Abstract
Visceral leishmaniasis (VL) is a tropical neglected disease caused by Leishmania donovani, results in significant mortality in the Indian subcontinent. The plasticity of T cell proliferation and differentiation depends on microRNA mediated gene regulation which leads Th1/Th2 or Th17/Treg type of immune response during human VL. This study depicts the identification of target immune signaling molecule and transcription factors, which play a role in T-cell proliferation and differentiation followed by the identification of miRNA controlling their gene expression using three web servers’ viz., TargetScan, miRPath and miRDB. This study provides the bioinformatics evidences that seed region present in the miRNAs miR-29-b, miR-29a, have the putative binding site in the 3′-untranslated region (UTR) of TBX21 transcription factor of CD4+ T helper (Th1), which may suppress the Th1 specific protective immune response. Development of Th2 type specific immune response can be suppressed by binding of miR-135 and miR-126 miRNAs over the 3′-UTR region of GATA-3 transcription factor of Th2 specific CD4+ T helper cells. MiRNA identified against Th2/Treg immune cells are important and their over expression or administration can be used for developing the Th1/Th17 type of protective immune response during VL infection. This study indicates that miRNAs have the capacity to regulate immune signaling, cytokine production and immune cell migration to control the VL infection in human. This observation warrants further investigation for the development of miRNA based therapy controlling T cell differentiation in human VL.
Keywords: MicroRNAs, Leishmania donovani, visceral leishmaniasis, Th1/Th2, CD4+ T cell, Th17/Treg
Introduction
Leishmaniasis is a tropical neglected diseases which affect 350 million people worldwide living in 98 countries and three territories of five continents (Alvar et al., 2012). The increasing incidence of leishmaniasis show different faces such as visceral, cutaneous and muco-cutaneous and post kala-azar dermal leishmaniasis. Among all these form of leishmaniasis, visceral leishmaniasis (VL) is most severe which affects 58,200 individuals yearly worldwide. It causes absolute death if untreated and is clearly a poverty-related disease which accounts over 30,000 deaths annually (Prajapati et al., 2012). During VL infection, serum plays an important role in binding of promastigotes to macrophage through adsorption of opsonins (antibody/complement protein) to the parasite and subsequent binding to Fc receptor of the macrophage followed by endocytosis (Sharma and Singh, 2009). Therefore, parasites escape the humoral immune response of host by residing in the phagolysosome of macrophage and subsequently antibodies have no effect on the infection which may be harmful to the host (Unanue and Allen, 1987).
Leishmania donovani infection and its presentation by conventional antigen presenting cells induce different pro-inflammatory cytokine response. Naive CD4+ T cell can initiates differentiation into specific lineages such as Th1, Th2, Th17 and regulatory T (Treg) cells depending upon pro-inflammatory response and expression of specific transcription factors. Each T cell lineage has distinct effectors mechanism along with specific cytokine profiles. In response of IFN-γ and IL-12 pro-inflammatory cytokines, naïve CD4+ T cells differentiate into Th1 cell through controlled process and activity of STAT1, STAT4 and T-bet transcription factors. The STAT4 transcription factor is required to initiate IL-12 signaling along with IFN-γ and inducible nitric oxide synthase expression. Th1 immune cells are the main player of parasite clearance during VL by producing a large amount of IFN-γ and play a critical role to protect human against intracellular parasite by the activation of macrophages (Romagnani, 1999). On the other way, IL-4 promotes the differentiation of naïve CD4+ T cell into Th2 type immune cell through the activation of STAT6 and transcription factor GATA3. In case of VL, Th2 cells help in parasite survival by down regulation of IL-12 via IL-4, to maintain the homeostasis between Th1 and Th2 (Sundar et al., 1997). The differentiation of CD4+ T cells to Th17 cells is carried out in response to IL6 and TGF-β. Initially, IL-6 activates STAT3, then the combined effect of IL-6 and TGF-β signaling leads to the expression of retinoid related orphan receptor (ROR-γt and ROR-α) transcription factor resulting in the initiation of Th17 differentiation. During VL, L. donovani induces differentiation of Th17 cells and the resulting IL-17 and IL-22 cytokines are associated with resistance to VL (Pitta et al., 2009). Treg cells differentiation requires same TGF-β to promote the expression of transcription factor Foxp3 but in the absence of IL-6, blocks the expression of ROR-γt and ROR-α transcription factor which results in differentiation of naïve T cell into Treg cells (Zhou et al., 2008).
MiRNAs are evolutionary conserved, non-protein coding, small silencing RNA with size ranges from 20 to 24 nucleotides. The seed sequence of miRNA (7–8 nucleotide) makes base pair with 3′-untranslated region (UTR) of target mRNA results in either translation inhibition or/and mRNA degradation (Bartel, 2009). Till date, several miRNAs have shown the capacity to regulate biological processes like cell differentiation, proliferation, and apoptosis (Miska, 2005; Ruan et al., 2009). It also contributes to the process of thymic T cell maturation and differentiation. The controlled expression of miR-181a contributes to clonal deletion of auto-reactive T cells by the modulation of TCR signaling threshold and survival of low affinity peptide-specific T cell (Li et al., 2007). MiR-155, miR-181c, miR-9, and miR-31 also play an important role in T cell activation by regulating IL-2 signaling pathway. MiR132/212 cluster induces Th17 cells differentiation and its deficiency lowers the frequencies of Th1 and Th17cells due to inhibition of experimental autoimmune encephalitis development (Nakahama et al., 2013). While the over-expression of miR-26a leads to increased Treg expression which play an important role in Th1 and Th17 differentiation (Zhang et al., 2015). In this article, we have presented whether miRNA expression can regulate the CD4+ T cell differentiation during VL infection. Here, we have identified transcription factors and important cytokines playing a role in T cell differentiation and favoring the condition for the development of VL disease in human. Using computational study, we predicted up-regulation of putative miRNAs regulating key transcription factor and guiding conversion of Th0 cell into Th2 type of immune cells which results in VL infection.
Materials and Methods
Identification of Target Immune Signaling Molecules
Visceral leishmaniasis infection in human is associated with activation of Th2 and suppression of Th1 immune response. Suppression of Th1 immune response is associated with decreased level of IFN-γ, IL-12 and higher production of IL-4 and IL-13 cytokines to maintain the Th1/Th2 plasticity. Involvement of Th17 immune cells for the production of IL-17, IL-22 cytokines and to recruit neutrophil is also important factor to provide the protection from VL infection. Development of protective immune response against L. donovani parasites depends upon the pro inflammatory immune response which is mediated by antigen presenting cells. In response to pro inflammatory cytokines CD4+ T cells converts into either Th1 or Th2 type of the immune cell. Conversion of immune cells in Th1/Th2 is dependent upon the formation of key transcription factors and important cytokines. We have identified key transcription factors and cytokines playing an important role in T cell differentiation and plasticity during the development of protection from VL disease (Table 1).
Table 1.
Key transcription factors regulating CD4+ T cell differentiation and maturation.
| S. No. | Name | Category | Function | Reference |
|---|---|---|---|---|
| 1 | TBX21 | Transcription factor | Induce IFN-γ production | Szabo et al., 2000 |
| GATA-3 | Transcription factor | Master regulator of Th2 cell differentiation | Zheng and Flavell, 1997 | |
| 2 | STAT-1 | STAT protein | Induces TBX21 expression | Afkarian et al., 2002 |
| 3 | STAT-4 | STAT protein | Induces IFN-γ production and expression of TBX21 | Usui et al., 2003 |
| 4 | STAT-5 | STAT protein | Th2 differentiation | Zhu et al., 2003 |
| 5. | STAT-6 | STAT protein | Induce GATA-3 | Takeda et al., 1996 |
| 6 | Runx-3/Eomes | Transcription repressor | Induces IFN-γ expression | Naoe et al., 2007 |
| 7 | IRF-1 | IFN regulated factor | Induces IFN-γ expression | Kano et al., 2008 |
| 8 | IRF-4 | IFN regulated factor | Up regulate GATA-3 expression | Lohoff et al., 2002 |
| 9 | HLX | Other factor | Enhances TBX21 mediated IFN-γ production | Mullen et al., 2002 |
| 10 | Ets-1 | Other factor | Cofactor for TBX21 | Grenningloh et al., 2005 |
| 11 | GFi-1 | Transcription repressor | Promote Th2 cell differentiation | Li et al., 1999 |
| 12 | IKaros | Other transcription factor | Suppress Th1 cell differentiation | Quirion et al., 2009 |
| 13 | cMaf | Other transcription factor | Enhances IL-4 production | Kim et al., 1999 |
| 14 | JunB | Other transcription factor | Enhances IL-4 production | Li et al., 1999 |
| 15 | Dec2 | Other transcription factor | Induce GATA-3 expression | Yang et al., 2009 |
| 16 | Blimp-1 | Other transcription factor | Suppresses expression of IL-2 and IFN-γ | Martins and Calame, 2008 |
| 17 | IL-12 | Cytokines | Induction of Th1 cell differentiation | Hsieh et al., 1993 |
| 18 | IFN-γ | Cytokines | Induction of Th1 cell differentiation | Lighvani et al., 2001 |
| 19 | IL-4 | Cytokines | Induction of Th2 cell differentiation | Swain et al., 1990; Seder et al., 1992 |
| 20 | IL-2 | Cytokines | Induction of Th1 cell differentiation | Le Gros et al., 1990 |
| 21 | ROR-γt | Transcription factor | Induction of Th17 cell differentiation | Park et al., 2005 |
| 22 | ROR-α | Transcription factor | Promote Th17 differentiation | Park et al., 2005 |
| 23 | STAT3 | STAT protein | Induction of Th17 cell differentiation | Park et al., 2005 |
| 24 | IL-6 | Cytokine | Induction of Th17 cell differentiation | Harrington et al., 2005 |
| 25 | TGF-β | Transcription factor | Induction of both Th17and Treg cell differentiation | Bettelli et al., 2006 |
| 26 | Foxp3 | Transcription factor | Induction of Treg cell differentiation | Zhou et al., 2008 |
Resources Used for the miRNA Prediction
MiRNAs are small RNA molecule which down regulate the target gene expression at post transcription level. There are many resources available which can be used to predict the most potential miRNAs against target genes. Here, we have tabulated the name of web servers which is freely available for public use and can be used to predict miRNAs (Table 2). For this study, we used three web servers viz., TargetScan, miRPath and miRDB to predict miRNAs against key transcription factors and important cytokines regulating CD4+ T cell differentiation during VL infection.
Table 2.
List of resources used for microRNA prediction.
| S. No. | Availability | Name of tool | Resource (URL) | Reference |
|---|---|---|---|---|
| 1 | Online search | TargetScan | http://www.targetscan.org/ | Thomson et al., 2011 |
| 2 | miRPath | http://diana.imis.athena-innovation.gr/DianaTools/index.php?r~$=$~mirpath/index | Huang et al., 2014 | |
| 3 | miRDB | http://mirdb.org/miRDB/ | Wang and El Naqa, 2008 | |
| 4 | miRTrail | http://mirtrail.bioinf.uni-sb.de/mirtrail.php | Yang et al., 2011 | |
| 5 | miRanda | http://www.microrna.org/microrna/home.do | Thomson et al., 2011 | |
| 6 | miRwalk | http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/ | Dweep et al., 2013 | |
| 7 | miRGen | http://carolina.imis.athena-innovation.gr/index.php?r=mirgenv3 | Al-Nakhle et al., 2010 | |
| 8 | Pictar | http://pictar.mdc-berlin.de | Thomson et al., 2011 |
MiRNAs Prediction Against Transcription Factors Controlling T Cell Differentiation During Visceral Leishmaniasis Infection
CD4+ T cell differentiation follows different pathways in human VL. When L. donovani parasites enters in human peripheral blood, CD4+ T cells (Th0) transformation started in response to pro inflammatory cytokines. Pro inflammatory cytokines released from antigen presenting cells play an important role in the conversion of Th0 cell into different lineages such as Th1, Th2, Th17 and Treg etc. Induction of IL-4/IL-13 cytokine signaling from antigen presenting cells induces the expression of GATA3 transcription factor which leads to conversion of Th0 cells into Th2 cells. Using potentially used webservers TargetScan, miRPath and miRDB web servers, we have identified putative miRNAs which subsequently down regulate the expression of transcription factor to control Th0 cell differentiation. In these web servers, we submitted the NCBI gene ID of key transcription factors and important genes to get the potential miRNAs.
Results
MiRNAs Regulating Th1/Th2 Differentiation and Plasticity
MiRNAs regulating Th1/Th2 signaling were predicted using three foremost web servers viz., TargetScan, (Agarwal et al., 2015) miRPath (Paraskevopoulou et al., 2013) and miRDB (Wong and Wang, 2015). TBX21 protein is a Th1 immune cell-specific transcription factor regulates Th1 immune signaling specific cytokines. By using TargetScan, we speculated that miR-29a, miR-29b, and miR-29c were found to be potential miRNA with the high probability of conserved targeting (PCT) value which can bind at position 245–251 of TBX21 3′-UTR. While, the miRNA obtained from miRPath and miRDB have shown that miR-548ah-5p, miR-526b-5p, and miR-4726-5p have the same target binding site at 3′-UTR of TBX21 gene. These miRNAs have the potential to down regulate the expression of TBX21 gene and subsequently inhibit the conversion of Th0 cells into Th1 cells. STAT1 and STAT4 transcription factors were also submitted for miRNA prediction since they play an important role in the development of Th1 cell and IFN-γ production. By using all three web servers, miR-1252, miR-4697, miR-4724, and miR-495 were found to be potential miRNA against STAT1 gene. Similarly, TargetScan and miRDB prediction have shown that miR-200a have the putative binding sites at position 237–244 of STAT4 gene with high PCT values (Table 3). Entry of L. donovani parasites inside human peripheral blood induces a number of signaling in the CD4+ T cells which develops an environment to induce above miRNAs to negatively regulate Th1 immune cell conversion.
Table 3.
MiRNAs regulating Th1 cell differentiation and plasticity.
| Serial No. | Gene | miRDB | miRPath | TargetScan |
|---|---|---|---|---|
| 1 | TBX21 | miR-548ah-5p miR-526b-5p miR-4726-5p |
miR-548ah-5p miR-526b-5p miR-4726-5p |
miR-29a miR-29b miR-29c |
| 2 | STAT1 | miR-1252 miR-4697 miR-4724 miR-495 |
miR-1252 miR-4697 miR-4724 miR-495 |
miR-1252 miR-4697 miR-4724 miR-495 |
| 3 | STAT4 | miR-200a | – | miR-200a |
| 4 | IFNG | miR-24 miR-29 |
miR-24 – |
miR-24 – |
| 5 | IL12 | miR-21 miR-590-5p |
miR-21 – |
miR-21 miR-590-5p |
| 6 | RunX3 | miR-130b miR-106b miR-130a |
miR-130b miR-106b miR-130a |
– – – |
| 7 | EOMES | miR-182 miR-29a miR-29b |
miR-182 miR-29a miR-29b |
miR-182 – – |
| 8 | IRF1 | – – miR-4708-3p – miR-4483 – |
miR-635 miR-4747-5p – miR-4667 – miR-23 |
miR-635 miR-4747-5p miR-4708-3p miR-4667 miR-4483 – |
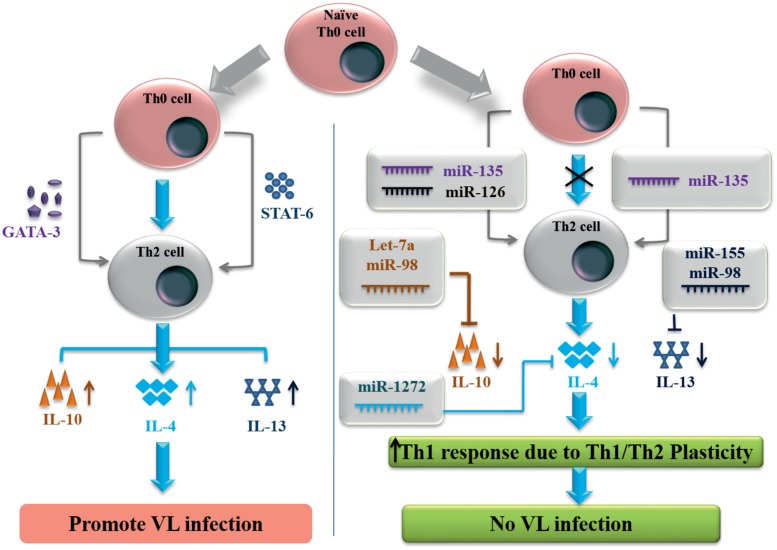
Simultaneously, we have identified miRNAs against Th2 immune signaling molecules, which is responsible for L. donovani replication and VL disease development. GATA3 protein is T cell specific transcription factor which induces the conversion of Th0 into Th2 immune cells and IL-4/IL-13 signaling for developing a Th2 immune response during VL disease. GATA3 protein has multiple transcripts but for NM_001002295 transcript, by using all three web servers we found that miR-135 has the binding site at the 3′-UTR of GATA3 gene (position 207–213). MiR-135 has shown high PCT value against GATA3 gene and has the potential to inhibit the expression of this gene and Th2 immune cell conversion. While miR-126 was predicted by miRPath and miRDB as a negative regulator of GATA3. STAT6 transcription factor induced by IL-4 has shown multiple transcripts with different 3′-UTR length. STAT6 gene with 3′-UTR length of 1168 nucleotides were found to show miR-135 binding at position 1100–1107. To control the VL infection, IL-4 and IL-13 pro-inflammatory cytokines released from antigen presenting cells can be inhibited by miR-1272 and miR-155 (Figure 1). Signature cytokines secreted by Th1/Th2 cell were also submitted for putative miRNA prediction, since they have important role in the maintenance of Th1/Th2 plasticity. MiRNAs against key transcription factor and important cytokines controlling Th1 and Th2 specific immune signaling events are listed in the Tables 3 and 4 respectively.
FIGURE 1.
MicroRNAs controlling Th2 signaling in human visceral leishmaniasis infection.
Table 4.
MiRNAs regulating Th2 cell differentiation and plasticity.
| S. No. | Gene | miRDB | miRPath | TargetScan |
|---|---|---|---|---|
| 1 | GATA3 | miR-135 miR-126 |
miR-135 miR-126 |
miR-135 - |
| 2 | IL4 | miR-1272 | miR-1272 | miR-1272 |
| 3 | STAT6 | miR-135 | - | miR-135 |
| 4 | IL13 | - - - - - - |
let-7d let-7e let-7f let-7g let-7i miR-98 |
miR-155 - - - - - |
| 5 | IL10 | - - - - - - - |
miR-98 miR 4500 miR-4458 let-7a let-7b let-7c let-7d |
miR-98 miR 4500 miR-4458 let-7a let-7b let-7c let-7d |
| 6 | IL2 | miR-181a miR-181b miR-181c miR-181d |
- - - - |
miR-181a-5p miR-181b-5p miR-181c-5p miR-181d-5p |
| 7 | STAT5 | mir-4663 - - - - - - |
- miR-548k miR-3941 miR-4672 miR-4719 miR-524-5p miR-520d-5p |
mir-4663 miR-548k miR-3941 miR-4672 miR-4719 miR-524-5p miR-520d-5p |
| 8 | IRF4 | miR-128 miR-125b |
miR-128 miR-125b |
miR-128 - |
| 9 | GFI1 | miR-142-3p | - | miR-142-3p |
| 10 | CMAF | - - - |
miR-182 miR-143 miR-301a |
miR-182 - - |
| 11 | JunB | miR-199a-5p miR-199b-5p |
- - |
miR-199a-5p miR-199b-5p |
MiRNAs Regulating Th17/Treg Differentiation and Plasticity
Subpopulations of CD4+ T cells producing IL-17 cytokines were capable to inducing IFN-γ in STAT4 dependent manner. During VL infection, it has been shown that IL-17 and IL-22 cytokine level increases in PBMC culture. Using TargetScan, miRPath and miRDB web servers, we have collected the series of miRNAs activated in response to L. donovani infection and controlling Th17/Treg differentiation. TargetScan and miRPath analysis have shown that miR-4500, let-7a, let-7b, and let-7c have the putative binding site at position 407-413 of 3′-UTR for RORC gene which can subsequently negatively regulate the expression of this gene for Th17 differentiation. RORC gene encodes for RORγt transcription factors. MiR-106a and miR-106b have shown putative binding site at the position 252–258 of 3′-UTR for STAT3 gene, when analyzed by all three web servers, which have the potential to down regulate the expression of this gene. MiR-124 was also found to be a negative regulator of STAT3 gene, by miRDB and miRPath and literature (Lu et al., 2013).
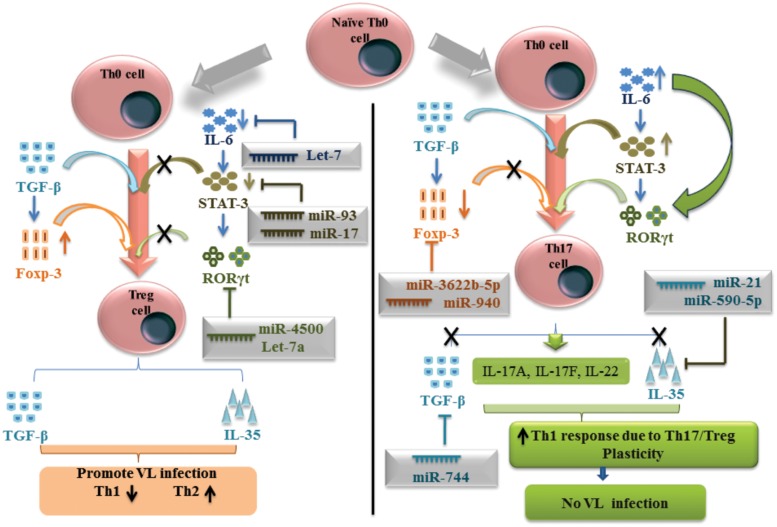
A small population of CD4+ T cells showing CD4+ CD25+ Foxp3+ characteristic termed as Treg cell and this subpopulation help in the Leishmania parasitic growth and development. TargetScan and miRDB prediction gave that miR-3622b-5p has shown putative binding site at position 796–802 of 3′-UTR for Foxp3 gene. MiR-940 and miR-1827 were predicted by TargetScan and miRPath, have shown binding site at position 413–419 and 305–315 respectively of 3′-UTR for the same gene, to down regulate the Treg-specific differentiation (Figure 2). Interestingly, all the predicted miRNAs for Foxp3 gene have shown different binding at 3′-UTR of Foxp3 gene. Th17/Treg plasticity is equally important for the L. donovani parasite to raise VL infection. In presence of Treg-specific miRNA environment, Th17 cell differentiation followed by IL-17 and IL-22 cytokine production, may further inhibit the growth of L. donovani parasites. Predicted miRNAs for the key transcription factors and important cytokines related to Th17 and Treg cells have been explained in the Table 5.
FIGURE 2.
MicroRNAs mediated plasticity between Thl7 and Treg cells in human visceral leishmaniasis.
Table 5.
MiRNAs regulating Th17 and Treg cell differentiation and plasticity.
| Serial No. | Gene | miRDB | miRPath | TargetScan |
|---|---|---|---|---|
| 1 | RORC | - - - - |
let-7a-5p let-7b-5p let-7c miR-4500 |
let-7a-5p let-7b-5p let-7c miR-4500 |
| 2 | STAT3 | miR-93 miR-519d miR-17 miR-106b miR-106a miR-124 |
miR-93 miR-519d miR-17 miR-106b miR-106a miR-124 |
miR-93 miR-519d miR-17 miR-106b miR-106a - |
| 3 | TGFβ | - | - | miR-744 |
| 4 | IL6 | - | - | let-7 |
| 5 | Foxp3 | miR-3622b-5p - - |
- miR-940 miR-1827 |
miR-3622b-5p miR-940 miR-1827 |
| 6 | IL35 | - - |
- - |
miR-21 miR-590-5p |
Discussion
Human VL is associated with increased Th2 and Treg immune responses which induce the elevation of disease signature cytokines such as IL-6, IL-8, IL-10 and TGF-β. While a protective immune response against L. donovani is associated with elevated IFN-γ and IL-12 level, which leads to Th1 immune response. T cells developed from lymphoid progenitor stem cell in bone marrow and their maturation takes place in the thymus. MiRNAs have shown significant role in CD4+ T cell differentiation, maturation and to develop plasticity during cancer and infectious diseases (Liu et al., 2013). An immunological study indicates that Th1/Th2 paradigm is not solely responsible to controlling or favoring VL disease in the human model. Suppression of signature cytokines associated with Th1 immune response and elevation of Th2 type immune response cytokines have opened the window for development of immunotherapy against VL disease. Here, in this study we have predicted miRNAs as the regulatory molecule to control the CD4+ T cell differentiation to protect or favor VL disease. MiRNAs are an important player at the molecular level in cells which controls immune response in the human by controlling immune signaling at the post-transcriptional level. MiRNAs are emerging as a key controller for CD4+ T cell differentiation and for maintaining plasticity during VL infection. We targeted CD4+ T cell differentiation controlling transcription factor molecule for miRNA prediction which can regulate T cell differentiation phenomenon.
In our computational study, we observed that miR-548ah, miR-526b, miR-4726-5p, miR-29C, miR-29b, and miR-29a putatively control the expression of TBX21 protein expression in CD4+ T cell. During VL infection in Th2 type immune environment, negative regulation of TBX21 transcription factor by aforementioned miRNAs leads to raise the VL infection. Our computational study states that the elevated level of miR-1252, miR-4697, miR-4724, and miR-495, negatively regulates the STAT1 signaling. It was also noted that miR-200a expression in CD4+ T cells has the binding sites at the position 237-244 of 3′-UTR for STAT4 gene which can subsequently negatively regulate the Th1 type of immune response during VL infection (Huang et al., 2011).
In human VL infection, Th2 type of immune response activates to favor the replication of L. donovani in macrophages cells. Using TargetScan, miRPath and miRDB web servers, we identified miRNAs against the key transcription factor which has shown the potential to down regulate Th2 immune response. MiR-135 has a putative binding site to regulate GATA3 transcription factor which control the Th2 immune cell differentiation from CD4+ T cells. We also identified that miR-135 have the great potential to down regulate the expression of the STAT6 transcription factor at the post-transcriptional level in CD4+ T cells. Our finding indicates that let-7e, let-7c and miR-98 have the putative binding site at the 3′-UTR of IL-10 gene and negatively regulates the functional expression of IL-10. Our study states that miR-1272 and miR-155 have the potential to down regulate the IL-4/IL-13 signaling to check the Th2 response during VL infection. In the Th1 type of immune response micro-environment miR-1272 and miR-155 activated which subsequently inhibit the IL-4/IL-13 signaling pathway to suppress the Th2 type of immune response in human. Immune signaling polarization toward Th2 immune response is one of the main streams for the development of VL disease. In the presence of miRNAs investigated against GATA3, STAT6 transcription factors and IL-4, IL-13, IL-10 cytokines have the great potential to down regulate the Th2 immune response which subsequently induces the production of IFN-γ by Th1/Th2 plasticity phenomenon in human during VL infection. Furthermore, in the presence of elevated IFN-γ response, reactive oxygen species and nitric oxide signaling induces the parasitic clearance and protection from L. donovani parasites.
Mature T cells in peripheral blood of human consist of Th17 immune cells characterized by the production of IL-17, IL-21 and IL-22 cytokines. Protective role of IL-17 and IL-22 cytokines have been associated with the protection of human in response to L. donovani infection to suppress VL infection. Th17 immune cell differentiation from CD4+ T cell takes place by the expression of ROR-γt transcription factor. MiR-4500 and let-7c were found to have a putative binding site at the 3′-UTR of RORC gene, which negatively regulates the maturation and differentiation of Th17 cells during VL infection. In the condition of elevated expression of miR-93 and miR-124, miR-106a, and miR-106b, Th17 immune diminished and L. donovani parasite finds the favorable condition to replicate in the macrophages of human. Regulatory T cells are the subpopulation of CD4+ T cells and characterized by the expression of CD4+ CD25+ Foxp3+ in human VL and associated with the production of TGF-β cytokines to favor the parasite replication in VL disease. Our study indicates that miR-744 suppresses the expression of TGF-β cytokine which subsequently have the potential to inhibit the Treg cell differentiation and maturation in VL disease (Butz et al., 2012).
Our study highlights the importance of miRNAs in the CD4+ T cell differentiation, maturation and their functional aspects toward the development of protection from L. donovani parasites. This computational study generates the signaling mechanism controlled and regulated by miRNAs in human VL infection. MiRNA developed against Th2 and Treg immune cells are important and their over expression can be used for developing Th1 and Th17 type of protection specific immune response during VL infection. In cancer disease, many miRNA based therapy is under clinical trial and have shown excited efficacious response by developing a protective immune response. By this study, we have postulated that miRNAs against key transcription factors and important cytokines can be used to design miRNA based therapy to develop Th1 and Th17 type of protective immune response to generate IFN-γ and nitric oxide signaling to kill L. donovani parasites. By understanding miRNA regulatory network to control CD4+ T cell differentiation, we can develop less toxic accurate and targeted therapy against VL disease. By this fascinating bioinformatics based study one can provide the intricate mechanism to control CD4+ T cell differentiation and to develop gene regulation mechanism to inhibit L. donovani growth and replication. This study further develops the deep concern for the development of miRNA based therapy in VL disease.
Author Contributions
Conceived and designed the experiments: RKP, SS, and VKP. Performed the experiments: RKP and VKP. Analyzed the data: RKP and VKP. Contributed reagents/materials/analysis tools: RKP and VKP. Wrote the paper: RKP, SS, and VKP.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to Radheshyam Maurya from University of Hyderabad, India for reading this manuscript and his suggestion.
Footnotes
Funding. RKP is thankful to the Department of science and Technology, India for providing INSPIRE fellowship. VKP is thankful to University Grant commission for providing UGC startup grant [grant number 30-66/2014 (BSR)] and SERB (YSS/2015/000716).
References
- Afkarian M., Sedy J. R., Yang J., Jacobson N. G., Cereb N., Yang S. Y., et al. (2002). T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat. Immunol. 3 549–557. 10.1038/ni794 [DOI] [PubMed] [Google Scholar]
- Agarwal V., Bell G. W., Nam J. W., Bartel D. P. (2015). Predicting effective microRNA target sites in mammalian mRNAs. Elife 4:e05005 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Nakhle H., Burns P. A., Cummings M., Hanby A. M., Hughes T. A., Satheesha S., et al. (2010). Estrogen receptor β1 expression is regulated by miR-92 in breast cancer. Cancer Res. 70 4778–4784. 10.1158/0008-5472.CAN-09-4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvar J., Velez I. D., Bern C., Herrero M., Desjeux P., Cano J., et al. (2012). Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE 7:e35671 10.1371/journal.pone.0035671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E., Carrier Y., Gao W., Korn T., Strom T. B., Oukka M., et al. (2006). Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441 235–238. 10.1038/nature04753 [DOI] [PubMed] [Google Scholar]
- Butz H., Rácz K., Hunyady L., Patócs A. (2012). Crosstalk between TGF-β signaling and the microRNA machinery. Trends Pharmacol. Sci. 33 382–393. 10.1016/j.tips.2012.04.003 [DOI] [PubMed] [Google Scholar]
- Dweep H., Sticht C., Gretz N. (2013). In-silico algorithms for the screening of possible microrna binding sites and their interactions. Curr. Genomics. 14 127–136. 10.2174/1389202911314020005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenningloh R., Kang B. Y., Ho I. C. (2005). Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J. Exp. Med. 201 615–626. 10.1084/jem.20041330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington L. E., Hatton R. D., Mangan P. R., Turner H., Murphy T. L., Murphy K. M., et al. (2005). Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 6 1123–1132. 10.1038/ni1254 [DOI] [PubMed] [Google Scholar]
- Hsieh C. S., Macatonia S. E., Tripp C. S., Wolf S. F., O’garra A., Murphy K. M. (1993). Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science 260 547–549. 10.1126/science.8097338 [DOI] [PubMed] [Google Scholar]
- Huang C., Xiao X., Chintagari N. R., Breshears M., Wang Y., Liu L. (2014). MicroRNA and mRNA expression profiling in rat acute respiratory distress syndrome. BMC Med. Genomics 7:46 10.1186/1755-8794-7-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Lei Y., Zhang H., Hou L., Zhang M., Dayton A. I. (2011). MicroRNA regulation of STAT4 protein expression: rapid and sensitive modulation of IL-12 signaling in human natural killer cells. Blood 118 6793–6802. 10.1182/blood-2011-05-356162 [DOI] [PubMed] [Google Scholar]
- Kano S., Sato K., Morishita Y., Vollstedt S., Kim S., Bishop K., et al. (2008). The contribution of transcription factor IRF1 to the interferon-gamma-interleukin 12 signaling axis and TH1 versus TH-17 differentiation of CD4+ T cells. Nat. Immunol. 9 34–41. 10.1038/ni1538 [DOI] [PubMed] [Google Scholar]
- Kim J. I., Ho I. C., Grusby M. J., Glimcher L. H. (1999). The transcription factor c-Maf controls the production of interleukin-4 but not other Th2 cytokines. Immunity 10 745–751. 10.1016/S1074-7613(00)80073-4 [DOI] [PubMed] [Google Scholar]
- Le Gros G., Ben-Sasson S. Z., Seder R., Finkelman F. D., Paul W. E. (1990). Generation of interleukin 4 (IL-4)-producing cells in vivo and in vitro: IL-2 and IL-4 are required for in vitro generation of IL-4-producing cells. J. Exp. Med. 172 921–929. 10.1084/jem.172.3.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Tournier C., Davis R. J., Flavell R. A. (1999). Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 18 420–432. 10.1093/emboj/18.2.420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. J., Chau J., Ebert P. J., Sylvester G., Min H., Liu G., et al. (2007). miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 129 147–161. 10.1016/j.cell.2007.03.008 [DOI] [PubMed] [Google Scholar]
- Lighvani A. A., Frucht D. M., Jankovic D., Yamane H., Aliberti J., Hissong B. D., et al. (2001). T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc. Natl. Acad. Sci. U.S.A. 98 15137–15142. 10.1073/pnas.261570598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wu C. P., Lu B. F., Jiang J. T. (2013). Mechanism of T cell regulation by microRNAs. Cancer Biol. Med. 10 131–137. 10.7497/j.issn.2095-3941.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohoff M., Mittrucker H. W., Prechtl S., Bischof S., Sommer F., Kock S., et al. (2002). Dysregulated T helper cell differentiation in the absence of interferon regulatory factor 4. Proc. Natl. Acad. Sci. U.S.A. 99 11808–11812. 10.1073/pnas.182425099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Yue X., Cui Y., Zhang J., Wang K. (2013). MicroRNA-124 suppresses growth of human hepatocellular carcinoma by targeting STAT3. Biochem. Biophys. Res. Commun. 441 873–879. 10.1016/j.bbrc.2013.10.157 [DOI] [PubMed] [Google Scholar]
- Martins G., Calame K. (2008). Regulation and functions of Blimp-1 in T and B lymphocytes. Annu. Rev. Immunol. 26 133–169. 10.1146/annurev.immunol.26.021607.090241 [DOI] [PubMed] [Google Scholar]
- Miska E. A. (2005). How microRNAs control cell division, differentiation and death. Curr. Opin. Genet. Dev. 15 563–568. 10.1016/j.gde.2005.08.005 [DOI] [PubMed] [Google Scholar]
- Mullen A. C., Hutchins A. S., High F. A., Lee H. W., Sykes K. J., Chodosh L. A., et al. (2002). Hlx is induced by and genetically interacts with T-bet to promote heritable T(H)1 gene induction. Nat. Immunol. 3 652–658. [DOI] [PubMed] [Google Scholar]
- Nakahama T., Hanieh H., Nguyen N. T., Chinen I., Ripley B., Millrine D., et al. (2013). Aryl hydrocarbon receptor-mediated induction of the microRNA-132/212 cluster promotes interleukin-17-producing T-helper cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 110 11964–11969. 10.1073/pnas.1311087110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoe Y., Setoguchi R., Akiyama K., Muroi S., Kuroda M., Hatam F., et al. (2007). Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J. Exp. Med. 204 1749–1755. 10.1084/jem.20062456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevopoulou M. D., Georgakilas G., Kostoulas N., Reczko M., Maragkakis M., Dalamagas T. M., et al. (2013). DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res. 41 D239–D245. 10.1093/nar/gks1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Li Z., Yang X. O., Chang S. H., Nurieva R., Wang Y. H., et al. (2005). A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 6 1133–1141. 10.1038/ni1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitta M. G., Romano A., Cabantous S., Henri S., Hammad A., Kouriba B., et al. (2009). IL-17 and IL-22 are associated with protection against human kala azar caused by Leishmania donovani. J. Clin. Invest. 119 2379–2387. 10.1172/JCI38813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapati V. K., Mehrotra S., Gautam S., Rai M., Sundar S. (2012). In vitro antileishmanial drug susceptibility of clinical isolates from patients with Indian visceral leishmaniasis–status of newly introduced drugs. Am. J. Trop. Med. Hyg. 87 655–657. 10.4269/ajtmh.2012.12-0022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirion M. R., Gregory G. D., Umetsu S. E., Winandy S., Brown M. A. (2009). Cutting edge: Ikaros is a regulator of Th2 cell differentiation. J. Immunol. 182 741–745. 10.4049/jimmunol.182.2.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagnani S. (1999). Th1/Th2 cells. Inflamm. Bowel Dis. 5 285–294. 10.1002/ibd.3780050410 [DOI] [PubMed] [Google Scholar]
- Ruan K., Fang X., Ouyang G. (2009). MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett. 285 116–126. 10.1016/j.canlet.2009.04.031 [DOI] [PubMed] [Google Scholar]
- Seder R. A., Paul W. E., Davis M. M., De St Groth B. (1992). The presence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of CD4+ T cells from T cell receptor transgenic mice. J. Exp. Med. 176 1091–1098. 10.1084/jem.176.4.1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U., Singh S. (2009). Immunobiology of leishmaniasis. Indian J. Exp. Biol. 47 412–423. [PubMed] [Google Scholar]
- Sundar S., Reed S. G., Sharma S., Mehrotra A., Murray H. W. (1997). Circulating T helper 1 (Th1) cell- and Th2 cell-associated cytokines in Indian patients with visceral leishmaniasis. Am. J. Trop. Med. Hyg. 56 522–525. [DOI] [PubMed] [Google Scholar]
- Swain S. L., Weinberg A. D., English M., Huston G. (1990). IL-4 directs the development of Th2-like helper effectors. J. Immunol. 145 3796–3806. [PubMed] [Google Scholar]
- Szabo S. J., Kim S. T., Costa G. L., Zhang X., Fathman C. G., Glimcher L. H. (2000). A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100 655–669. [DOI] [PubMed] [Google Scholar]
- Takeda K., Tanaka T., Shi W., Matsumoto M., Minami M., Kashiwamura S., et al. (1996). Essential role of Stat6 in IL-4 signalling. Nature 380 627–630. 10.1038/380627a0 [DOI] [PubMed] [Google Scholar]
- Thomson D. W., Bracken C. P., Goodall G. J. (2011). Experimental strategies for microRNA target identification. Nucleic Acids Res. 39 6845–6853. 10.1093/nar/gkr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unanue E. R., Allen P. M. (1987). The basis for the immunoregulatory role of macrophages and other accessory cells. Science 236 551–557. 10.1126/science.2437650 [DOI] [PubMed] [Google Scholar]
- Usui T., Nishikomori R., Kitani A., Strober W. (2003). GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rbeta2 chain or T-bet. Immunity 18 415–428. 10.1016/S1074-7613(03)00057-8 [DOI] [PubMed] [Google Scholar]
- Wang X., El Naqa I. M. (2008). Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics 24 325–332. 10.1093/bioinformatics/btm595 [DOI] [PubMed] [Google Scholar]
- Wong N., Wang X. (2015). miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 43 D146–D152. 10.1093/nar/gku1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Dai Z., Chen S., Chen L. (2011). MicroRNA-mediated gene regulation plays a minor role in the transcriptomic plasticity of cold-acclimated zebrafish brain tissue. BMC Genomics 12:605 10.1186/1471-2164-12-605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. O., Angkasekwinai P., Zhu J., Peng J., Liu Z., Nurieva R., et al. (2009). Requirement for the basic helix-loop-helix transcription factor Dec2 in initial TH2 lineage commitment. Nat. Immunol. 10 1260–1266. 10.1038/ni.1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Tian A., Wang J., Shen X., Qi G., Tang Y. (2015). miR26a modulates Th17/T reg balance in the EAE model of multiple sclerosis by targeting IL6. Neuromol. Med. 17 24–34. 10.1007/s12017-014-8335-5 [DOI] [PubMed] [Google Scholar]
- Zheng W., Flavell R. A. (1997). The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89 587–596. 10.1016/S0092-8674(00)80240-8 [DOI] [PubMed] [Google Scholar]
- Zhou L., Lopes J. E., Chong M. M., Ivanov I. I., Min R., Victora G. D., et al. (2008). TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature 453 236–240. 10.1038/nature06878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Cote-Sierra J., Guo L., Paul W. E. (2003). Stat5 activation plays a critical role in Th2 differentiation. Immunity 19 739–748. 10.1016/S1074-7613(03)00292-9 [DOI] [PubMed] [Google Scholar]