Abstract
We herein report an association between TMEM132D and panic disorder (PD) in a Japanese population, evaluating the effects of HLA-DRB1*13:02, which we previously reported as a susceptibility genetic factor for PD. SNPs in TMEM132D showed significant associations with PD in subjects without HLA-DRB1*13:02 (rs4759997; P=5.02×10−6, odds ratio=1.50) but not in those with the HLA allele. TMEM132D might have a role in the development of PD in subjects without HLA-DRB1*13:02.
Panic disorder (PD) is an anxiety disorder characterized by panic attacks and anticipatory anxiety. PD is relatively common; the lifetime prevalence is reported to be 1–3%.1 According to a previous twin study, the heritability of PD is estimated to be 0.43,2 which suggests that both genetic and environmental factors have a role in the pathogenesis of PD. To date, several studies that applied a candidate-gene approach have reported susceptibility genes of PD, but many of them have not been successfully replicated in subsequent studies.3 Recently, a genome-wide association study (GWAS) of European ancestry identified single-nucleotide polymorphisms (SNPs) in the transmembrane protein 132D gene (TMEM132D) associated with PD.4 This result was supported by a replication study and meta-analyses of European subjects, which confirmed that TMEM132D is a susceptibility gene of PD.5,6 However, in a Japanese GWAS of PD, SNPs in TMEM132D did not show a positive association with PD.7,8
We previously found associations between PD and human leukocyte antigen (HLA), especially the HLA-B and HLA-DRB1 genes, based on pathway analyses using the results from our Japanese GWAS of PD.8 HLA is the human version of the major histocompatibility complex, which presents endogenous antigens to CD8+ and CD4+ T cells. There is a great number of polymorphisms in the HLA genes. HLA genes have been reported to be involved in not only immune-related diseases9 but also several psychiatric disorders.10 We genotyped the HLA-B and HLA-DRB1 genes, and confirmed that the frequency of HLA-DRB1*13:02 was significantly higher in PD patients than in healthy individuals (case positivity: 18.1%; control positivity: 11.5%; P=2.62×10−5; odds ratio (OR)=1.70).11
Previous studies have reported that the genetic factors and clinical features of several HLA-associated diseases differ between HLA allele-positive and -negative patients. Narcolepsy, with and without cataplexy, was associated with HLA-DQB1*06:02,12 and the severity of narcolepsy without cataplexy was higher in HLA-DQB1*06:02-positive patients than in HLA-DQB1*06:02-negative patients.12,13 HLA-B*51 was strongly associated with risk factors for Behçet’s disease,14 and a significant association between one SNP in the ERAP1 locus was observed only in HLA-B*51-positive patients.14 Hence there is a possibility that the genetic backgrounds might differ in PD subjects with or without HLA-DRB1*13:02. To account for these effects of HLA alleles, we focused on a candidate PD gene, TMEM132D, and investigated the SNPs in the TMEM132D region in both HLA-DRB1*13:02-positive and -negative subjects. In this analysis, genotyping data for the SNPs were generated using the Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA). Inclusion criteria for quality control were SNP call rate >0.95, Hardy–Weinberg equilibrium (HWE) test P>0.001, and minor allele frequency (MAF)>0.05. We defined ‘gene region’ as the region located 50 kb upstream to 50 kb downstream of TMEM132D (chr12: 129556271–130388212 (GRCh37/hg19)). The SNP genotype data were subdivided into two data sets, those of HLA-DRB1*13:02-positive subjects (cases: N=103; controls: N=198) and those of HLA-DRB1*13:02-negative subjects (cases: N=438; controls: N=1,341). An imputation analysis was also performed to evaluate the potential association of ungenotyped SNPs in the TMEM132D region of both subgroups. IMPUTE2 software15 was used to estimate SNP genotypes using the reference data set from 1000 Genomes Phase 3 haplotypes.15 We filtered out low-quality imputed SNPs by applying the following conditions: SNP call rate ⩾0.95, HWE test P>0.0001, and probability of imputation certainty ⩾0.9. After filtering, a total of 8,070 SNPs remained for subsequent analysis. Using the genotype data of these SNPs, case–control association tests were performed to examine whether SNPs in TMEM132D showed an association with PD in each subgroup. We set the significance level after multiple testing correction to α=1.26×10−5, which was calculated from 0.05 divided by the number of SNPs (N=3,978) pruned by high linkage disequilibrium (LD; r 2>0.8) with PLINK SNP pruning procedure (window size in SNPs=100, the number of SNPs to shift the window=1).16
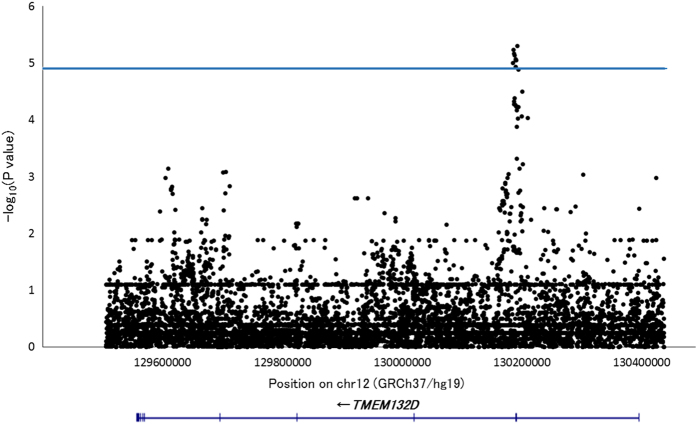
In the analysis of the HLA-DRB1*13:02-negative subgroup, nine SNPs in the TMEM132D region showed significant associations, and SNP rs4759997 had the lowest P value (P=5.02×10−6, OR=1.50; Table 1 and Figure 1). In contrast, these SNPs were found to have no association with PD in the HLA-DRB1*13:02-positive group (Table 1 and Supplementary Figure 1). To find other SNPs potentially associated with PD in the HLA-DRB1*13:02-negative group, logistic regression analysis adjusting for the effect of rs4759997 was also performed. The analysis showed that none of the SNPs in the TMEM132D region had an association that reached the threshold level of significance, which suggested that the nominal associations of SNPs in this region were derived from LD with rs4759997 (Supplementary Figure 2).
Table 1. SNPs with P-value <10−4 in the TMEM132D region.
| Positiona | SNP |
HLA-DRB1*13:02 negative |
HLA-DRB1*13:02 positive |
||||||
|---|---|---|---|---|---|---|---|---|---|
|
MAF |
P-value | OR |
MAF |
P-value | OR | ||||
| PD | Control | PD | Control | ||||||
| 130185851 | rs1567509 | 0.283 | 0.210 | 1.01×10−5 b | 1.49 | 0.211 | 0.203 | 0.820 | 1.05 |
| 130186374 | rs7311162 | 0.279 | 0.205 | 5.87×10−6 b | 1.50 | 0.199 | 0.198 | 0.975 | 1.01 |
| 130187014 | rs264463 | 0.105 | 0.064 | 4.79×10−5 | 1.73 | 0.050 | 0.054 | 0.854 | 0.93 |
| 130187283 | rs1397504 | 0.281 | 0.208 | 6.92×10−6 b | 1.49 | 0.199 | 0.200 | 0.989 | 1.00 |
| 130187566 | rs264464 | 0.104 | 0.063 | 5.30×10−5 | 1.73 | 0.050 | 0.054 | 0.854 | 0.93 |
| 130188352 | rs264465 | 0.105 | 0.063 | 4.19×10−5 | 1.73 | 0.058 | 0.061 | 0.908 | 0.96 |
| 130188504 | rs7962650 | 0.279 | 0.206 | 7.32×10−6 b | 1.49 | 0.194 | 0.200 | 0.876 | 0.97 |
| 130189452 | rs67208922 | 0.104 | 0.063 | 5.46×10−5 | 1.72 | 0.050 | 0.054 | 0.833 | 0.92 |
| 130189478 | rs264468 | 0.104 | 0.063 | 5.46×10−5 | 1.72 | 0.050 | 0.054 | 0.833 | 0.92 |
| 130189868 | rs10773696 | 0.279 | 0.206 | 8.65×10−6 b | 1.49 | 0.194 | 0.200 | 0.876 | 0.97 |
| 130190130 | rs7312812 | 0.279 | 0.207 | 1.19×10−5 b | 1.48 | 0.194 | 0.199 | 0.888 | 0.97 |
| 130190285 | rs1510820 | 0.279 | 0.207 | 9.10×10−6 b | 1.48 | 0.194 | 0.200 | 0.876 | 0.97 |
| 130191111 | rs7132791 | 0.279 | 0.207 | 9.10×10−6 b | 1.48 | 0.194 | 0.200 | 0.876 | 0.97 |
| 130191332 | rs264472 | 0.104 | 0.063 | 5.90×10−5 | 1.72 | 0.050 | 0.056 | 0.745 | 0.88 |
| 130191567 | rs2398467 | 0.104 | 0.063 | 5.90×10−5 | 1.72 | 0.049 | 0.056 | 0.725 | 0.87 |
| 130191851 | rs529395389 | 0.104 | 0.063 | 6.92×10−5 | 1.71 | 0.049 | 0.056 | 0.716 | 0.87 |
| 130192489 | rs588761 | 0.104 | 0.063 | 5.90×10−5 | 1.72 | 0.049 | 0.056 | 0.716 | 0.87 |
| 130193038 | rs4759997 | 0.282 | 0.208 | 5.02×10−6 b | 1.50 | 0.199 | 0.200 | 0.989 | 1.00 |
| 130193940 | rs663071 | 0.104 | 0.064 | 9.67×10−5 | 1.69 | 0.049 | 0.056 | 0.716 | 0.87 |
| 130195133 | rs67408383 | 0.104 | 0.063 | 6.03×10−5 | 1.72 | 0.049 | 0.056 | 0.716 | 0.87 |
| 130195225 | rs7304093 | 0.279 | 0.208 | 1.31×10−5 | 1.47 | 0.194 | 0.200 | 0.876 | 0.97 |
| 130199905 | rs6486497 | 0.356 | 0.286 | 8.73×10−5 | 1.38 | 0.257 | 0.293 | 0.356 | 0.84 |
| 130201128 | rs10744430 | 0.366 | 0.292 | 3.19×10−5 | 1.41 | 0.277 | 0.296 | 0.630 | 0.91 |
| 130210550 | rs76801035 | 0.055 | 0.027 | 9.36×10−5 | 2.07 | 0.025 | 0.020 | 0.738 | 1.21 |
Abbreviations: MAF, minor allele frequency; OR, odds ratio; PD, panic disorder; SNP, single-nucleotide polymorphism.
Physical position (according to GRCh37/hg19).
The significance level after multiple testing correction was set as α=1.26×10−5.
Figure 1.
Results of the HLA-DRB1*13:02-negative subgroup analysis in the TMEM132D region. Physical positions are based on GRCh37/hg19. The blue line represents the significance threshold (α=1.26×10−5).
A previous study identified two SNPs, rs7309727 and rs11060369, in TMEM132D as susceptibility variants for PD in populations of European ancestry.4 The two SNPs were also associated with higher anxiety and larger amygdala volumes.17 In addition, the risk genotype of rs11060369 was found to enhance TMEM132D mRNA expression in the brain.4 These two SNPs identified in populations of European ancestry were located in intron 3 of TMEM132D, while the SNPs found in our study, rs4759997 and the surrounding SNPs with significant P values, were located in intron 1. The SNP with the lowest P value, rs4759997, was not in LD with either rs7309727 or rs11060369 in individuals of Japanese ancestry (Japanese; rs7309727, r 2=0.001; rs11060369, r 2=0.003), while in individuals of European ancestry, SNP rs4759997 had very low frequency (MAF=0.009) according to HapMap data.18,19 In addition, imputation analysis revealed that the two SNPs, rs7309727 and rs11060369, were not associated with PD in HLA-DRB1*13:02-negative Japanese subjects (rs7309727: case MAF=0.36, control MAF=0.39, P=0.124; rs11060369: case MAF=0.46, control MAF=0.46, P=0.826). Such results, showing that different SNPs in TMEM132D are associated with PD in individual populations, might be derived from differences in the LD structure between the populations of Japanese and European ancestry (Supplementary Figure 3). Therefore, targeted resequencing of this gene is required in a future study.
Our study provides initial evidence that SNPs in TMEM132D show significant associations with PD in a HLA-DRB1*13:02-negative group of Japanese individuals. Specifically, TMEM132D might affect PD in HLA-DRB1*13:02-negative individuals. Further replication studies in independent and larger HLA-typed population samples are required to confirm these associations.
Acknowledgments
We thank all the participants in this study. This study was supported by JSPS KAKENHI (No. 25461723; No. 26461712) and Grants-in-Aid for Scientific Research on Priority Areas ‘Comprehensive Genomics’ and ‘Applied Genomics’ (No. 17019029), and Innovative Areas (No. 22133008), from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Supplementary Information for this article can be found on the Human Genome Variation website (http://www.nature.com/hgv)
The authors declare no conflict of interest.
References
- Eaton WW , Kessler RC , Wittchen HU , Magee WJ . Panic and panic disorder in the United States. Am J Psychiatry 1994; 151: 413–420. [DOI] [PubMed] [Google Scholar]
- Hettema JM , Neale MC , Kendler KS . A review and meta-analysis of the genetic epidemiology of anxiety disorders. Am J Psychiatry 2001; 158: 1568–1578. [DOI] [PubMed] [Google Scholar]
- Na HR , Kang EH , Lee JH , Yu BH . The genetic basis of panic disorder. J Korean Med Sci 2011; 26: 701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt A , Czibere L , Roeske D , Lucae S , Unschuld PG , Ripke S et al. TMEM132D, a new candidate for anxiety phenotypes: evidence from human and mouse studies. Mol Psychiatry 2011; 16: 647–663. [DOI] [PubMed] [Google Scholar]
- Erhardt A , Akula N , Schumacher J , Czamara D , Karbalai N , Müller-Myhsok B et al. Replication and meta-analysis of TMEM132D gene variants in panic disorder. Transl Psychiatry 2012; 2: e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe AS , Buttenschøn HN , Bani-Fatemi A , Maron E , Otowa T , Erhardt A et al. Candidate genes in panic disorder: meta-analyses of 23 common variants in major anxiogenic pathways. Mol Psychiatry (e-pub ahead of print 22 September 2015; doi:10.1038/mp.2015.138). [DOI] [PubMed]
- Otowa T , Yoshida E , Sugaya N , Yasuda S , Nishimura Y , Inoue K et al. Genome-wide association study of panic disorder in the Japanese population. J Hum Genet 2009; 54: 122–126. [DOI] [PubMed] [Google Scholar]
- Otowa T , Kawamura Y , Nishida N , Sugaya N , Koike A , Yoshida E et al. Meta-analysis of genome-wide association studies for panic disorder in the Japanese population. Transl Psychiatry 2012; 2: e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoshitz J . The quest for better understanding of HLA-disease association: scenes from a road less travelled by. Discov Med 2013; 16: 93–101. [PMC free article] [PubMed] [Google Scholar]
- Corvin A , Morris DW . Genome-wide association studies: findings at the major histocompatibility complex locus in psychosis. Biol Psychiatry 2014; 75: 276–283. [DOI] [PubMed] [Google Scholar]
- Shimada-Sugimoto M , Otowa T , Miyagawa T , Khor SS , Kashiwase K , Sugaya N et al. Immune-related pathways including HLA-DRB1(∗)13:02 are associated with panic disorder. Brain Behav Immun 2015; 46: 96–103. [DOI] [PubMed] [Google Scholar]
- Sasai T , Inoue Y , Komada Y , Sugiura T , Matsushima E . Comparison of clinical characteristics among narcolepsy with and without cataplexy and idiopathic hypersomnia without long sleep time, focusing on HLA-DRB1(*)1501/DQB1(*)0602 finding. Sleep Med 2009; 10: 961–966. [DOI] [PubMed] [Google Scholar]
- Sasai-Sakuma T , Inoue Y . Differences in electroencephalographic findings among categories of narcolepsy-spectrum disorders. Sleep Med 2015; 16: 999–1005. [DOI] [PubMed] [Google Scholar]
- Kirino Y , Bertsias G , Ishigatsubo Y , Mizuki N , Tugal-Tutkun I , Seyahi E et al. Genome-wide association analysis identifies new susceptibility loci for Behçet’s disease and epistasis between HLA-B*51 and ERAP1. Nat Genet 2013; 45: 202–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN , Donnelly P , Marchini J . A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S , Neale B , Todd-Brown K , Thomas L , Ferreira MA , Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaker J , Lonsdorf TB , Raczka KA , Mechias ML , Gartmann N , Kalisch R . Higher anxiety and larger amygdala volumes in carriers of a TMEM132D risk variant for panic disorder. Transl Psychiatry 2014; 4: e357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A haplotype map of the human genome. Nature 2005; 437: 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. The International HapMap Project. Nature 2003; 426: 789–796. [DOI] [PubMed] [Google Scholar]
Data Citations
- Otowa Takeshi.HGV Database. 2015. 10.6084/m9.figshare.hgv.771. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Otowa Takeshi.HGV Database. 2015. 10.6084/m9.figshare.hgv.771. [DOI]