Abstract
Glucosinolates (GSLs) are secondary metabolites that have anticarcinogenic activity and play defense roles in plants of the Brassicaceae family. MYB28 is known as a transcription factor that regulates aliphatic GSL biosynthesis in Arabidopsis thaliana. Brassicaceae plants have three orthologous copies of AtMYB28 derived from recent genome triplication. These BrMYB28 genes have a high level of sequence homology, with 81–87 % similarities in the coding DNA sequence compared to Arabidopsis. Overexpression of three paralogous BrMYB28 genes in transgenic Chinese cabbage increased the total GSL content in all T1 generation plants and in two inbred lines of homozygous T2 plants. The highest total GSL contents were detected in homozygous T2 lines overexpressing BrMYB28.1, which showed an approximate fivefold increase compared to that of nontransgenic plants. The homozygous T2 lines with overexpressed BrMYB28.1 also showed an increased content of aliphatic, indolic, and aromatic GSLs compared to that of nontransgenic plants. Furthermore, all of the three BrMYB28 genes were identified as negative regulators of BrAOP2 and positive regulators of BrGSL-OH in the homozygous T2 lines. These data indicate the regulatory mechanism of GSL biosynthesis in B. rapa is unlike that in A. thaliana. Our results will provide useful information for elucidating the regulatory mechanism of GSL biosynthesis in polyploid plants.
Electronic supplementary material
The online version of this article (doi:10.1007/s11103-016-0437-z) contains supplementary material, which is available to authorized users.
Keywords: Brassica rapa, Glucosinolates, Transcription factor BrMYB28s, Chinese cabbage, Transgenic plats
Introduction
Plants produce various secondary metabolites that play roles in plant defense against environmental change or stress, but are unrelated to the primary functions of plants, such as development, reproduction, and photosynthesis. More than 200,000 plant secondary metabolites are known, which perform useful, although not necessarily essential, functions in plant survival (Verpoorte et al. 2002; Yonekura-Sakakibara and Saito 2009). One of the largest known groups of secondary metabolites in the Brassicaceae family is the glucosinolates (GSLs)—compounds derived from amino acids and sugars (Hayes et al. 2008). GSLs are classified into three groups: aliphatic, derived from Met, Ala, Leu, Ile, or Val; indolic, derived from Trp; and aromatic, derived from Phe or Tyr (Kliebenstein et al. 2001). The biosynthesis of the three types of GSLs generally occurs via three stages. The precursor amino acids are chain-elongated in the early stage, and then converted into the core GSL structure during the second stage. The final stage involves side chain modification of the GSL structure (Zang et al. 2009). The compounds produced by the GSL biosynthesis pathway have recently attracted a high level of research interest, owing to their various beneficial activities, such as anticarcinogenic and antioxidative activity in mammals, and defense against pests and pathogens in plants (Sawada et al. 2009; Yonekura-Sakakibara and Saito 2009). Therefore, an understanding of the regulation of GSL biosynthesis will provide useful information for the study of plant secondary metabolism, and thereby improvements in the value of agricultural crops.
Studies of the mechanisms underlying GSL biosynthesis have mostly been performed in the model plant Arabidopsis thaliana using a molecular biology approach (Kliebenstein et al. 2001; Skirycz et al. 2006; Sønderby et al. 2010). Recently, genes controlling GSL biosynthesis have been identified in Arabidopsis. Certain R2R3 MYB transcription factors are known to participate in GSL regulation in Arabidopsis (Sønderby et al. 2010). For example, MYB28 has been identified as a positive regulator of aliphatic GSLs and high transcription levels of the MYB28 are associated with the production of large amounts of aliphatic GSLs (Hirai et al. 2007). Furthermore, the transcription factors MYB29, MYB51, and MYB76 have recently been shown to regulate the biosynthesis of indolic GSLs (Gigolashvili et al. 2007; Sønderby et al. 2007).
Recently, studies on GSL content regulation have investigated candidate genes using a genetic approach. The RsMAM3, RsIPMDH1, and RsBCAT4 genes as candidate genes controlling 4-MTB-GSL content were identified in radish roots by QTL analysis using SNP markers developed by next-generation sequencing (Zou et al. 2013). The transcription factor MYB28 (HAG1), which participates in aliphatic GSL biosynthesis, was also identified by QTL analysis using associative transcriptomics by association between genetic variation and trait variation in the seeds of Brassica napus (Harper et al. 2012). However, although some studies have identified a few candidate genes and QTLs controlling GSL contents, functional validation of the candidate genes is very limited in Brassica crops owing to the difficulty of genetic modification.
The B. rapa genome has undergone genome triplication. A representative late flowering BrFLCs genes was obtained by selecting five BrFLC genes for sequence comparisons of triplication blocks for At5_3Mb (Kim et al. 2006; Yang et al. 2006). In addition, the genome sequence project for B. rapa as model for polyploid genome studies was undertaken (Wang et al. 2011). The B. rapa genome sequence provides an important resource for studying the evolution of polyploid genomes and a foundation for the genetic improvement of Brassica oil and vegetable crops.
Using the B. rapa genome sequence, many genes related to GSL biosynthesis pathways were identified by comparative genome analysis with Arabidopsis (Zang et al. 2009). The results predictably suggested that the complexity of GSL biosynthesis regulation in B. rapa is greater than that in Arabidopsis because of the polyploid B. rapa genome. Polyploidy of genomes is a very important event for plant evolution and diversity. Polyploidy leads to amplification or redundancy of genes, and duplicated genes with different expression patterns, such as silencing or strong expression, have arisen compared with the ancestral function. Allopolyploid plants also show various plant-specific expression patterns of genes duplicated by polyploidy (Udall et al. 2006).
The existence of multiple paralog genes in the polyploid B. rapa genome suggests the possibility for variation of expression and function among the paralogs of candidate genes. Although functional studies for candidate GSL biosynthesis genes have been performed in a few Brassica crops, the functional analysis of paralog genes in B. rapa is currently still very limited (Augustine et al. 2013). Moreover, no transgenic B. rapa has been produced for paralog genes related to GSL biosynthesis because of technical difficulties associated with the genetic transformation of B. rapa.
The MYB28 transcription factor, which is known to be a key regulator of aliphatic GSL biosynthesis, has been determined to have three paralogs, as a result of genome triplication in B. rapa (Zang et al. 2009). In the present study, we identified the three BrMYB28 transcription factors (designated BrMYB28.1, BrMYB28.2, and BrMYB28.3) and characterized their gene structure and variation in expression. Furthermore, in a functional study, we successfully over-expressed the three paralogous BrMYB28 transcription factors in transgenic B. rapa. Understanding the structure and function of the BrMYB28 transcription factors will facilitate analysis of the mechanisms underlying GSL biosynthesis in B. rapa using a molecular approach.
Materials and methods
Isolation of three BrMYB28 genes from B. rapa
Genomic information on the GSL biosynthesis-related transcription factors of B. rapa was obtained from the National Center for Biotechnology Information (NCBI) and the Brassica Database (http://brassicadb.org/brad/). PCR amplification was performed on total genomic DNA of the ‘Chiifu’ cultivar of B. rapa using gene-specific primers designed based on genomic information. The gene-specific primers of the three BrMYB28 genes were designed for gateway cloning (Invitrogen, USA). PCR was performed with 30 cycles (94 °C for 30 s, 52 °C for 30 s, and 72 °C for 1.5 min) and a final extension at 72 °C for 10 min. PCR products were cloned into a pDONR221 vector to prepare an entry clone through the use of BP clonase (Invitrogen, USA). After transformation into Escherichia coli DH5α competent cells, plasmids were isolated and genomic sequences were validated using an ABI 3730xl sequencer. The gene structures were predicted by sequence comparison with the Arabidopsis sequence. The BrMYB28 genes with complete coding sequence were analyzed for molecular characterization, and the R2R3 binding domain was predicted by SMART analysis (http://smart.embl-heidelberg.de/). A multiple alignment analysis of BrMYB28 and AtMYB28 was performed using ClustalW2. A phylogenetic tree was obtained using MEGA6 software based on the previously published MYB transcription factors related to the GSL biosynthesis pathway in B. rapa and A. thaliana.
Expression analysis of BrMYB28 transcription factors
Total RNA was isolated from various organs of B. rapa ‘Chiifu’ using an RNeasy mini kit (Qiagen, USA), and treated with RNase-free DNase I (Takara, Japan) to eliminate contaminated genomic DNA. One microgram of total RNA was used as a template for RT-PCR using an AMV one-step RT-PCR kit (Takara, Japan). The primers for member-specific detection of the expression of BrMYB28s were respectively designed for the 5′ and 3′ UTR regions. The BrActin gene primer was used as a control for all expression analyses. All the specific primers used in this study are listed in supplementary Table S1. The PCR reaction comprised predenaturating at 94 °C for 5 min, followed by 35 cycles of denaturation (94 °C for 30 s), annealing (52 °C for 30 s), and extension (72 °C for 1.5 min), and with a final extension of 10 min at 72 °C. The PCR products were visualized on a 1.2 % agarose gel stained with ethidium bromide. The RT-PCRs were performed with two replicates as a check for experimental fidelity. The expression profile of BrMYB28 genes related to the developmental stages in B. rapa was examined by analyzing the microarray and unigene databases (http://nabic.rda.go.kr).
Genetic transformation of Chinese cabbage
Two inbred lines (NW and CT001) of Chinese cabbage (B. rapa ssp. pekinensis), i.e., plant materials for which successful transformation was previously reported (Min et al. 2007; Park et al. 2011), were used. These lines were kindly provided by the Nong Woo Bio Co. (NW) and Carrotop Seed Company (CT001) in Korea. Mature seeds were surface-sterilized in 70 % ethanol for 1 min and in 2 % sodium hypochlorite for 20 min, and then rinsed 3 times with sterile distilled water. Seeds were placed on 1/2 MS medium (Murashige and Skoog 1962) solidified with 7.5 g/L plant agar. The plates were incubated at 25 °C in the dark for 5 days. The hypocotyls germinated from the mature seeds were cut to a length of approximately 8 mm and placed on MS medium supplemented with 3 mg/L BA (6-benzylaminopurine), 1 mg/L NAA (1-naphthalene acetic acid) and solidified with 7 g/L plant agar. Hypocotyls were incubated at 22 °C under a 16/8 h light/dark photoperiod for 3 days.
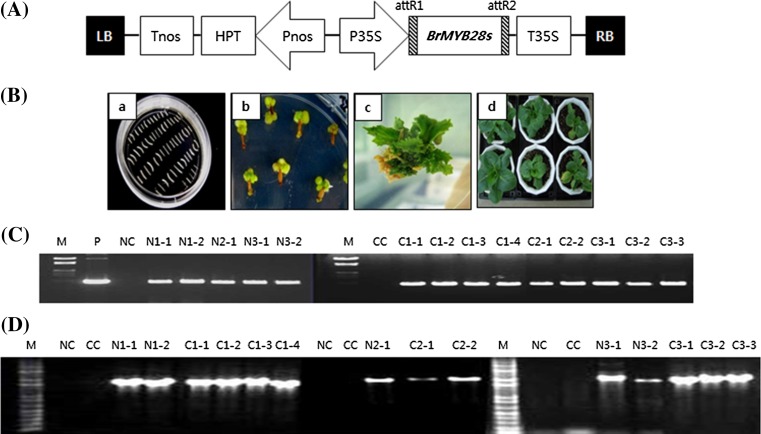
For transformation with the three BrMYB28 genes, the clones within the pDONR221 entry vector (obtained as described above) were inserted into a pH2GW7 binary vector using LR clonase (Invitrogen, USA). The binary vector contains a selectable hygromycin resistance marker gene. The disarmed Agrobacterium tumefaciens strain GV3101 containing the BrMYB28 constructs were used for transformation (Fig. 4A). Agrobacterium carrying BrMYB28 TFs were grown at 28 °C in the dark for 24 h on LB medium supplemented with 100 mg/L spectinomycin and harvested by centrifugation. The bacterial pellets were resuspended in MS suspension medium containing 36 g/L glucose and 5 mg/L acetosyringone, and then the hypocotyl explants were immersed in the bacterial suspension for 20 min. The explants were co-cultivated on co-culture medium (MS medium containing 3 mg/L BA, 1 mg/L NAA, 1 mg/L AgNO3, and 5 mg/L acetosyringone) for 3 days at 23 °C in the dark. After co-cultivation, the explants were rinsed three times in distilled water containing 250 mg/L cefotaxime to kill the Agrobacterium. The explants were transferred to selection medium (MS medium containing 3 mg/L BA, 1 mg/L NAA, and 1 mg/L AgNO3, 250 mg/L cefotaxime, and 10 mg/L hygromycin) and then incubated at 23 °C under a 16/8 h light/dark photoperiod. Hygromycin-resistant shoots that regenerated from the hypocotyls were transferred to an MS regeneration medium (MS medium containing 3 mg/L BA, 0.1 mg/L NAA, 1 mg/L AgNO3, 250 mg/L cefotaxime, and 10 mg/L hygromycin) for shoot elongation. The regenerated hygromycin-resistant plantlets were transferred to MS hormone-free medium containing 150 mg/L cefotaxime, and 10 mg/L hygromycin for further root development. The regeneration was carried out at 23 °C under a 16/8 h light/dark photoperiod. The hygromycin-resistant plantlets were transferred to soil in pots and grown to maturity in a greenhouse. Genomic DNA was extracted from the leaves of nontransformant and hygromycin-resistant plants. Hygromycin-resistant T0 plants were identified from the insertion of hpt and BrMYB28 genes by PCR using gene-specific primers (Supplementary Table 1). PCR for the three BrMYB28 genes was performed using the gene-specific forward and reverse primers of the 35S terminator. The PCR was performed in a thermal cycler using the following amplification conditions: 35 cycles of 30 s at 94 °C, 30 s at 52 °C, and 2 min at 72 °C. The primers used for the 757-bp hpt gene fragment were 5′-ATTCCGGAAGTGCTTGACAT-3′(forward) and 5′-CGGCGAGTACTTCTACACAGC-3′(reverse). Thermal cycling conditions for the hpt gene were as follows: 30 cycles of 30 s at 94 °C, 30 s at 58 °C, and 1 min at 72 °C.
Fig. 4.
Schematic diagram of part of the T-DNA region of the binary vector and PCR analysis of transgenic Chinese cabbage. A T-DNA region of binary vector construct used for Agrobacterium-mediated transformation. LB, left border; RB, right border; 35S Pro, CaMV 35S promoter; Pnos, Nos promoter; Tnos, Nos terminator, HPT, hygromycin resistance gene. B Agrobacterium-mediated transformation of BrMYB28 genes in Chinese cabbage. a Hypocotyl explants of B. rapa used for transformation. b Hygromycin-resistant callus induced from hypocotyl in selection medium containing 10 mg/L hygromycin. c Hygromycin resistance shoot regenerated from callus in regeneration medium containing 10 mg/L hygromycin. d The hygromycin-resistant plantlets were transferred to soil in pots and grown to maturity in a greenhouse with non-transgenic plants (leftmost panel). C, D Detection of the hpt gene (C) and the three BrMYB28 genes (D) in hygromycin-resistant plants (T1) by PCR analysis. The PCR products were identified at 757 bp for the hpt gene and 1454 bp for BrMYB28.1, 1581 bp for BrMYB28.2, and 1821 bp for BrMYB28.3. M, molecular weight marker; P, plasmid DNA; NC, CC, nontransgenic NW line (NC) and CT001 line (CC); N1-1,2, NW BrMYB28.1 gene transgenic plants; N2-1, NW BrMYB28.2 gene transgenic plant; N3-1,2, NW BrMYB28.3 gene transgenic plants; C1-1–4, CT001 BrMYB28.1 gene transgenic plants; C2-1–2; CT001 BrMYB28.2 gene transgenic plants; C3-1–3, CT001 BrMYB28.3 gene transgenic plants
Production of T1 and T2 progeny
T0 plants were vernalized at 4 °C and the T1 seeds were obtained by self-crossing. The T1 seeds were surface-sterilized and then grown on MS medium supplemented with hygromycin (20 mg/L). Hygromycin-resistant T1 transgenic seedlings were transplanted into soil. The T1 progenies with a segregation ratio of 3 (resistant):1(sensitive) determined by the χ2 test were self-crossed and T2 seeds were obtained. The T2 seeds were germinated on MS medium containing hygromycin (20 mg/L) for the selection of homozygous transgenic lines, and homozygous T2 transgenic hygromycin-resistant seedlings geminated from all of the seeds. All of the T1 and T2 plants were identified to contain the inserted hpt gene and BrMYB28 genes by PCR analysis.
Gene expression in transgenic plants
Real-time PCR was carried out to investigate the transcription levels of BrMYB28 genes and the GSL structural genes in transgenic Chinese cabbage. Total RNA was isolated as described above. Approximately 2 µg of total RNA was reverse transcribed into cDNA with oligo-dT primers using a first-strand cDNA synthesis kit (Genedepot). The synthesized cDNAs were diluted 10 times in sterilized water and real-time PCR was performed using 2 µL of diluted cDNA in 20 µL using SYBR Green mix (Geneall). The primers for member-specific detection of the expression of BrMYB28s were designed for the 3′ terminal region. The gene-specific primers used for PCR analysis are shown in Supplementary Table S1. We used the thermal cycler conditions recommended by the manufacturer as follows: 95 °C for 10 min, 55 °C for 30 s, and 72 °C for 30 s, with a final extension at 72 °C for 3 min.
HPLC analysis of GSL contents
Desulfo (DS)-GSLs were extracted according to the procedure of Kim et al. (2007) and ISO 9167-1 (1992). Fresh leaves of 50-day-old plants were ground with a mortar and pestle into fine powder, and freeze-dried materials (100 mg) were extracted twice in 70 % methanol. As an external standard, we used 0.5 mg of sinigrin dissolved in 5 mL ultrapure water. The crude extracts were loaded on Sephadex A25 columns and desulfated overnight using aryl sulfatase (E.C.3.1.6.1) prior to HPLC. DS-GSLs were analyzed using a 1200 series HPLC system (Agilent Technologies, CA, USA) equipped with an Inertsil ODS-3 column [150 × 3.0 mm ID, particle size 3 µm (GL Science, Tokyo, Japan)]. The HPLC analysis was carried out using a flow rate of 0.4 mL/min at a column oven temperature of 35 °C and a wavelength of 227 nm. The individual GSLs were quantified by comparison with the external standard sinigrin, and the values for total GSLs were obtained by summing the values of the individual GSLs identified (Supplementary Table S3).
Results
Genomic sequence analysis of three MYB28s isolated from B. rapa
The three orthologous copies in B. rapa corresponding to AtMYB28 identified by comparative analysis with Arabidopsis using the Brassica Database (http://brassicadb.org/brad/) in Table 1. The sequences of the three genes were as follows: 1350 nucleotides long for BrMYB28.1, 1378 for BrMYB28.2, and 1618 for BrMYB28.3. These are all longer than the sequence of AtMYB28. The three BrMYB28 transcription factors of B. rapa showed 81–87 % sequence homology with AtMYB28 (Supplementary Fig. 1). The structures of three BrMYB28 genes comprise 3 exons and 2 introns, which is comparable to AtMYB28. The first and second exons have a very similar size (133 and 130 nucleotides, respectively) and show orthologous and paralogous sequence variation. These three paralogous genes anchored BAC clones with KBrB034G03, KBrB051M06, and KBrH005L20, respectively, and are located on the A03, A09, and A02 chromosomes of B. rapa. After divergence of Arabidopsis and Brassicaceae and triplication of the Brassica genome, these genes may have undergone sequence substitutions, as well as insertion and deletion.
Table 1.
Comparison of the BrMYB28 TFs related glucosinolate biosynthesis with the Arabidopsis orthologs
| Gene name | Length (bp) | No. of exsons [length (bp)] | No. of introns [length (bp)] | Chromosome position of B. rapa | Corresponding B. rapa | ||
|---|---|---|---|---|---|---|---|
| Gene | CDS | BAC clone | EST clone | ||||
| AtMYB28(AT5G61420a) | 1321 | 1101 | 3 (133,130,838) | 2 (80,140) | – | – | – |
| BrMYB28.1(Bra012961) | 1350 | 1065 | 3 (133,130,802) | 2 (89,196) | A03 | KBrB034G03 | KBLS-095C01 |
| BrMYB28.2(Bra035929) | 1378 | 1074 | 3 (133,130,811) | 2 (166,138) | A09 | KBrB051M06 | KBFL-120H07 |
| BrMYB28.3(Bra029311) | 1618 | 1119 | 3 (133,130,856) | 2 (83,416) | A02 | KBrH005L20 | KFFB-103G11 |
aArabidopsis genome ID of NCBI and Brassica rapa gene ID of BRAD
Amino acid sequence comparison of BrMYB28 genes
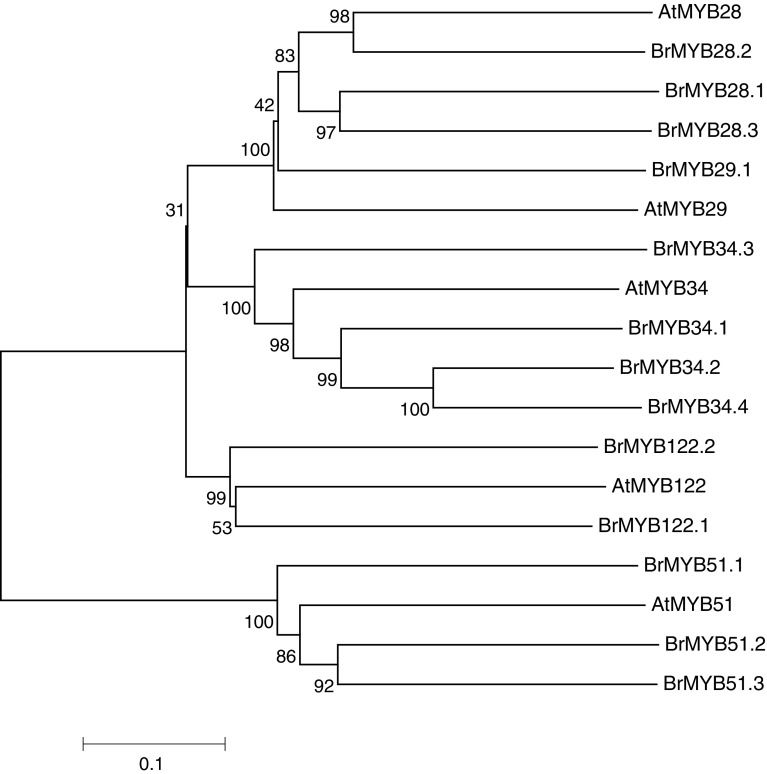
Alignment and phylogenetic analyses of the three BrMYB28 proteins indicated that they are highly conserved. Multi-alignment revealed that they have two typical R2R3 MYB-DNA-binding domains (Fig. 1). An analysis of the deduced amino acid sequences indicated that BrMYB28.1, BrMYB28.2, and BrMYB28.3 contain conserved R2R3 repeat MYB-DNA binding domains that share high amino acid sequence similarities of 94–99 % with AtMYB28. In contrast, the C-terminal region was shown to be highly polymorphic. These results indicate that amino acid sequence variations in the C-terminal region have led to the structural divergence of the BrMYB28 transcription factors. The structural divergence of the three BrMYB28 genes suggests the possibility of functional divergence of these genes in B. rapa. A phylogenetic tree was constructed using the deduced amino acid sequences of 14 MYB transcription factors related to the GSL biosynthesis pathways in A. thaliana and B. rapa (Fig. 2). In this tree, BrMYB28.1 is more related to BrMYB28.3 than to BrMYB28.2. BrMYB28.1 and BrMYB28.3 proteins were clustered in a small subgroup, whereas AtMYB28 was clustered with BrMYB28.2. The subgroup with AtMYB29 and BrMYB29.1 forms a distinct large group. Therefore, the high homology of the BrMYB28 and BrMYB29 transcription factors in this large group indicates that they are evolutionary conserved and closely related.
Fig. 1.
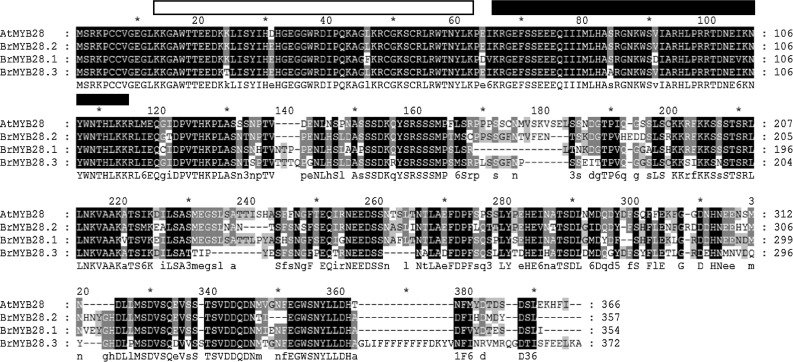
Amino acid sequence alignment of MYB28 proteins that regulate the glucosinolate biosynthesis pathway of B. rapa and A. thaliana. The R2 and R3 binding domains are boxed in white and black, respectively
Fig. 2.
Phylogenetic analysis of MYB TFs related to the glucosinolate biosynthesis pathway in B. rapa and Arabidopsis. This tree was constructed using MEGA, version 6, software. Bootstrap values with 1000 replicates are denoted as percentages
Differential expression of BrMYB28 genes in B. rapa
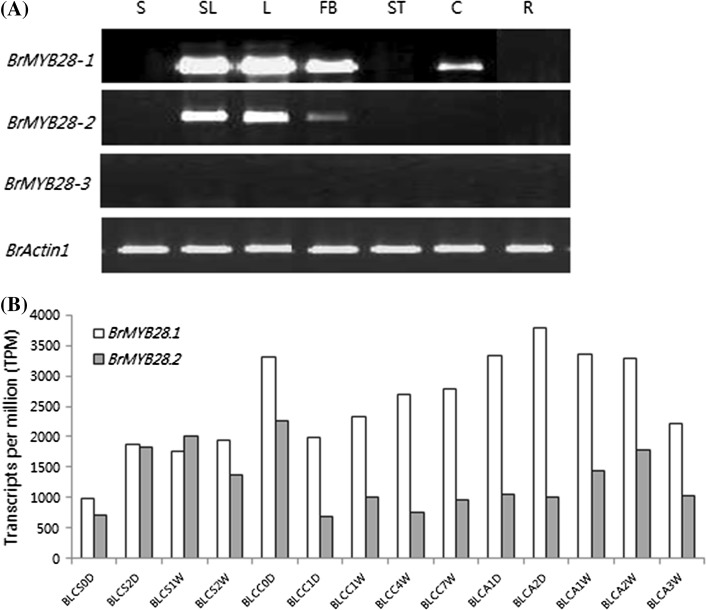
To identify the transcription levels of BrMYB28 genes in various organs and developmental stages of B. rapa, the expression patterns of these genes were analyzed by RT-PCR and microarray (Fig. 3). The BrMYB28.3 gene was not expressed in all organs and growth stages. The expression patterns of BrMYB28.1 and BrMYB28.2 were similar in all organs except the carpels (Fig. 3A). These two genes were most highly expressed in leaves, but showed no expression in seeds, stamens, and roots. The BrMYB28.1 gene showed the highest transcription levels in various organs of B. rapa. The expression profiles of BrMYB28.1 and BrMYB28.2 in different developmental stages of B. rapa analyzed using the microarray database is shown in Fig. 3B. The expression of both these genes was observed in most developmental stages. Maximum expression was detected at 72 days for BrMYB28.1 and 21 days for BrMYB28.2. BrMYB28.1 was more highly expressed than BrMYB28.2 in various developmental stages, consistent with the results of RT-PCR analysis. Expression of the two genes was markedly decreased by chilling treatment; however, the expression levels of BrMYB28.1 increased with time. Higher levels of BrMYB28.2 expression were detected in vegetative stages (BLCS2D–BLCC0D) than in reproductive stages (BLCA1D–BLCA3W), whereas for BrMYB28.1, higher expression levels were detected in the reproductive stages. Therefore, these paralogous BrMYB28 genes have different expression patterns in various organs and developmental stages.
Fig. 3.
Expression analysis of BrMYB28 TFs in various organs and developmental stages. A RT-PCR analysis of BrMYB28 TFs in different types of tissues. S, seed; SL, seedling; L, mature leaf (3-week-old vegetative stage); FB, floral bud; ST, stamen; C, carpel; R, root. The PCR products are approximately 1 kb for BrMYB28 genes. BrActin1 is approximately 500 bp and serves as an internal control. B Microarray expression analysis of BrMYB28 TFs in different growth stages. BLCS0D, seeds; BLCS2D, seedling (2 days old); BLCS1W, whole plant, 1-week-old vegetative stage (7 days old); BLCS2W, whole plant, 2-week-old vegetative stage (14 days old); BLCC0D, whole plant, 3-week-old vegetative stage (21 days old); BLCC1D, whole plant, 1 day after light chilling at 4 °C (22 days old); BLCC1W, whole plant, 1 week after light chilling at 4 °C (28 days old); BLCC4W, whole plant, 4 weeks after light chilling at 4 °C (56 days old); BLCC7W, whole plant, 7 weeks after light chilling at 4 °C (70 days old); BLCA1D, whole plant, 1 day after greenhouse growth (71 days old); BLCA2D, whole plant, 2 days after greenhouse growth (72 days old); BLCA1W, whole plant, 1 week after greenhouse growth (77 days old); BLCA2W, whole plant, 2 weeks after greenhouse growth (84 days old); BLCA3W, whole plant, 3 weeks after greenhouse growth (91 days old)
Genetic transformation of the three BrMYB28 genes in Chinese cabbage
The three BrMYB28 genes were transferred into two different Chinese cabbage inbred lines using Agrobacterium-mediated transformation (Fig. 4B). The hypocotyls were infected by co-cultivation with Agrobacterium containing each of the three BrMYB28 genes, and shoots were regenerated in selection medium containing 10 mg/L hygromycin. The hygromycin-resistant plants were obtained after 3 months in regeneration medium and grown to maturity in a greenhouse. T1 seeds were obtained from the self-crossed T0 plants, and the segregation ratio was determined in MS medium containing 20 mg/L hygromycin (Supplementary Table 2). To identify gene integration into T1 plants with a segregation ratio of 3:1, PCR amplification was performed (Fig. 4C). PCR amplification of the 757-bp hpt gene products was observed in all hygromycin-resistant plants, whereas this band was not detected in nontransgenic plants. T1 plants containing hpt were also identified by PCR amplification of BrMYB28 genes containing the 35S terminator (Fig. 4D). The T1 plants were self-crossed and T2 seeds were germinated on MS medium containing hygromycin for selection of transgenic homozygous lines (Supplementary Table S2). The T2 homozygous lines were obtained as nonsegregated resistant plants in all T1 plants with a segregation ratio of 3:1. T1 and T2 transgenic plants overexpressing BrMYB28 genes showed normal phenotypes compared to the nontransgenic plants (Fig. 4B–d). However, the homozygous T2 plants obtained from C2-1 and C2-2 transgenic T1 plants did not grow well. In our future studies, we will further attempt to obtain homozygous T2 plants from C2-1 and C2-2.
GSL content variation in T1 transgenic plants containing overexpressed BrMYB28 genes
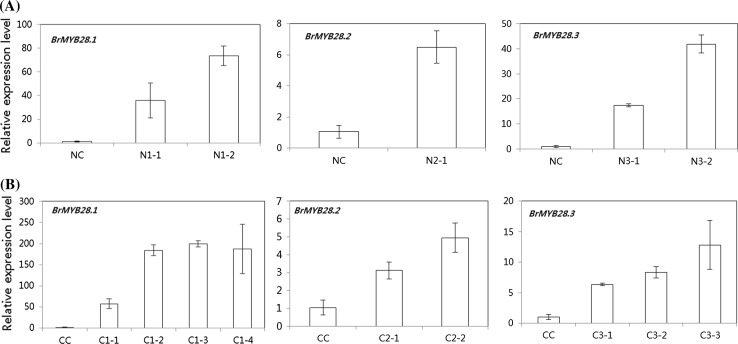
Six-week-old leaves were used for analysis of transcript levels and GSL content in BrMYB28 transgenic plants. Real-time PCR demonstrated that the BrMYB28 genes were overexpressed efficiently in all of the T1 transgenic Chinese cabbage plants (Fig. 5). Significantly up-regulated expression of BrMYB28 genes was observed in all transgenic plants compared to nontransgenic plants; moreover, the expression levels varied in the transgenic plants. The relative gene expression levels were increased by approximately 35- to 185-fold for BrMYB28.1, 5- to 8.5-fold for BrMYB28.2, and 8- to 41-fold for BrMYB28.3 compared with the nontransgenic plants. To determine which GSLs are increased in transgenic plant, we measured GSL accumulation in T1 transgenic plants by HPLC analysis. The overexpression of BrMYB28 genes in transgenic plants elevated the accumulation of total GSLs by 1.8- to 5.6-fold for NW lines and 1.2- to 2.4-fold for CT001 lines compared with the nontransgenic plants (Fig. 6, Supplementary Table S4). Both aliphatic and indolic GSLs were found to be elevated in NW transgenic plants. In contrast, the aliphatic GSLs were decreased in some CT001 transgenic plants.
Fig. 5.
Expression of genes involved in glucosinolate biosynthesis in the 6-week-old leaves of T1 transgenic NW (A) and CT001 (B) plants. Relative expression was determined in triplicate measurements in three independent biological replicates. The Bractin gene was used as a quantitative control. NC, CC, non-transgenic plants; N1-1,2, NW BrMYB28.1 gene transgenic plants; C1-1–4, CT001 BrMYB28.1 gene transgenic plants; N2-1, NW BrMYB28.2 gene transgenic plant; C2-1–3, CT001 BrMYB28.2 gene transgenic plants; N3-1,2, NW BrMYB28.3 gene transgenic plants; C3-1–3, CT001 BrMYB28.3 gene transgenic plants
Fig. 6.
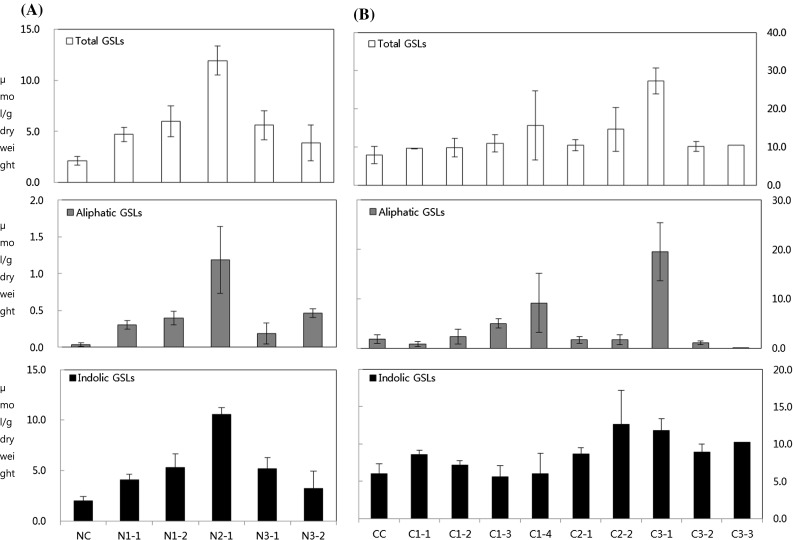
HPLC analysis of GSL content in leaves of T1 transgenic NW (A) and CT001 (B) plants. Values are the means of 3 replications. Bars represent the standard error of the mean. NC, CC, non-transgenic plants; N1-1, 2, NW BrMYB28.1 gene transgenic plants; C1-1–5, CT001 BrMYB28.1 gene transgenic plants; N2-1, NW BrMYB28.2 gene transgenic plant; C2-1, 2, CT001 BrMYB28.2 gene transgenic plants; N3-1,2, NW BrMYB28.3 gene transgenic plants; C3-1–3, CT001 BrMYB28.3 gene transgenic plants
Expression analysis of genes related to GSL biosynthesis in homozygous T2 lines
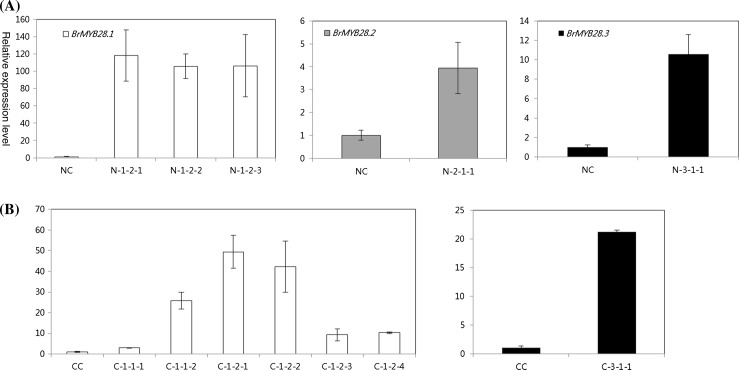
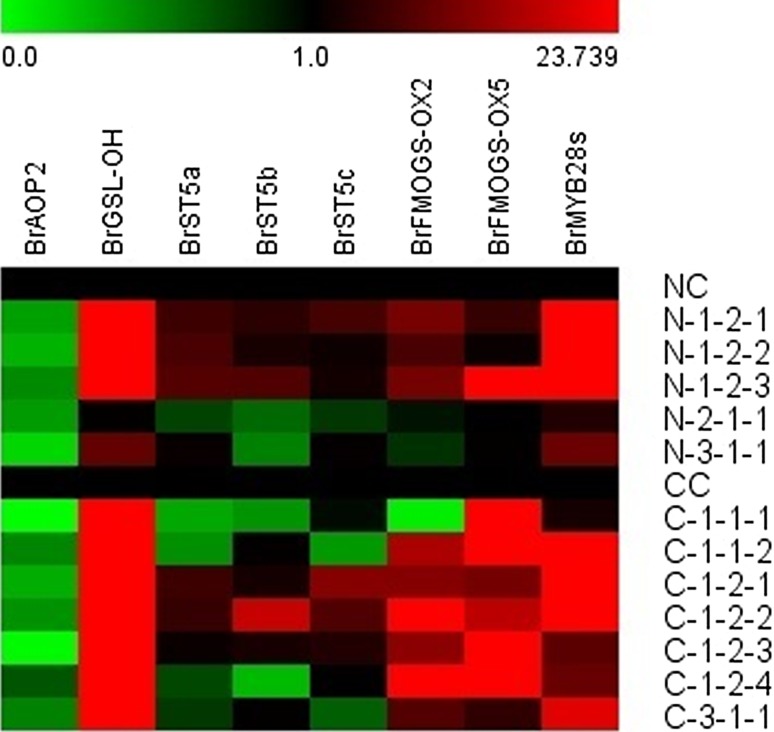
To assess the different expression of genes for GSL biosynthesis, the expression profiles of genes were investigated in 6-week-old leaves of homozygous T2 lines obtained from T1 transgenic plants with a segregation ratio of 3:1 by real-time PCR. As shown in Fig. 7, the BrMYB28.1, BrMYB28.2, and BrMYB28.3 genes were all significantly up-regulated in the leaves of homozygous transgenic plants. The T2 lines showed a significant increase in transcript levels of BrMYB28 genes, resulting in a significant change in the expression level of GSL biosynthetic genes (Fig. 8, Supplementary Table S5). The expression of BrGSL-OH genes was significantly increased in transgenic plants with high transcript levels of the BrMYB28 genes. Expression of the BrAOP2 gene was significantly decreased compared to that in nontransgenic plants. The BrMYB28.1-overexpressing transgenic T2 lines N1-2-1, N1-2-2, and N-1-2-3 all showed up-regulation of six GSL biosynthetic genes, except the BrAOP2 gene. Furthermore, the BrFMO GS-OX2 and BrFMO GS-OX5 genes were up-regulated in all BrMYB28.1-overexpressing CT001 transgenic plants, except C-1-1-1. These results apparently indicate that all three BrMYB28 genes can function as negative regulators of the BrAOP2 gene and as positive regulators of the BrGSL-OH gene.
Fig. 7.
Expression of genes involved in glucosinolate biosynthesis in the 6-week-old leaves of T2 transgenic NW (A) and CT001 (B) plants. Relative expression was determined in triplicate measurements in three independent biological replicates. The Bractin gene was used as a quantitative control. NC, CC, non-transgenic plants; N-1-2-1–N-1-2-3, NW BrMYB28.1 gene transgenic plants; C-1-1-1–C-1-2-4, CT001 BrMYB28.1 gene transgenic plants; N-2-1-1, NW BrMYB28.2 gene transgenic plant; N-3-1-1, NW BrMYB28.3 gene transgenic plant; C-3-1-1, CT001 BrMYB28.3 gene transgenic plants
Fig. 8.
Expression profiling for glucosinolate biosynthesis-related genes in T2 homozygous transgenic lines. NC, CC, non-transgenic plants; NC, CC, non-transgenic plants; N-1-2-1–N-1-2-3, NW BrMYB28.1 gene transgenic plants; C-1-1-1–C-1-2-4, CT001 BrMYB28.1 gene transgenic plants; N-2-1-1, NW BrMYB28.2 gene transgenic plant; N-3-1-1, NW BrMYB28.3 gene transgenic plant; C-3-1-1, CT001 BrMYB28.3 gene transgenic plants
Accumulation of GSLs in homozygous T2 lines
Homozygous T2 transgenic plants with up-regulated expression of all three BrMYB28 genes showed significant increases in the accumulation of GSLs in leaves. As shown in Table 2, the total GSL content increased in all transgenic plants with overexpression of the three BrMYB28 genes. All transgenic plants showed markedly increased contents of both aliphatic and indolic GSLs compared to nontransgenic plants. The highest accumulation of total GSLs was observed in two inbred lines with BrMYB28.1 overexpression: 8.9 µmol/g in N-1-2-3 and 46.8 µmol/g in C-1-2-3. Total GSLs in N-1-2-3 and C-1-2-3 were significantly increased by approximately 4.7- and 4.2-fold, respectively, compared with nontransgenic plants. The total GSL content of N-2-1-1 with BrMYB28.2 overexpression showed a small increase compared with BrMYB28.1 and BrMYB28.3 overexpression. The content of major aliphatic and indolic GSLs was found to be increased in transgenic plants. In contrast, glucoraphanin and sinigrin were only found in the BrMYB28.1 or BrMYB28.3 -overexpressing transgenic lines. Furthermore, these homozygous transgenic T2 plants showed significantly increased levels of all GSLs compared with T1 transgenic plants. Thus, analysis of GSL content and gene expression in transgenic plants suggested that all three BrMYB28 genes participate in the regulation of aliphatic, indolic, and aromatic GSL biosynthesis.
Table 2.
GSLs content (μmol g−1 dw) in the transgenic T2 plants
| Trivial name | NC | N-l-2-1 | N-l-2-2 | N-l-2-3 | N-2-1-1 | N-3-1-1 |
|---|---|---|---|---|---|---|
| Progoitrin | 0 ± 0 | 0.059 ± 0.00 | 0.050 ± 0.036 | 0.053 ± 0.043 | 0.160 ± 0.014 | 0.147 ± 0.034 |
| Glucoraphanin | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Sinigrin | 0 ± 0 | 0.070 ± 0.02 | 0.047 ± 0.034 | 0.053 ± 0.038 | 0 ± 0 | 0 ± 0 |
| Glucoalyssin | 0 ± 0 | 0.10 ± 0.00 | 0.074 ± 0.016 | 0.091 ± 0.013 | 0 ± 0 | 0.036 ± 0.051 |
| Gluconapoleiferin | 0 ± 0 | 0.047 ± 0.00 | 0.036 ± 0.005 | 0.044 ± 0.006 | 0 ± 0 | 0.084 ± 0.080 |
| Gluconapin | 0 ± 0 | 0.08 ± 0.02 | 0.087 ± 0.012 | 0.100 ± 0.028 | 0 ± 0 | 0.041 ± 0.031 |
| Glucocochlearin | 0 ± 0 | 0.422 ± 0.37 | 0 ± 0 | 0.671 ± 0.444 | 0.151 ± 0.008 | 0.084 ± 0.060 |
| Glucobrassicanapin | 0.034 ± 0.024 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0.116 ± 0.084 | 0.058 ± 0.082 |
| Aliphatic GSLs | 0.034 ± 0.024 | 0.786 ± 0.37 | 0.295 ± 0.084 | 1.011 ± 0.460 | 0.427 ± 0.084 | 0.450 ± 0.275 |
| Glucobrassicin | 1.194 ± 0.274 | 2.88 ± 0.50 | 2.954 ± 0.176 | 3.299 ± 0.194 | 3.223 ± 0.331 | 4.119 ± 1.846 |
| 4-Methoxyglucobrassicin | 0.488 ± 0.117 | 2.708 ± 0.43 | 3.435 ± 0.211 | 3.199 ± 0.531 | 0.505 ± 0.246 | 1.480 ± 0.346 |
| Neoglucobrassicin | 0.335 ± 0.068 | 1.10 ± 0.09 | 0.875 ± 0.062 | 1.295 ± 0.183 | 1.102 ± 0.197 | 0.921 ± 0.272 |
| Indolic GSLs | 2.017 ± 0.430 | 6.694 ± 0.93 | 7.264 ± 0.430 | 7.793 ± 0.723 | 4.829 ± 0.435 | 6.519 ± 2.401 |
| Gluconasturtiin | 0.074 ± 0.003 | 0.07 ± 0.01 | 0.124 ± 0.026 | 0.073 ± 0.018 | 0.043 ± 0.061 | 0.239 ± 0.102 |
| Total GSLs | 2.124 ± 0.433 | 7.555 ± 1.23 | 7.683 ± 0.454 | 8.876 ± 1.027 | 5.300 ± 0.365 | 7.208 ± 2.695 |
| Trivial name | CC | C-l-1-1 | C-l-1-2 | C-l-2-1 | C-l-2-2 | C-l-2-3 | C-l-2-4 | C-3-1-1 |
|---|---|---|---|---|---|---|---|---|
| Progoitrin | 0.094 ± 0.03 | 2.743 ± 0.075 | 2.40 ± 0.78 | 1.16 ± 0.26 | 3.524 ± 0.727 | 2.844 ± 0.410 | 2.509 ± 0.27 | 1.259 ± 0.65 |
| Glucoraphanin | 0 ± 0 | 0 ± 0 | 0.045 ± 0.003 | 0.03 ± 0.02 | 0.065 ± 0.018 | 0.049 ± 0.003 | 0.042 ± 0.00 | 0.015 ± 0.02 |
| Sinigrin | 0.017 ± 0.02 | 0.13 ± 0.03 | 0.146 ± 0.024 | 0.13 ± 0.03 | 0.140 ± 0.014 | 0.184 ± 0.037 | 0.138 ± 0.01 | 0.098 ± 0.02 |
| Glucoalyssin | 0.087 ± 0.03 | 0.15 ± 0.03 | 0.163 ± 0.047 | 0.14 ± 0.04 | 0.130 ± 0.028 | 0.229 ± 0.011 | 0.176 ± 0.01 | 0.096 ± 0.01 |
| Gluconapoleiferin | 0.025 ± 0.03 | 0.32 ± 0.10 | 0.696 ± 0.078 | 0.59 ± 0.13 | 1.119 ± 0.363 | 0.719 ± 0.132 | 0.747 ± 0.09 | 0.595 ± 0.20 |
| Gluconapin | 1.126 ± 0.20 | 6.65 ± 2.48 | 8.383 ± 2.992 | 6.74 ± 2.96 | 5.988 ± 0.166 | 10.328 ± 2.091 | 5.764 ± 0.58 | 2.637 ± 0.89 |
| Glucocochlearin | 0 ± 0 | 0.09 ± 0.01 | 0.067 ± 0.009 | 0.07 ± 0.01 | 0.062 ± 0.016 | 0.066 ± 0.011 | 0.077 ± 0.01 | 0.113 ± 0.09 |
| Glucobrassicanapin | 2.519 ± 0.29 | 12.55 ± 3.29 | 17.572 ± 4.936 | 14.88 ± 5.79 | 16.488 ± 0.840 | 21.846 ± 3.047 | 14.924 ± 1.19 | 14.715 ± 4.12 |
| Aliphatic GSLs | 3.868 ± 0.21 | 21.04 ± 5.85 | 29.815 ± 7.774 | 24.98 ± 9.67 | 27.516 ± 1.150 | 36.265 ± 5.321 | 24.376 ± 2.08 | 19.528 ± 5.88 |
| Glucobrassicin | 4.774 ± 0.87 | 5.93 ± 0.57 | 3.368 ± 0.895 | 3.87 ± 1.29 | 5.103 ± 2.090 | 4.904 ± 1.588 | 6.342 ± 0.78 | 2.304 ± 0.52 |
| 4-Methoxyglucobrassicin | 1.448 ± 0.17 | 2.76 ± 0.07 | 2.513 ± 0.386 | 2.81 ± 0.24 | 2.743 ± 0.344 | 3.305 ± 0.476 | 3.178 ± 0.22 | 2.718 ± 0.70 |
| Neoglucobrassicin | 0.764 ± 0.34 | 1.50 ± 0.16 | 2.931 ± 0.575 | 1.38 ± 0.21 | 1.572 ± 0.819 | 1.387 ± 0.278 | 1.948 ± 0.61 | 6.784 ± 2.08 |
| Indolic GSLs | 6.986 ± 1.11 | 10.20 ± 0.74 | 8.813 ± 0.645 | 8.06 ± 1.38 | 9.419 ± 3.088 | 9.595 ± 1.815 | 11.468 ± 1.02 | 11.806 ± 1.56 |
| Gluconasturtiin | 0.296 ± 0.16 | 0.93 ± 0.06 | 0.678 ± 0.216 | 0.52 ± 0.03 | 0.811 ± 0.256 | 0.971 ± 0.181 | 0.574 ± 0.17 | 0.671 ± 0.31 |
| Total GSLs | 11.151 ± 1.30 | 32.17 ± 5.56 | 39.305 ± 8.165 | 33.56 ± 9.67 | 37.746 ± 4.365 | 46.831 ± 3.713 | 36.419 ± 2.99 | 32.005 ± 5.17 |
Each value is mean ± standard error (n = 3)
Discussion
The whole-genome sequencing of B. rapa was recently reported (Wang et al. 2011). The results of a comparative analysis with Arabidopsis suggest that the triplicated genes of B. rapa result from genome triplication via genome evolution following polyploidy. MYB transcription factors are a large gene family of transcription factors in plants and their N termini contain a highly conserved MYB domain (Dias et al. 2003). In particular, the R2R3-MYB transcription factors are known as regulatory proteins in the secondary metabolism of plants (Boddu et al. 2006; Du et al. 2009). R2R3-MYB transcription factors related to the regulation of GSL biosynthesis were recently reported in Arabidopsis (Celenza et al. 2005; Gigolashvil et al. 2007). Three members of MYB28, which is known to be a transcription factor that participates in the regulation of aliphatic GSL biosynthesis, were identified in B. rapa. The three BrMYB28 members share 84–85 % nucleotide sequence identity with AtMYB28 and are located on different chromosomal loci of BAC clones. These results indicate that MYB28 was triplicated by Brassica genome triplication. In the Brassica family, three members of the MYB28 transcription factors are found in B. rapa, two in B. oleracea, and four in B. juncea (Augustine et al. 2013). All MYB28 genes of the Brassica family also share high levels of sequence conservation with the AtMYB28 gene of A. thaliana and close evolutionary relationships.
The duplication of genes by polyploidy in plants has led to functional diversity as pseudogenes or the gain of additional or novel functions (Adams 2007). In this study, we investigated the expression of three BrMYB28 genes in different organization and developmental stages of Chinese cabbage by RT-PCR and microarray. BrMYB28.1 and BrMYB28.2 showed high levels of expression in seedlings or leaves, with expression patterns differing according to developmental stages. In contrast, BrMYB28.3 showed no expression in any of the investigated organization and developmental stages of B. rapa. Although BrMYB28.3 showed a high level of sequence homology with Arabidopsis, it might be a pseudogene or be subject to epigenetic silencing by randomly occurring polyploidy in the genome. The results of protein alignment analysis shown in Fig. 1 suggested the possibility that insertion of a repetitive sequence of ‘F’ amino acid in the C-terminal region might lead to the pseudogenization of the BrMYB28.3 gene. These results also support recent reports that gene duplication in B. rapa and Arabidopsis may allow functional diversification during evolution by changing protein structures (Du et al. 2012). However, it is uncertain whether BrMYB28.3 is a pseudogene resulting from the insertion of repetitive sequences with no expression in any of the organizational or developmental stages of B. rapa. In addition, the differential and various expression patterns between BrMYB28.1 and BrMYB28.2 suggest that internal sequence divergence of paralogs may result in the functional change of duplicated genes.
We observed changes in the function of the three BrMYB28 transcription factors by polyploidization of the B. rapa genome. The BrMYB28 genes were identified to increase in expression levels during GSL accumulation in all of T1 and T2 lines of transgenic Chinese cabbage. Although transgenic plants contained the same construct, the expression levels of three BrMYB28 genes differed markedly in independent T1 and T2 transgenic plants. Consequently, the differences in the increase of expression level of BrMYB28 genes induced high GSL accumulation compared to nontransgenic plants. Additionally, there was no similar correlation between the expression of BrMYB28 genes and glucosinolate content. Such a lack of correlations between mRNA levels and levels of the final target product has also been shown in other studies (Emani et al. 2003; Takahashi et al. 2010). Some studies have demonstrated that variation in expression levels are caused by difference in transgene integration sites (Matzke and Matzke 1998; Kohli et al. 2006). Thus, we can speculate that the different expression levels of the BrMYB28 genes are attributable to differences in the transgene integration site.
We also found that the total GSL content in homozygous T2 lines of transgenic CT001 was significantly increased compared to the heterozygous parental transgenic lines, whereas the total GSL content of T2 lines of transgenic NW were similar to their heterozygous parental transgenic lines. In particular, five homozygous T2 lines overexpressing BrMYB28.1 exhibited a high content of total GSLs and expression of glucoraphanin. Duan et al. (1996) reported that the homozygous transgenic plants produced a higher content of foreign protein compared to heterozygous transgenic plants in some transgenic plants. The similar GSL content in T1 and T2 of NW transgenic lines indicates that the variation in GSL content by the overexpression of BrMYB28 genes can differ depending on the genotype in B. rapa. Overexpression of BrMYB28 genes regulated the mRNA levels of BrAOP2 and BrGSL-OH in transgenic T2 plants. Although MYB28 and MYB29 have been reported as positive regulators of aliphatic GSL biosynthetic genes in Arabidopsis (Sønderby et al. 2010), in the present study, all three BrMYB28 genes were shown to function as negative regulators of the BrAOP2 gene. This suggests that regulation of GSL biosynthesis by MYB28 may differ depending on the plant. Furthermore, accurate observation of changes in the expression levels of more paralog genes will be required to determine the expression network of the GSL biosynthesis genes regulated by the three BrMYB28 transcription factors. Analysis of the transgenic plants showed that BrMYB28.3 is fully functional for GSL biosynthesis, similar to BrMYB28.1 and BrMYB28.2. Zou et al. (2009) found evidence of expression for a few pseudogenes, although with lower expression levels compare to functional genes in Arabidopsis and rice. Our findings indicate that BrMYB28.3 has a unique role as an expressed pseudogene that regulates GSL biosynthesis.
Gene silencing of the BnAOP2 gene has been reported to cause a reduction in progoitrin and an increase in glucoraphanin in the seeds of B. napus (Liu et al. 2012). Furthermore, the GSL-OH gene was shown to be involved in the accumulation of progoitrin in Arabidopsis (Hansen et al. 2008). Therefore, in the present study, the increased level of progoitrin in all T2 transgenic plants and only glucoraphanin in overexpressed CT001 transgenic plants caused by overexpression of BrMYB28.1 and BrMYB28.3 may be due to the decreased expression level of the BrAOP2 gene and the increased expression level of the BrGSL-OH gene. We can consider various possibilities, such as total GSL content, different genotype, interaction with paralog genes of BrAOP2 and BrGSL-OH, or other functions of BrMYB28s, as reasons for the detection of glucoraphanin only in the BrMYB28 genes-overexpressing lines of CT001 T2 transgenic plants.
The three BrMYB28 proteins are involved in regulating the biosynthesis of all aliphatic, indolic, and aromatic GSLs in transgenic plants of two inbred lines of B. rapa, although there are sequence differences among the paralogs. The overexpressing transgenic plants clearly showed that all of the three BrMYB28 participate in controlling the accumulation of both short- and long-chain GSLs. Recent studies on GSL biosynthesis-related genes have reported that MYB28 is involved only in the aliphatic GSL biosynthesis pathway (AtMYB28 of A. thaliana and two BjMYB28 genes of B. juncea). This finding indicates that the two BjMYB28 genes resulting from polyploidization of the B. juncea genome showed no function other than regulation of aliphatic GSL biosynthesis in transgenic A. thaliana. Therefore, the different accumulation patterns of GSL biosynthesis between B. rapa and A. thaliana suggests that some different mechanisms may operate in the GSL biosynthesis pathway of these plant species. The constant functionalization of BjMYB28 paralog genes may also be related to the functional analysis of GSL biosynthesis using transgenic Arabidopsis.
We have successfully developed Chinese cabbage containing high levels of GSL by overexpressing BrMYB28 genes in B. rapa, which is recalcitrant to Agrobacterium-mediated transformation. Our results clearly indicate that all of the three BrMYB28 genes are related to GSL biosynthetic processes in B. rapa. The high GSL contents of homozygous T2 plants promoted by increased expression of BrMYB28 genes will prove useful for studying the regulation of complex GSL biosynthesis pathways in polyploid plants and in subsequent functional studies that will examine the anti-carcinogenic activity and defense properties of GSLs.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Mrs. Bun-Ja Baek and Mrs. Chang-Duck Jang for assistance with tissue culture. This work was supported by a Grant from the National Academy of Agricultural Science (PJ008673 and PJ010157), Rural Development of Administration, Korea.
Author contribution statements
SMS and KJS charge of planning and conducting to the experiment and write a manuscript. JM, JHC, and SJK charge of HPLC analysis of GSL contents. PBS and SSH are planning of this project and assist on breeding on transgenic plants.
Contributor Information
Mi-Suk Seo, Email: sms1030@korea.kr.
Mina Jin, Email: genemina@korea.kr.
Jin-Hyuk Chun, Email: slaldinv2@gmail.com.
Sun-Ju Kim, Email: kimsunju@cnu.ac.kr.
Beom-Seok Park, Email: pbeom@korea.kr.
Seong-Han Shon, Email: sohnseonghan@korea.kr.
Jung Sun Kim, Phone: 82-63-238-4559, Email: jsnkim@korea.kr.
References
- Adams KL. Evolution of duplicate gene expression in polyploidy and hybrid plants. J Hered. 2007;98:136–141. doi: 10.1093/jhered/esl061. [DOI] [PubMed] [Google Scholar]
- Augustine R, Majee M, Gershenzon J, Bisht NC. Four genes encoding MYB28, a major transcriptional regulator of the aliphatic glucosinolate pathway, are differentially expressed in the allopolyploid Brassica juncea. J Exp Bot. 2013;64:4907–4921. doi: 10.1093/jxb/ert280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddu J, Jiang C, Sangar V, Olson T, Peterson T, Chopra S. Comparative structural and functional characterization of sorghum and maize duplications containing orthologous Myb transcription regulators of 3-deoxyflavonoid biosynthesis. Plant Mol Biol. 2006;60:185–199. doi: 10.1007/s11103-005-3568-1. [DOI] [PubMed] [Google Scholar]
- Celenza JL, Quiel JA, Smolen GA, Merrikh H, Silvestro AR, Normanly J, Bender J. The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 2005;137:253–262. doi: 10.1104/pp.104.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias AP, Braun EL, McMullen MD, Grotewold E. Recently duplicated maize R2R3 myb genes provide evidence for distinct mechanisms of evolutionary divergence after duplication. Plant Physiol. 2003;131:610–620. doi: 10.1104/pp.012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Zhang L, Liu L, Tang XF, Yang WJ, Wu YM, Huang YB, Tang YX. Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry. 2009;74:1–11. doi: 10.1134/s0006297909010015. [DOI] [PubMed] [Google Scholar]
- Du H, Feng BR, Yang SS, Huang YB, Tang YX. The R2R3-MYB transcription factor gene family in maize. PLoS One. 2012;7:e37463. doi: 10.1371/journal.pone.0037463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan XL, Li XG, Xue QZ, Abo-EI-Saad M, Xu DP, Wu R. Transgenic rice plants harboring an introduced potato proteinase inhibitor II gene are insect resistant. Nat Biotechnol. 1996;14:494–498. doi: 10.1038/nbt0496-494. [DOI] [PubMed] [Google Scholar]
- Emani CJM, Garcia ELF, Pozo MJ, Uribe P, Kim DJ, Sunikumar G, Cook DR, Kenerley CM, Rathore KS. Enhanced fungal resistance in transgenic cotton expressing an endochitinase gene from Trichoderma virens. Plant Biotechnol J. 2003;1:321–336. doi: 10.1046/j.1467-7652.2003.00029.x. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Engqvist M, Yatusevich R, Muller C, Flugge UI. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007;50:886–901. doi: 10.1111/j.1365-313X.2007.03099.x. [DOI] [PubMed] [Google Scholar]
- Hansen BG, Kerwin RE, Ober JA, Lambrix VM, Mitchell-Olds T, Gershenzon J, Halkier BA, Kliebenstein DJ. A novel 2-oxoacid-dependent dioxygenase involved in the formation of the goiterogenic 2-hydroxybut-3-enyl glucosinolate and generalist insect resistance in Arabidopsis. Plant Physiol. 2008;148:2096–2108. doi: 10.1104/pp.108.129981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper AL, Trick M, Higgins J, Fraser F, Clissold L, Wells R, Hattori C, Werner P, Bancroft I. Associative transcriptomics of traits in the polyploidy crop species Brassica napus. Nat Biotechnol. 2012;30:798–802. doi: 10.1038/nbt.2302. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Kelleher MO, Eggleston IM. The cancer chemopreventive actions of phytochemicals derived from GSLs. Eur J Nutr. 2008;47:73–88. doi: 10.1007/s00394-008-2009-8. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci USA. 2007;104:6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E, Kim SJ, Kim GH. Identification and quantitative determination of GSLs in seeds and edible parts of Korean Chinese cabbage. Food Chem. 2011;128:1115–1120. doi: 10.1016/j.foodchem.2010.11.102. [DOI] [Google Scholar]
- Kim JS, Chung TY, King GJ, Jin M, Yang TJ, Jin YM, Kim HI, Park BS. A sequence-tagged linkage map of Brassica rapa. Genetics. 2006;174:29–39. doi: 10.1534/genetics.106.060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Kawaharada C, Jin S, Hashimoto M, Ishii G, Yamauchi H. Structural elucidation of 4-(Cystein-S-yl) butyl glucosinolate from the leaves of Eruca sativa. Biosci Biotechnol Biochem. 2007;71:114–121. doi: 10.1271/bbb.60400. [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, Mitchell-Olds T. Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 2001;126:811–825. doi: 10.1104/pp.126.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli A, Melendi PG, Abranches R, Capell T, Stoger E, Christou P. The quest to understand the basis and mechanisms that control expression of introduced transgenes in crop plants. Plant Signal Behav. 2006;1:185–195. doi: 10.4161/psb.1.4.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Hirani AH, Mcvetty PB, Daayf F, Quiros CF, Li G. Reducing progoitrin and enriching glucoraphanin in Brassica napus seeds through silencing of the GSL-ALK gene family. Plant Mol Biol. 2012;79:179–189. doi: 10.1007/s11103-012-9905-2. [DOI] [PubMed] [Google Scholar]
- Matzke AJM, Matzke MA. Position effects and epigenetic silencing of plant transgenes. Curr Opin Plant Biol. 1998;1:142–148. doi: 10.1016/S1369-5266(98)80016-2. [DOI] [PubMed] [Google Scholar]
- Min BW, Cho YN, Song MJ, Noh TK, Kim BK, Chae WK, Park YS, Choi YD, Harn CH. Successful genetic transformation of Chinese cabbage using phosphomannose isomerase as a selection marker. Plant Cell Rep. 2007;26:337–344. doi: 10.1007/s00299-006-0247-x. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Park JH, Lee SJ, Kim BR, Woo ET, Lee JS, Han EH, Lee YH, Park YD. Isolation of myrosinase and glutathione S-transferase genes and transformation of these genes to develop phenylethylisothiocynate enriching Chinese cabbage. Kpr J Hort Sci Technol. 2011;29:623–632. [Google Scholar]
- Sawada Y, Toyooka K, Kuwahara A, Sakata A, Nagano M, Saito K, Hirai MY. Arabidopsis bile acid: sodium symporter family protein 5 is involved in methionine-derived glucosinolate biosynthesis. Plant Cell Physiol. 2009;50:1579–1586. doi: 10.1093/pcp/pcp110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Reichelt M, Burow M, et al. DOF transcription factor AtDof1.1(OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 2006;47:10–24. doi: 10.1111/j.1365-313X.2006.02767.x. [DOI] [PubMed] [Google Scholar]
- Sønderby IE, Hansen BG, Bjarnholt N, Ticconi C, Halkier BA, Kliebenstein DJ. A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic GSLs. PLoS One. 2007;2:e1322. doi: 10.1371/journal.pone.0001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby IE, Burow M, Rowe HC, Kliebenstein DJ, Halkier BA. A complex interplay of three R2R3 MYB transcription factors determines the profile of aliphatic GSLs in Arabidopsis. Plant Physiol. 2010;153:348–363. doi: 10.1104/pp.109.149286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi W, Oishi H, Ebina M, Komatsu T, Takamizo T. Production of transgenic Italian ryegrass expressing the betaine aldehyde dehydrogenase gene of zoysiagrass. Breed Sci. 2010;60:279–285. doi: 10.1270/jsbbs.60.279. [DOI] [Google Scholar]
- Udall JA, Swanson JM, Nettleton D, Percifield RJ, Wendel JF. A novel approach for characterizing expression levels of genes duplicated by polyploidy. Genetics. 2006;173:1823–1827. doi: 10.1534/genetics.106.058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpoorte R, Contin A, Memelink J. Biotechnology for the production of plant secondary metabolites. Phytochem Rev. 2002;1:13–25. doi: 10.1023/A:1015871916833. [DOI] [Google Scholar]
- Wang XW, Wang HZ, Wang J, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011;43:1035–1157. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- Yang TJ, Kim JS, Kwon SJ, et al. Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell. 2006;18:1339–1347. doi: 10.1105/tpc.105.040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Saito K. Functional genomics for plant natural product biosynthesis. Nat Prod Rep. 2009;26:1466–1487. doi: 10.1039/b817077k. [DOI] [PubMed] [Google Scholar]
- Zang YX, Kim HU, Kim JA, et al. Genome-wide identification of glucosinolate synthesis genes in Brassica rapa. FEBS J. 2009;276:3559–3574. doi: 10.1111/j.1742-4658.2009.07076.x. [DOI] [PubMed] [Google Scholar]
- Zou C, Lehti-Shiu MD, Thibaud-Nissen F, Prakash T, Buell CR, Shiu SH. Evoulutionary and expression signatures of pseudogenes in Arabidopsis and rice. Plant Physiol. 2009;151:3–15. doi: 10.1104/pp.109.140632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Ishida M, Li F, Kakizaki T, Suzuki S, Kitashiba H, Nishio T. QTL analysis using SNP markers developed by next-generation sequencing for identification of candidate genes controlling 4-methylthio-3-butenyl glucosinolate contents in roots of Radish, Raphanus sativus L. PLoS One. 2013;8:e5354. doi: 10.1371/journal.pone.0053541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.