-4159.1994.63051987.x. [DOI] [PubMed] [Google Scholar]
- 5.Bai J, Nakamura H, Ueda S, Kwon YW, Tanaka T, Ban S, Yodoi J. Proteasome-dependent degradation of cyclin D1 in 1-methyl-4-phenylpyridinium ion (MPP+)-induced cell cycle arrest. J Biol Chem. 2004;279:38710–38714. doi: 10.1074/jbc.M403329200. [DOI] [PubMed] [Google Scholar]
- 6.Bloem BR, Irwin I, Buruma OJ, Haan J, Roos RA, Tetrud JW, Langston JW. The MPTP model: versatile contributions to the treatment of idiopathic Parkinson’s disease. J Neurol Sci. 1990;97:273–293. doi: 10.1016/0022-510x(90)90225-c. [DOI] [PubMed] [Google Scholar]
- 7.Langston JW, Irwin I. MPTP: current concepts and controversies. Clin Neuropharmacol. 1986;9:485–507. [PubMed] [Google Scholar]
- 8.Albani D, Polito L, Batelli S, De Mauro S, Fracasso C, Martelli G, Colombo L, Manzoni C, Salmona M, Caccia S, Negro A, Forloni G. The SIRT1 activator resveratrol protects SK-N-BE cells from oxidative stress and against toxicity caused by alpha-synuclein or amyloid-beta (1-42) peptide. J Neurochem. 2009;110:1445–1456. doi: 10.1111/j.1471-4159.2009.06228.x. [DOI] [PubMed] [Google Scholar]
- 9.Alvira D, Yeste-Velasco M, Folch J, Verdaguer E, Canudas AM, Pallas M, Camins A. Comparative analysis of the effects of resveratrol in two apoptotic models: inhibition of complex I and potassium deprivation in cerebellar neurons. Neuroscience. 2007;147:746–756. doi: 10.1016/j.neuroscience.2007.04.029. [DOI] [PubMed] [Google Scholar]
- 10.Pallas M, Pizarro JG, Gutierrez-Cuesta J, Crespo-Biel N, Alvira D, Tajes M, Yeste-Velasco M, Folch J, Canudas AM, Sureda FX, Ferrer I, Camins A. Modulation of SIRT1 expression in different neurodegenerative models and human pathologies. Neuroscience. 2008;154:1388–1397. doi: 10.1016/j.neuroscience.2008.04.065. [DOI] [PubMed] [Google Scholar]
- 11.Ferretta A, Gaballo A, Tanzarella P, Piccoli C, Capitanio N, Nico B, Annese T, Di Paola M, Dell’aquila C, De Mari M, Ferranini E, Bonifati V, Pacelli C, Cocco T. Effect of resveratrol on mitochondrial function: implications in parkin-associated familiar Parkinson’s disease. Biochim Biophys Acta. 2014;1842:902–915. doi: 10.1016/j.bbadis.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 12.Ye J, Liu Z, Wei J, Lu L, Huang Y, Luo L, Xie H. Protective effect of SIRT1 on toxicity of microglial-derived factors induced by LPS to PC12 cells via the p53-caspase-3-dependent apoptotic pathway. Neurosci Lett. 2013;553:72–77. doi: 10.1016/j.neulet.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 13.Donmez G, Outeiro TF. SIRT1 and SIRT2: emerging targets in neurodegeneration. EMBO Mol Med. 2013;5:344–352. doi: 10.1002/emmm.201302451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang F, Wang S, Gan L, Vosler PS, Gao Y, Zigmond MJ, Chen J. Protective effects and mechanisms of sirtuins in the nervous system. Prog Neurobiol. 2011;95:373–395. doi: 10.1016/j.pneurobio.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grayson DR, Kundakovic M, Sharma RP. Is there a future for histone deacetylase inhibitors in the pharmacotherapy of psychiatric disorders? Mol Pharmacol. 2010;77:126–135. doi: 10.1124/mol.109.061333. [DOI] [PubMed] [Google Scholar]
- 16.Feng Y, Jankovic J, Wu YC. Epigenetic mechanisms in Parkinson’s disease. J Neurol Sci. 2015;349:3–9. doi: 10.1016/j.jns.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 17.Konsoula Z, Barile FA. Epigenetic histone acetylation and deacetylation mechanisms in experimental models of neurodegenerative disorders. J Pharmacol Toxicol Methods. 2012;66:215–220. doi: 10.1016/j.vascn.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Feng Y, Liu T, Dong SY, Guo YJ, Jankovic J, Xu H, Wu YC. Rotenone affects p53 transcriptional activity and apoptosis via targeting SIRT1 and H3K9 acetylation in SH-SY5Y cells. J Neurochem. 2015;134:668–676. doi: 10.1111/jnc.13172. [DOI] [PubMed] [Google Scholar]
- 19.Sharp FR, Bernaudin M. HIF1 and oxygen sensing in the brain. Nat Rev Neurosci. 2004;5:437–448. doi: 10.1038/nrn1408. [DOI] [PubMed] [Google Scholar]
- 20.Permenter MG, Dennis WE, Sutto TE, Jackson DA, Lewis JA, Stallings JD. Exposure to cobalt causes transcriptomic and proteomic changes in two rat liver derived cell lines. PLoS One. 2013;8:e83751. doi: 10.1371/journal.pone.0083751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Correia SC, Moreira PI. Hypoxia-inducible factor 1: a new hope to counteract neurodegeneration? J Neurochem. 2010;112:1–12. doi: 10.1111/j.1471-4159.2009.06443.x. [DOI] [PubMed] [Google Scholar]
- 22.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutt DM, Roth DM, Vignaud H, Cullin C, Bouchecareilh M. The histone deacetylase inhibitor, Vorinostat, represses hypoxia inducible factor 1 alpha expression through translational inhibition. PLoS One. 2014;9:e106224. doi: 10.1371/journal.pone.0106224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ke Q, Costa M. Hypoxia-inducible factor-1 (HIF-1) Mol Pharmacol. 2006;70:1469–1480. doi: 10.1124/mol.106.027029. [DOI] [PubMed] [Google Scholar]
- 25.Lim S, Janzer A, Becker A, Zimmer A, Schule R, Buettner R, Kirfel J. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31:512–520. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- 26.Wu Y, Li X, Zhu JX, Xie W, Le W, Fan Z, Jankovic J, Pan T. Resveratrol-activated AMPK/SIRT1/autophagy in cellular models of Parkinson’s disease. Neurosignals. 2011;19:163–174. doi: 10.1159/000328516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krug AK, Gutbier S, Zhao L, Pöltl D, Kullmann C, Ivanova V, Förster S, Jagtap S, Meiser J, Leparc G, Schildknecht S, Adam M, Hiller K, Farhan H, Brunner T, Hartung T, Sachinidis A, Leist M. Transcriptional and metabolic adaptation of human neurons to the mitochondrial toxicant MPP+ Cell Death Dis. 2014;5:e1222. doi: 10.1038/cddis.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patten DA, Germain M, Kelly MA, Slack RS. Reactive oxygen species: stuck in the middle of neurodegeneration. J Alzheimers Dis. 2010;20:S357–S367. doi: 10.3233/JAD-2010-100498. [DOI] [PubMed] [Google Scholar]
- 29.Seffer I, Nemeth Z, Hoffmann G, Matics R, Seffer AG, Koller A. Unexplored potentials of epigenetic mechanisms of plants and animals-theoretical considerations. Genet Epigenet. 2013;5:23–41. doi: 10.4137/GEG.S11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuda KI. Epigenetic changes in the estrogen receptor alpha gene promoter: implications in sociosexual behaviors. Front Neurosci. 2014;8:344. doi: 10.3389/fnins.2014.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graff J, Tsai LH. Histone acetylation: molecular mnemonics on the chromatin. Nat Rev Neurosci. 2013;14:97–111. doi: 10.1038/nrn3427. [DOI] [PubMed] [Google Scholar]
- 32.Chen PS, Peng GS, Li G, Yang S, Wu X, Wang CC, Wilson B, Lu RB, Gean PW, Chuang DM, Hong JS. Valproate protects dopaminergic neurons in midbrain neuron/glia cultures by stimulating the release of neurotrophic factors from astrocytes. Mol Psychiatry. 2006;11:1116–1125. doi: 10.1038/sj.mp.4001893. [DOI] [PubMed] [Google Scholar]
- 33.Zhu M, Li WW, Lu CZ. Histone decacetylase inhibitors prevent mitochondrial fragmentation and elicit early neuroprotection against MPP+ CNS Neurosci Ther. 2014;20:308–316. doi: 10.1111/cns.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]