Dysregulation of cholesterol synthesis is implicated in Huntington’s disease. Boussicault et al. show that expression of CYP46A1, the rate-limiting enzyme in cholesterol degradation, is reduced in patients and a mouse model. Restoration of CYP46A1 re-establishes normal cholesterol levels and is neuroprotective, suggesting that targeting cholesterol degradation may have therapeutic potential.
Keywords: CYP46A1, cholesterol, striatum, Huntington’s disease, neuroprotection
Dysregulation of cholesterol synthesis is implicated in Huntington’s disease. Boussicault et al. show that expression of CYP46A1, the rate-limiting enzyme in cholesterol degradation, is reduced in patients and a mouse model. Restoration of CYP46A1 re-establishes normal cholesterol levels and is neuroprotective, suggesting that targeting cholesterol degradation may have therapeutic potential.
Abstract
Huntington’s disease is an autosomal dominant neurodegenerative disease caused by abnormal polyglutamine expansion in huntingtin (Exp-HTT) leading to degeneration of striatal neurons. Altered brain cholesterol homeostasis has been implicated in Huntington’s disease, with increased accumulation of cholesterol in striatal neurons yet reduced levels of cholesterol metabolic precursors. To elucidate these two seemingly opposing dysregulations, we investigated the expression of cholesterol 24-hydroxylase (CYP46A1), the neuronal-specific and rate-limiting enzyme for cholesterol conversion to 24S-hydroxycholesterol (24S-OHC). CYP46A1 protein levels were decreased in the putamen, but not cerebral cortex samples, of post-mortem Huntington’s disease patients when compared to controls. Cyp46A1 mRNA and CYP46A1 protein levels were also decreased in the striatum of the R6/2 Huntington’s disease mouse model and in ST hdh Q111 cell lines. In vivo , in a wild-type context, knocking down CYP46A1 expression in the striatum, via an adeno-associated virus-mediated delivery of selective shCYP46A1, reproduced the Huntington’s disease phenotype, with spontaneous striatal neuron degeneration and motor deficits, as assessed by rotarod. In vitro , CYP46A1 restoration protected ST hdh Q111 and Exp-HTT-expressing striatal neurons in culture from cell death. In the R6/2 Huntington’s disease mouse model, adeno-associated virus-mediated delivery of CYP46A1 into the striatum decreased neuronal atrophy, decreased the number, intensity level and size of Exp-HTT aggregates and improved motor deficits, as assessed by rotarod and clasping behavioural tests. Adeno-associated virus-CYP46A1 infection in R6/2 mice also restored levels of cholesterol and lanosterol and increased levels of desmosterol. In vitro , lanosterol and desmosterol were found to protect striatal neurons expressing Exp-HTT from death. We conclude that restoring CYP46A1 activity in the striatum promises a new therapeutic approach in Huntington’s disease.
Introduction
Huntington’s disease is a dominantly inherited genetic disease characterized by an abnormal expansion of a CAG trinucleotide repeat in the 5’ region of the huntingtin gene ( HTT ) leading to a polyglutamine expansion in the huntingtin protein (Exp-HTT) ( The Huntington’s Disease Collaborative Group, 1993 ). Individuals with 36 CAG repeats or more in the HTT gene will develop clinical symptoms of Huntington’s disease including neuropsychiatric, motor and cognitive deficits ( Aylward et al. , 2000 ). Once the first symptoms have appeared, the disease progresses and leads to death within ∼15 to 20 years. Brain atrophy in Huntington’s disease predominates in the caudate-putamen (striatum) and, to a lesser extent, in the cerebral cortex.
Exp-HTT leads to many cellular dysfunctions, including transcriptional dysregulation ( Moumné et al. , 2013 ), altered energy metabolism ( Cui et al. , 2006 ; Tsunemi et al. , 2012 ), excitotoxicity ( Milnerwood and Raymond, 2010 ), impaired axonal growth ( Li et al. , 2001 ) and transport ( Gauthier et al. , 2004 ). It also induces protein aggregation, a mechanism involved in several neurological disorders, including Alzheimer’s disease and other CAG repeat-associated diseases ( Ross and Poirier, 2004 ). Each of these cellular alterations can be involved, either separately or in concert, in striatal neuron dysfunction and death ( Roze et al. , 2008 a ).
Huntington’s disease is also associated with abnormalities in cholesterol metabolism ( Karasinska et al. , 2011 ; Valenza et al. , 2011 ), as observed in other neurodegenerative diseases including Niemann-Pick disease Type C, Smith-Lemli-Opitz ( Korade and Kenworthy, 2008 ) and Alzheimer’s disease ( Puglielli et al. , 2003 ; Shobab et al. , 2005 ). Cholesterol plays a critical role in brain development, synaptogenesis, neuronal activity, and neuron survival ( Bjorkem et al. , 2004 ; Dietschy et al. , 2004 ). In the CNS, the blood–brain barrier is not permeable to cholesterol and the brain sterol pool comes from in situ synthesis, mostly from astrocytes ( Pfrieger et al. , 2003 ; Bjorkem et al. , 2004 ). Normal cholesterol levels are strictly maintained by de novo synthesis and degradation and cells can sense their level of cholesterol through the membrane-bound sterol regulatory element-binding protein (SREBP) ( Brown and Goldstein, 1997 ). Brain cholesterol is catabolized to 24S-hydroxycholesterol (24S-OHC) by the neuron-specific enzyme CYP46A1 ( Lund et al. , 1999 ). 24S-OHC in turn crosses the blood–brain barrier and is metabolized in the periphery ( Bjorkhem et al. , 1997 ). Of note, cholesterol synthesis is also influenced by BDNF, which stimulates the transcription of enzymes involved in the cholesterol pathway in neuronal, but not glial cells, in culture ( Suzuki et al. , 2007 ; Valenza, 2007 a , 2010 ). Hence, reduced transcription and cortico-striatal release of BDNF may also account for decreased striatal cholesterol synthesis in Huntington’s disease striatal neurons ( Zuccato et al. , 2001 ; Gauthier et al. , 2004 ). Finally it must be emphasized that Exp-HTT impairs cholesterol metabolism and expression levels of MBP in oligodendrocytes, which may account for developmental myelination deficits ( Xiang et al. , 2011 ).
Altered expression of a number of genes involved in cholesterol and fatty acid metabolism has been described for Huntington’s disease cell lines and mouse models ( Sipione et al. , 2002 ; Valenza et al. , 2005 ; Valenza and Cattaneo, 2011 ). Enzymes involved in cholesterol biosynthesis, such as HMG-CoAR, CYP51 (lanosterol 14-α demethylase) and 7-DHC (7-dehydrocholesterol reductase), show reduced expression in mouse models, along with decreased levels of brain sterols, such as lanosterol and lathosterol. The levels of 24S-OHC are also reduced in the plasma of patients with Huntington’s disease ( Leoni et al. , 2008 , 2013 ), and in the brain of several yeast artificial chromosome (YAC) and knock-in mouse models ( Valenza et al. , 2010 ). Findings regarding cholesterol content in Huntington’s disease mouse models are less clear, with studies indicating either a decrease ( Valenza et al. , 2007 a , b , 2010 ), or an accumulation ( Trushina et al. , 2006 ), depending on the model and the methodology used ( Marullo et al. , 2012 ). In the caudate of patients and in Huntington’s disease neuronal cell lines, cholesterol showed an accumulation ( del Toro et al. , 2010 ). Therefore, increased cholesterol content can persist in Huntington’s disease, despite reduced synthesis. One important way to account for this apparent paradox is by reduced catabolism of cholesterol.
In the present work, we show that the expression levels of CYP46A1, the brain-specific rate-limiting enzyme in the degradation of cholesterol ( Lund et al. , 1999 ; Bjorkhem and Meaney, 2004 ), are decreased in the putamen of patients with Huntington’s disease, in the striatum of R6/2 mice, and in striatal Huntington’s disease cell lines. In vivo, in a wild-type context, CYP46A1 knocking-down in the mouse striatum, induced a decrease of 24S-OHC levels and produced a spontaneous striatal neurodegeneration associated to abnormal balance and motor coordination. We further show that restoring CYP46A1 in vitro reduces striatal neuron dysfunctions and aggregate formation. In the R6/2 Huntington’s disease mouse model, adeno-associated virus (AAV)-mediated delivery of CYP46A1 into the striatum decreased neuronal atrophy and protein aggregates and improved motor deficits, as assessed by rotarod and clasping behavioural tests. As expected, CYP46A1 restoration in the striatum of R6/2 mice increased 24S-OHC levels. It also restored normal cholesterol and lanosterol levels, and increased desmosterol, which proved to be neuroprotective. Hence, our study provides the first evidence that restoring normal levels of CYP46A1 may be neuroprotective in Huntington’s disease.
Materials and methods
Human extracts
Human brain samples were obtained from INSERM U289 Brain Bank at the Salpétrière hospital (Paris) according to standard legislation. Cerebral cortex and putamen samples were obtained from five control subjects with no history of neurological or psychiatric disorders (age: 72 ± 7.3 years; post-mortem delay: 10 ± 1.78 h), and six patients with Huntington’s disease clinically and neuropathologically diagnosed as grade 2, or 2/3 according to the classification of Vonsattel et al. (1985) (age: 64 ± 3.38 years; post-mortem delay: 25.7 ± 6.08 h). These patients displayed choreic movements at the end of the life.
Mice
Two-month-old female wild-type C57Bl/6 (Janvier) were used (average weight 20–25 g). R6/2 [B6CBA-Tg (HDexon1) 62Gpb/1J] mice, which express exon 1 of the human mutant Huntington’s disease gene containing 160 CAG repeats, under the control of the human HTT (IT15) promoter, were obtained from Jackson Laboratories by crossing ovarian transplant hemizygous females with B6CBAFI/J males. All mice used in the study were from the first offspring and the genotype was verified by polymerase chain reaction (PCR) using genomic DNA extracted from tail. The number of CAG repeat length varies very little in the progeny from the first generation ( http://chdifoundation.org/wp-content/uploads/HD_Field_Guide_040414.pdf ), and can therefore be considered around 160 CAG for all the mice that were used in the study. The mice were housed in groups with a 12-h light/dark cycle and provided with food and water ad libitum . Animal care was conducted in accordance with standard ethical guidelines (U.S. National Institutes of Health publication no. 85–24, revised 1985, and European Committee Guidelines on the Care and Use of Laboratory Animals) and the local ethics committee approved the experiments.
RNA extraction and LightCycler® real time polymerase chain reaction
Mice were sacrificed by decapitation, and heads were snap frozen in liquid nitrogen. Cerebral cortex, striata and hippocampus were punched on dry ice and stored at −80°C until RNA isolation. Samples were homogenized in QIAzol® reagent (Qiagen). RNA isolation was performed with RNeasy® Lipid Tissue Midi kit (Qiagen) according to manufacturer’s instructions. Reverse transcription was performed with Maxima First Strand cDNA synthesis kit for RT-qPCR (Fermentas). Quantitative real-time PCR reactions were performed using LightCycler® 480 SYBR® Green I Master according to manufacturer’s protocol and run on LightCycler® 480 (Roche Diagnostics). Expression of hypoxanthine guanine phosphoribosyltransferase 1 ( Hprt1 ) transcript was used to normalize the cDNA amounts. The cycle threshold values were calculated automatically by LightCycler® 480 SW 1.5 software with default parameters. For primers sequences see the Supplementary material .
Cultures of striatal neurons and STHdh (Q111/Q7) striatal cells, transfection, sterol treatment and cell survival assays
Primary striatal neurons were dissected from Day 14 embryos from pregnant Swiss mice (Janvier) as previously described ( Charvin et al. , 2005 ; Deyts et al. , 2009 ). After 7 days in vitro , transient transfection of striatal cells was performed with Lipofectamine™ 2000 (Invitrogen). At this stage, very few glial cells were observed (<5%; data not shown). Green fluorescent protein (GFP)-tagged constructs encoding the first exon of human HTT containing either 25 (25Q-HTT) or 103 (103Q-HTT) continuous CAA or CAG repeats were provided by the Huntington’s Disease Foundation Resource Bank, UCLA, USA. The human CYP46A1 cDNA was tagged with the haemagglutinin (HA) epitope (CYP46A1-HA). Conditionally immortalized wild-type (STHdhQ7) and mutant (STHdhQ111) striatal neuronal progenitor cell lines expressing HTT with either seven or 111 glutamines were grown as previously described ( Gines et al. , 2003 ). These cells were transfected with cDNA encoding CYP46A1-HA for 48 h, serum starved for 24 h and then fixed with 4% paraformaldehyde in phosphate-buffered saline.
Endogenous mouse CYP46A1 was detected owing to immunocytochemistry with anti-rabbit CYP46A1 antibody (1:1000, Abcam), neurons with anti-mouse MAP2 antibody (1:1000, Millipore), CYP46A1-HA transfected neurons with anti-rat HA antibody (1:1000, Roche). Appropriate anti-rabbit, anti-mouse, or anti-rat cy3, cy5, Alexa Fluor® 488 fluorophore-conjugated secondary antibodies were used. Lanosterol, desmosterol, cholesterol or 24S-OHC (1 µM in dimethyl sulphoxide) (Avanti Polar Lipids) was added for 48 h in the cell culture medium just after transfection. Nuclei were stained with Hoechst 1:20 000.
Immunoblotting
Human brain samples were homogenized in Nonidet™ P-40 lysis buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 2 mM EGTA, 1% Nonidet™ P-40, 10 mM β-glycerophosphate, 5 mM NaF, 1 mM NaPPi, 2 mM dithiothreitol, 1 mM sodium vanadate, and 100 μM PMSF) and cleared by centrifugation at 6000 g (15 min; 4°C). Supernatants were collected and analysed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis. Mouse tissues and striatal cells were homogenized as previously described ( Roze et al. , 2008 b ). Primary antibody rabbit polyclonal anti-CYP46A1 (1:1000) (Abcam), and mouse monoclonal anti-β-tubulin 1:5000 (Sigma-Aldrich) were revealed with appropriate anti-mouse or anti-rabbit peroxidase-conjugated secondary antibodies (Jackson Laboratories) and the ECL chemiluminescent reaction (Pierce).
Production and stereotaxic injection of AAV5-GFPshCYP46A1, AAV5-GFPshscramble, AAVrh10-GFP and AAVrh10-CYP46A1
All AAV vectors were produced and purified by Atlantic Gene therapies (Inserm U1089, Nantes, France). The viral constructs AAV5-shScramble-GFP and AAV5-shCYP46A1-GFP contained the expression cassette consisting of either short hairpin (sh) CYP46A1 (shCyp46a1) or scrambled sequence (shScramble), driven by a PU6 promoter and a GFP reporter gene, driven by the phosphoglycerate kinase 1 ( PGK1 ) promoter in the AAV5 vector ( Djelti et al. , 2015 ). The viral constructs for AAVrh10-GFP and AAVrh10-CYP46A1-HA contained the expression cassette consisting of either the GFP or the human CYP46A1 , driven by a CMV/β-actin hybrid promoter (CAG) surrounded by inverted terminal repeats of AAV2. The stereotaxic coordinates were: 0.5 mm rostral to the bregma, 2.1 mm lateral to midline and 3.35 mm ventral to the skull surface. The rate of injection was 0.2 μl/min with a total volume of 2 μl per striatum (equivalent to 3 × 10 9 genomic particles).
Animal behaviour
The rotarod protocol was slightly adapted from Menalled et al. (2009) : mice were tested over three consecutive days, each daily session included a 5 min training trial at 4 rpm on the rotarod apparatus. One hour later, the animals were tested for three consecutive accelerating trials of 5 min with the rotarod speed linearly increasing from 4 to 45 rpm over 300 s. The latency to fall from the rod was recorded for each trial. Mice remaining on the rod for more than 300 s were removed and their time scored as 300 s. Data from the training trial were not included. For hind limb clasping, mice were tested once a week from 6 to 11 weeks old. They were suspended by the tail for 30 s and the clasping phenotype was graded according to the following scale: level 0, no clasping; level 1, clasping of the forelimbs only or both fore- and hindlimbs once or twice; and level 2, clasping of both fore- and hindlimbs more than three times or more than 5 s.
Immunohistochemistry and neuropathology
At the end of the behavioural studies, groups of mice were rapidly anaesthetized by intraperitoneal injection of pentobarbital 250 mg/kg (Sanofi). Intracardiac perfusion of 4% paraformaldehyde in 0.1 M Na 2 HPO 4 /NaH 2 PO 4 buffer, pH 7.5 was performed and brains were post-fixed overnight in the same solution at 4°C. Brain sections (30 μm) were incubated with the following primary antibodies overnight: mouse-anti NeuN (1:500; Millipore); mouse anti-HA (1:500; Covance), mouse anti-DARPP32 (1/5000; generous gift from Pr P. Greengard) and rabbit anti-expHTT (EM48) (1:1000; Millipore). For immunofluorescence, secondary antibodies were incubated for 2 h: anti-mouse Alexa Fluor® 488 (1:400), anti-rabbit Alexa Fluor® 488 (1:400), anti-mouse Alexa Fluor® 568 (1:400), or anti-rabbit Alexa Fluor® 546 (1:400; Invitrogen) in Tris-buffered saline. Sections were mounted under cover slips using Vectashield (Vector Laboratories). For 3,3′ diaminobenzidine staining, the sections were then incubated with anti-mouse biotinylated antibody (1:250; Vector Laboratories Inc.) and visualized by the ABC amplification system (Vectastain ABC kit, Vector Laboratories) and 3,3′-diaminobenzidine tetrahydrochloride (peroxidase substrate kit, DAB, Vector Laboratories) as the substrate.
A systematic evaluation of the transduced region was performed by GFP fluorescence (expression of shScramble or shCYP46A1) or by haemagglutinin immunostaining (expression of CYP46A1 transgene). Adjacent sections were then stained by Cresyl violet or immuno-labelled with NeuN, DARPP-32 and EM48 antibodies, which are specific for neurons, medium spiny neurons and Exp-HTT aggregates, respectively. Sections were mounted under cover slips using Vectashield (Vector Laboratories).
Image acquisition
For DARPP32 depletion studies in shCYP46A1 experiments, images of immunostained sections were acquired with LAS V3.8 (Leica) software, with a bright-field Leica DM 4000B microscope equipped with 40×/0.75 and 100×/1.30 lenses and a Leica DFC500 digital camera. Confocal images were acquired with an Olympus BX 61 microscope equipped with 60×/1.35 lens and Fluoview FV-1000 image acquisition system. The extent of shCYP46A1-induced lesions in the striatum was analysed in six DARPP32-stained sections per animal (320 µm between 40-µm thick sections), selected to obtain a complete rostrocaudal sampling of the striatum and by quantifying the area of the lesion with a semi-automated image-analysis program (ImageJ software, NIH). Sections throughout the entire striatum were analysed. The area of the striatum showing a loss of DARPP32 staining was measured for each animal with an operator-independent macro. The volume was then estimated using the following formula: volume = d ( a 1 + a 2 + a 3 + · · ·), where d is the distance between serial sections (320 µm), and a 1, a 2, a 3 etc. are DARPP32-depleted areas for individual serial sections. The average grey value of all pixels measured in the lesioned area was recorded for each depleted area. Results are presented as the calculated value for volume of the DARPP32 depleted region for each animal.
For studies in R6/2 mice, the analysis of DARPP32 and EM48 staining, images were acquired from a confocal laser scanning microscope (TCS, SP5, Leica Microsystems). DARPP32-stained neurons were manually traced and MSN area quantified using ImageJ software. For the counting of aggregates, EM48-stained images were segmented using the same global threshold and quantified using ImageJ software. An x , y pixel size of 150 nm and a z -step of 500 nm were chosen, laser intensity and detector gain were constant for all images. Aggregates were segmented in 3D using the same global threshold. The ImageJ plugin 3D object counter was used to measure volume and mean intensity of each aggregate. For each aggregate, mean intensity is the mean of all voxel intensities in an 8-bit scale.
Cholesterol and oxysterol measurements
Cholesterol and oxysterol analysis followed the ‘gold standard’ method ( Dzeletovic et al. , 1995 ) to minimize the formation of autoxidation artefacts. Briefly, mouse striatal tissue samples were weighed and homogenized with a TissueLyser II apparatus (Qiagen) in a 500 μl solution containing butylated hydroxytoluene (BHT, 50 μg/ml) and EDTA (0.5 M). At this point, a mix of internal standards was added [epicoprostanol, 2 H 7 -7-lathosterol, 2 H 6 -desmosterol, 2 H 6 -lanosterol and 2 H 7 -24(R/S)-hydroxycholesterol] (Avanti Polar Lipids). Alkaline hydrolysis was performed under Ar using 0.35 M ethanolic KOH for 2 h at room temperature. After neutralization of the solution with phosphoric acid, sterols were extracted in chloroform. The lower phase was collected, dried under a stream of nitrogen and the residue was dissolved in toluene. Oxysterols were then separated from the cholesterol and its precursors on a 100 mg Isolute silica cartridge (Biotage); cholesterol was eluted in 0.5% propan-2-ol in hexane followed by oxysterols in 30% propan-2-ol in hexane.
The sterol and oxysterol fractions were independently silylated with Regisil® + 10% TMCS [bis(trimethylsilyl) trifluoro-acetamide + 10% trimethylchlorosilane] (Regis technologies) as described previously ( Chevy et al. , 2005 ). The trimethylsilylether derivatives of sterols and oxysterols were separated by gas chromatography (Hewlett-Packard 6890 series) in a medium polarity capillary column RTX-65 (65% diphenyl 35% dimethyl polysiloxane, length 30 m, diameter 0.32 mm, film thickness 0.25 μm; Restesk). The mass spectrometer (Agilent 5975 inert XL) in series with the gas chromatography was set up for detection of positive ions. Ions were produced in the electron impact mode at 70 eV. They were identified by the fragmentogram in the scanning mode and quantified by selective monitoring of the specific ions after normalization and calibration with the appropriate internal and external standards [epicoprostanol m/z 370, 2 H 7 -7-lathosterol m/z 465, 2 H 6 -desmosterol m/z 358, 2 H 6 -lanosterol m/z 504, 2 H 7 -24(R/S)-hydroxycholesterol m/z 553, cholesterol m/z 329, 7-lathosterol m/z, 7-dehydrocholesterol m/z 325, 8-dehydrocholesterol m/z 325, desmosterol m/z 343, lanosterol m/z 393 and 24(R/S)-hydroxycholesterol m/z 413].
Statistical analysis
Statistical analyses were performed with GraphPad Prism 5. All data are represented as mean ± SEM. For studies on expression levels in Huntington’s disease patient tissues, R6/2 mice and striatal neurons in culture, the significance was assessed by unpaired Student’s t -test with Welch correction. For survival studies in vitro , one-factor ANOVA corrected by the post hoc Bonferroni test was performed. For behavioural studies and neuronal areas in mice, a non-parametric two-way ANOVA analysis corrected by the post hoc Bonferroni was performed with time and treatment as independent factors. For mass spectrometry and aggregate studies, Mann-Whitney test was applied.
Results
CYP46A1 expression levels are reduced in Huntington’s disease
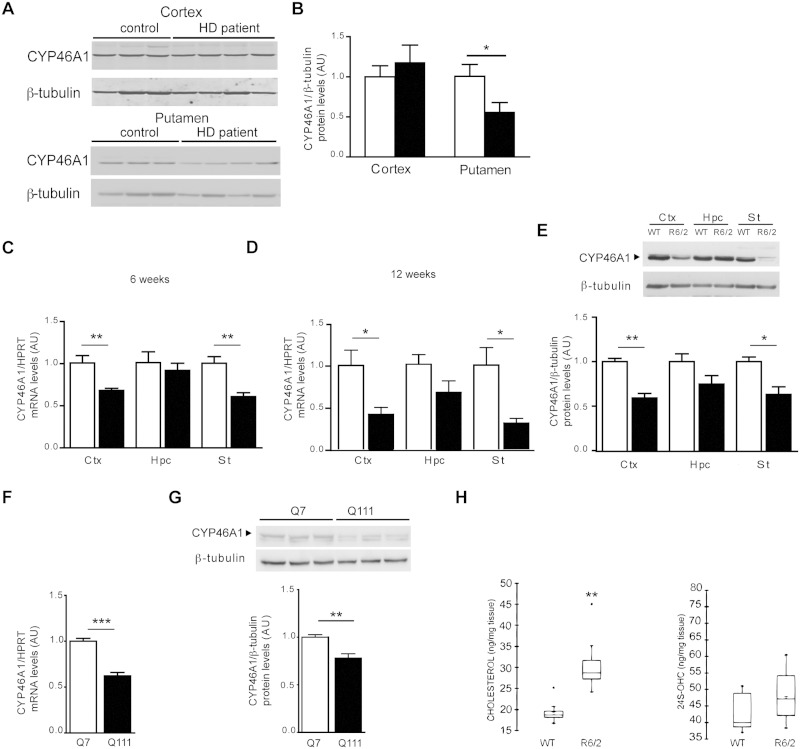
To evaluate the relevance of CYP46A1 in the context of Huntington’s disease, we analysed its protein expression levels in post-mortem samples of striatum, which comprises caudate nucleus and putamen, the most severely affected region in Huntington’s disease. By immunoblotting the samples with an antibody against CYP46A1, and anti-β-tubulin as a control of protein loading, we found a significant reduction of CYP46A1 over anti β-tubulin ratio (−44%) in the putamen of patients with Huntington’s disease when compared with control individuals ( Fig. 1 A and B). This reduction was not observed in cortex samples of Huntington’s disease patients ( Fig. 1 A and B). The post-mortem samples came from symptomatic patients (grade 2, 2/3). However, no significant modification in the levels of calbindin, a selective marker of medium spiny neurons, were found in these extracts ( Benchoua et al. , 2006 ) indicating that reduced CYP46A1 expression levels in post-mortem putamen tissues are not merely a reflection of cell death. Microarray data from human samples (caudate) of pathological grade 0–2 previously showed that transcripts for CYP46A1 were reduced when compared to control patients ( Kuhn et al. , 2007 ). Altogether, these data strongly support that reduced CYP46A1 protein levels result from transcriptional dysregulation, which occurs as a consequence of Exp-HTT.
Figure 1.
CYP46A1 expression levels in human, R6/2 mice and striatal cell lines. ( A ) Cortex and putamen extracts from control ( n = 5) and Huntington’s disease patients (cortex n = 5, putamen n = 6) were processed for western blotting with antibodies directed against CYP46A1 and β-tubulin. ( B ) Quantification of CYP46A1 protein levels normalized to β-tubulin from western blots in ( A ). ( C and D ) Cyp46a1 transcript levels were measured by quantitative PCR relative to Hprt from wild-type (WT, n = 6) and R6/2 ( n = 5) mice. Ctx = cortex, Hpc = hippocampus, St = striatum at 6 ( C ) or 12 ( D ) weeks (wk) of age. ( E ) Top : Extracts from wild-type and R6/2 mice at 12 weeks of age were processed for western blotting with CYP46A1 and β-tubulin antibodies. Bottom : Quantification of CYP46A1 protein levels normalized to β-tubulin from wild-type ( n = 3) and R6/2 ( n = 5) extracts. ( F ) Cyp46a1 transcript levels from ST Hdh Q7 (Q7) and ST Hdh Q111 (Q111) striatal cell line ( n = 5). ( G ) Top : Western blotting with CYP46A1 and β-tubulin from Q7 and Q111. Bottom : Quantification of CYP46A1 protein levels normalized to β-tubulin from Q7 and Q111 extracts ( n = 6). ( H ) Cholesterol and 24S-OHC levels were measured in striatum from wild-type and R6/2 at 12 week of age. ( B–G ) All data are represented as mean ± SEM; statistical analysis: unpaired Student's t -test. * P < 0.05; ** P < 0.01; *** P < 0.001 . ( H ) Data are shown as a box plot. Statistics Mann-Whitney test ** P < 0.01; for wild-type versus R6/2 mice. HD = Huntington’s disease.
We next studied CYP46A1 levels in the R6/2 transgenic Huntington’s disease mouse model, which expresses the first exon of the human gene containing 150 CAGs. Cyp46A1 mRNA levels were decreased in the striatum and cortex at 6 weeks (−40% and −32%, respectively) and 12 weeks (−69% and −59%, respectively) ( Fig. 1 C and D), but not in the hippocampus. Decreased CYP46A1 protein levels (−37% and −41%, respectively) were found in these brain areas at 12 weeks ( Fig. 1 E).
We next used immortalized wild-type (STHdhQ7) and mutant (STHdhQ111) striatal neuronal progenitor cell lines expressing HTT with either seven or 111 glutamines, respectively ( Gines et al. , 2003 ). The ST Hdh Q111 striatal cell line that expresses most of the cellular characteristics of Huntington’s disease ( Trettel et al. , 2000 ), also showed a significant reduction of CYP46A1 mRNA ( Fig. 1 F) and protein ( Fig. 1 G) levels when compared to ST Hdh Q7 striatal cells with seven CAG repeats. To determine the impact of CYP46A1 deficiency in Huntington’s disease, we measured cholesterol and 24S-OHC in the striatum of R6/2 mice (12 weeks old). We found a 1.5-fold increase of cholesterol in the striatum ( Fig. 1 H), therefore reproducing previous observations in the caudate of patients with Huntington’s disease ( Del Toro et al. , 2010 ). Comparison of cellular cholesterol localization in the striatum of wild-type and R6/2 mice was assessed by filipin staining, a naturally fluorescent polyene probe that binds to cholesterol but not to esterified sterols. Cholesterol labelling showed a marked staining of plasma membrane in both wild-type and R6/2 striatal cells. Cholesterol accumulation in internal organelles was not observed in R6/2 mice ( Supplementary Fig. 1 ). Despite decreased expression of CYP46A1, 24S-OHC was not altered in the striatum of R6/2 mice ( Fig. 1 H) .
Altogether, these data indicate that CYP46A1 levels are reduced in the striatum from Huntington’s disease patients, mouse and cellular models.
CYP46A1 knock-down in a wild-type context induces a Huntington’s disease-like phenotype
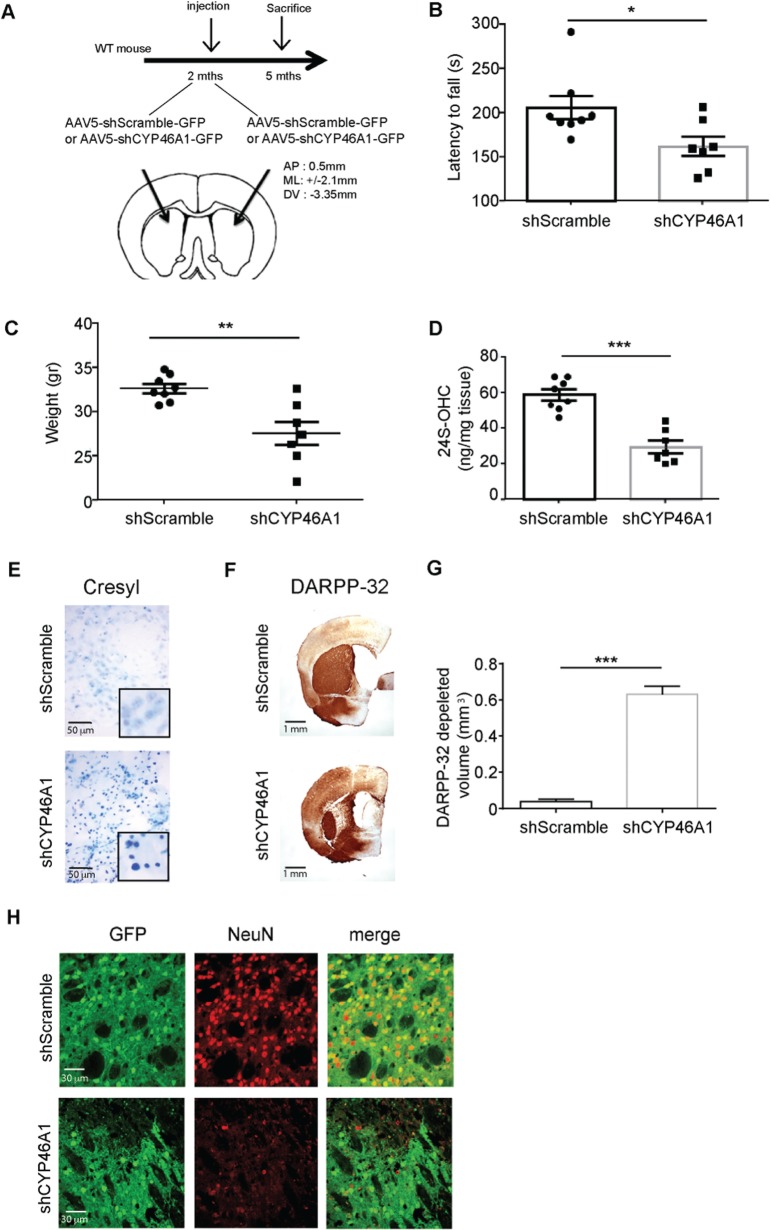
To assess the impact of CYP46A1 deficiency in a wild-type context, we used a shRNA sequence that was shown to inhibit CYP46A1 expression in vitro and in vivo ( Djelti et al. , 2015 ). Two-month-old mice were bilaterally injected into the striatum with either AAV5-shCyp46A1-GFP or AAV5-shScramble-GFP (control sequence that does not target Cyp46a1 mRNA sequence) ( Fig. 2 A). A GFP tag was used as a marker of viral infection. To evaluate whether the deficiency in CYP46A1 could be translated into behavioural dysfunction, the animals ( n = 7 for shCyp46a1 and n = 8 for shScramble) underwent a test of balance and motor coordination—the rotarod—at 3 months post-injection. All C57 mice had similar performances in rotarod before the viral infections. Three months post-injection, shCYP46A1-injected mice had a significant decrease in their latency to fall when compared to shScramble-injected mice ( Fig. 2 B), and their weight was significantly reduced (∼16%) ( Fig. 2 C). These mice were then sacrificed, and one striatum from each mouse was extracted, immediately frozen and used to measure 24S-OHC levels by mass spectrometry. A 50% reduction in the levels of 24S-OHC was found in the striata of shCYP46A1-injected mice, relative to striata of shScramble-injected mice ( Fig. 2 D), thus indicating that shCYP46A1 was efficient in reducing CYP46A1 activity. The second brain hemisphere was fixed and processed for neuropathological studies including Cresyl violet staining ( Fig. 2 E), DARPP32 ( Fig. 2 F) and NeuN ( Fig. 2 G) immunocytochemistry. Cresyl violet staining indicated increased condensed nuclei (degeneration) in the shCYP46A1-injected compared to shScramble-injected striata, where minimal condensed nuclei were observed around the needle tract region ( Fig. 2 E). DARPP32 (a selective marker of medium spiny neurons) immunostaining showed a strong depletion in the striatum of shCYP46A1 injected mice, whereas no DARPP-32 loss was observed in shScramble injected mice ( Fig. 2 F and G). Then we used immunodetection of NeuN (a neuronal marker) and found a strong decrease in animals injected with shCYP46A1 when compared to shScramble ( Fig. 2 H). Altogether these data indicate that motor dysfunction and neuronal death occurred as a consequence of CYP46A1 deficiency.
Figure 2.
ShCYP46A1 produces spontaneous motor deficits and striatal neuron dysfunctions in a wild-type context. ( A ) Two-month-old C57BL6J mice were stereotactically injected in the striatum with either AAV5-GFP-shCyp46A1 or AAV5-GFP-shScramble (control sequence that does not target Cyp46a1 mRNA). ( B ) All mice ( n = 8 for each group) were blindly submitted to behavioural experiments. The Rotarod test used to evaluate mouse balance and coordination revealed that the latency to fall was decreased in mice injected with shCYP46A1, compared to shScramble-injected mice. ( C ) Two months later, the weight of mice injected with shCYP461 showed a statistically significant reduction at the time of sacrifice (∼16%). ( D–G ) Mice were sacrificed 3 months later and for each mouse, the striatum from one hemisphere was processed for biochemical studies while the other was used for neuropathological studies. ( D ) 24S-OHC measurements were performed by mass spectrometry. A ∼50% reduction in the levels of 24S-OHC was shown in the striatum of shCYP46A1-injected mice, relative to shScramble-injected mice. ( E ) Cresyl violet staining from striata injected with shScramble ( top ) and shCYP46A1 ( bottom ). Note the increased condensed nuclei (degeneration) in the striatum injected with shCyp46A1 when compared to shScramble, where minimal condensed nuclei were observed around the needle tract region. ( F ) DARPP32 immunostaining shows a strong depletion in the striatum of shCYP46A1 injected mice ( lower panel), when compared to shScramble injected mice. ( G ) Quantification of DARPP32 depleted area ( n = 6 for each group). ( H ) Co-labelling of GFP (used as a tag for shRNA sequences) and NeuN immunostaining shows a strong co-localization (merge panel) in shScramble injected neurons. In contrast, no co-localization is observed between GFP and NeuN in animals injected with shCYP46A1, indicating neuronal dysfunction in the transduced area. ( B – G ): Data are represented as mean ± SEM; statistical analyses: Mann whitney’s test. * P < 0.01; ** P < 0.05; *** P < 0.001 shScramble versus shCYP46A1. WT = wild-type.
CYP46A1 restoration protects from Exp-HTT-induced striatal dysfunctions in vitro
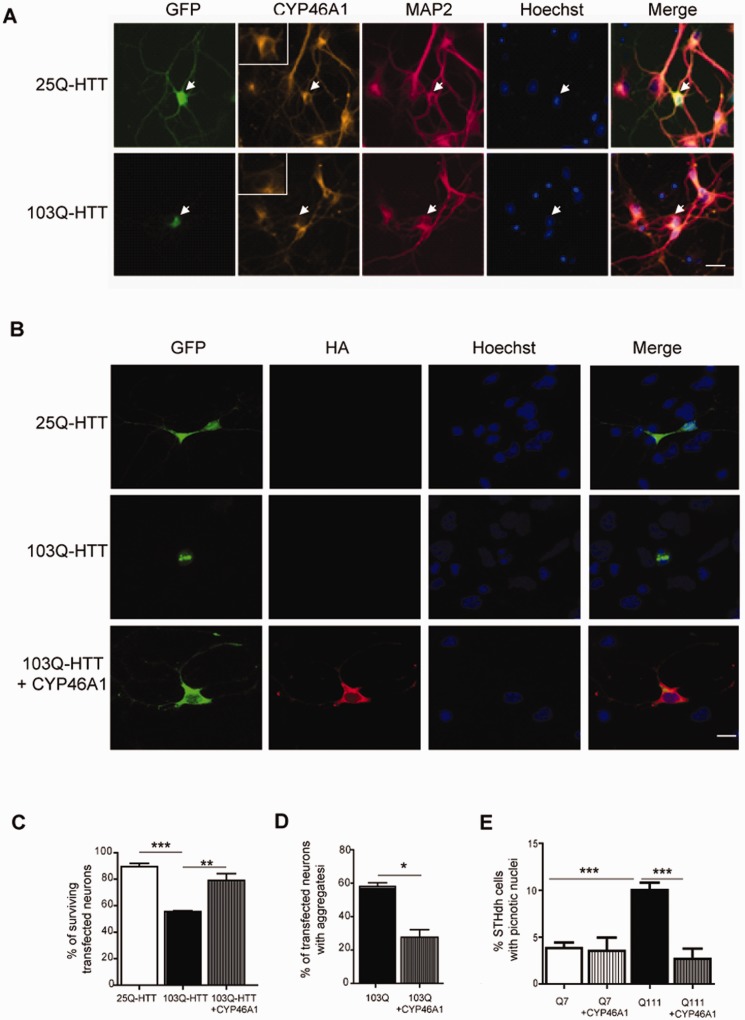
We then asked whether CYP46A1 restoration could protect from striatal neuron dysfunctions induced by Exp-HTT. Primary striatal neurons in culture were transiently transfected with cDNAs encoding the first exon of HTT containing either 25 (25Q-HTT) or 103 (103Q-HTT) CAG repeats, and fused to the GFP coding sequence. Expression levels of CYP46A1 were decreased in 103Q-HTT- when compared to 25Q-HTT-transfected neurons or non-transfected neurons ( Fig. 3 A). 103Q-HTT-expressing neurons showed decreased survival (−34%, evaluated by nuclear morphology after Hoechst staining) 48 h after transfection ( Fig. 3 B and C). When striatal neurons were co-transfected with cDNAs encoding 103Q-HTT and a CYP46A1-HA, a significant neuroprotection was found (55% versus 79%) ( Fig. 3 B and C). CYP46A1-HA overexpression also resulted in a significant reduction of 103Q-HTT-induced aggregate formation (58% versus 27%) ( Fig. 3 B and D).
Figure 3.
CYP46A1 decreases 103Q-HTT-induced toxicity in primary striatal neurons and STHdhQ111 striatal cells . ( A ) Striatal neurons were transfected with plasmids coding for either 25Q-HTT or 103Q-HTT. Transfected neurons were revealed after fixation 24 h later by GFP staining (green), and co-labelled after CYP46A1 (orange), MAP2 (red) immunostaining, and Hoechst staining (blue). White arrow indicates transfected neuron (25Q top , 103Q bottom ), that is shown at higher magnification in the insert. Note in the merge panel that CYP46A1 signal is decreased in 103Q-HTT expressing neurons when compared to 25Q-HTT-transfected neurons. Scale bar = 20 µm. ( B ) Striatal neurons were co-transfected with either 25Q-HTT or 103Q-HTT (GFP, green) in the presence or not of HA-CYP46A1. Haemagglutinin (HA, red) and Hoechst (blue) stainings were performed 48 h later after fixation for detection of transfected neurons and nuclear labelling, respectively. Scale bar = 40 µm. ( C ) Quantification of surviving transfected neurons based on nuclear integrity after Hoechst labelling and ( D ) quantification of aggregates based on GFP fluorescence. ( E ) ST Hdh Q7 and Q111 striatal cell lines were transfected with plasmids coding CYP46A1-HA and serum starved for 24 h. ( B – D ) Data were analysed from at least three independent experiments (100 transfected neurons per condition and per experiment) and expressed as mean ± SEM. One-factor analysis of variance (ANOVA) corrected by the Bonferroni’s post hoc test was performed.* P < 0.05; ** P < 0.01; *** P < 0.001.
For the STHdhQ111 striatal cell line, which shows higher susceptibility than STHdhQ7 under serum deprivation ( Tourette et al. , 2014 ), CYP46A1-HA was also neuroprotective ( Fig. 3 E). A lower percentage of picnotic nuclei was found in STHdhQ111 transfected with CYP46A1-HA when compared to STHdhQ111 control cells (2.67% versus 10.63%) ( Fig. 3 E).
Taken together, these results are consistent with a neuroprotective role for CYP46A1 against Exp-HTT-induced striatal dysfunction.
CYP46A1 restoration in the striatum improves the motor phenotype of R6/2 mice
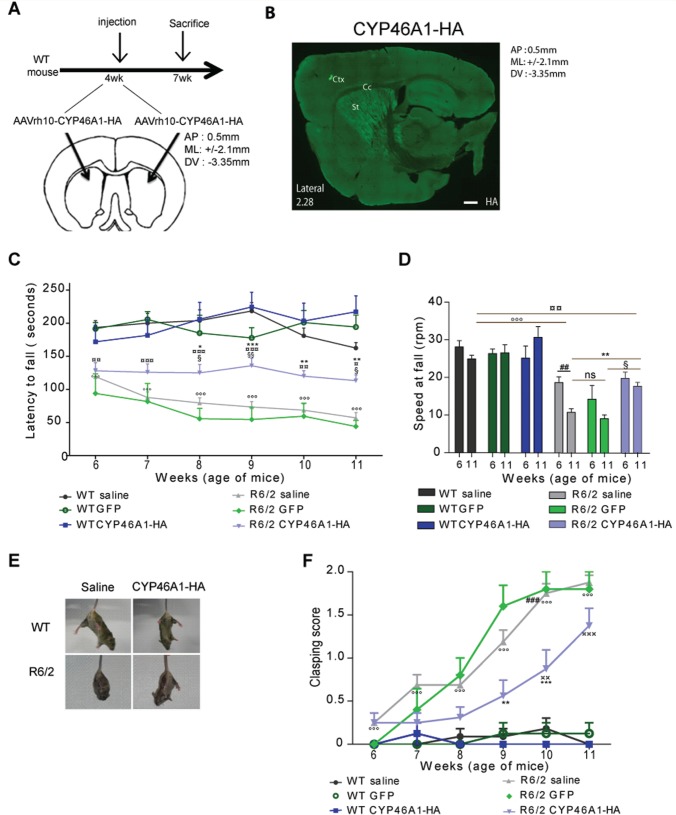
Next we investigated a possible neuroprotective role for CYP46A1 in vivo . We used AAVrh10 for CYP46A1 delivery in the striatum of mice. AAVrh10 shows widespread transduction efficiency and a strong level of gene expression in the striatum ( Sondhi et al. , 2007 ) (see also Supplementary Fig. 2B and Supplementary Data ). AAVrh10 encoding CYP46A1-HA was injected bilaterally in the striatum of 4-week-old R6/2 mice ( Fig. 4 A). Three weeks later, haemagglutinin immunolabelling showed widespread expression in the dorsal striatum ( Fig. 4 B). Double labelling showed no expression of CYP46A1-HA in astrocytes (GFAP), microglia (isolectin), or oligodendrocytes (OLIG2) ( Supplementary Fig. 2D ). In contrast, we found perfect co-localization in neuronal cells (NeuN, Supplementary Fig. 2D ). In addition, Cyp46a1 mRNA levels were rescued on CYP46A1-HA injection in these mice ( Supplementary Fig. 3 ). Therefore, this approach was deemed appropriate for further in vivo studies.
Figure 4.
Striatal injection of AAV-rh10-CYP46A1-HA improves behavioural phenotypes in R6/2 mice. ( A ) Stereotaxic injection of AAVrh10-CYP46A1-HA was performed at 4 weeks in the striatum, according to the indicated coordinates (AP = antero-posterior; ML = medio-lateral; and DV = dorso-ventral), and sacrificed 3 weeks later. ( B ) Haemagglutinin (HA) immunostaining was performed on a sagittal section from 7-week-old mice. Note the widespread expression of CYP46A1-HA within the striatum (Ctx = cortex; Cc = corpus callosum; St = striatum). Scale bar = 440 µm. ( C and D ) Rotarod performance of wild-type and R6/2 mice injected with saline, AAVrh10-GFP or AAVrh10-CYP46A1-HA. ( C ) Latency to fall was measured from 6 to 11 weeks for each group. ( D ) Rotarod speed when mice fall. ( E ) Representative pictures of postures in wild-type (WT, top ) or R6/2 ( bottom ) injected with saline ( left ) or AAVrh10-CYP46A1-HA ( right ; CYP46A1-HA) during tail suspension. Note the clasping posture of R6/2 saline ( bottom left ). ( F ) Clasping score of the six groups of mice. ( C – F ) Wild-type saline ( n = 11), wild-type AAVrh10-GFP ( n = 6), wild-type AAVrh10-CYP46A1-HA ( n = 8), R6/2 saline ( n = 16), R6/2 AAVrh10-GFP ( n = 5), R6/2 AAVrh10-CYP46A1-HA ( n = 16). Data are represented as mean ± SEM, statistical analysis: two-way ANOVA followed by Bonferroni’s post hoc test. ° P < 0.05; °° P < 0.01; °°° P < 0.001, wild-type saline versus R6/2 saline; * P < 0.05; ** P < 0.01; *** P < 0.001 R6/2 saline versus R6/2 AAVrh10-CYP46A1-HA; §P < 0.05; §§P < 0.01; R6/2 AAVrh10-GFP versus R6/2 AAVrh10-CYP46A1-HA; ¤P < 0.05; ¤¤P < 0.01; ¤¤¤P < 0.001 wild-type saline versus R6/2 AAVrh10-CYP46A1-HA; ##P < 0.01; ###P < 0.001; R6/2 saline at 6 and 11 weeks. ns = non-significant.
The R6/2 mouse model recapitulates many features of the human disease, including motor impairment appearing before the age of 6 weeks ( Menalled et al. , 2009 ). Thus, the early and reproducible phenotype of this mouse line has made it an ideal model for screening compounds, performing genetic crosses, and testing gene therapy approaches. No significant improvement in body weight was observed in R6/2 mice injected with AAVrh.10-CYP46A1-HA (CYP46A1-HA R6/2 mice; Supplementary Fig. 4 ). Neurological performance, tested using the Rotarod test, reliably declined in control R6/2 mice (saline or AAVrh10-GFP injected) as assessed every week from 6 to 11 weeks of age ( Fig. 4 C and D). In CYP46A1-HA R6/2 mice no significant improvement in performance was observed 2 weeks after injection ( Fig. 4 C and D). However, although this behaviour progressively declined from 7 to 11 weeks in control R6/2 mice, it remained stable in CYP46A1-HA injected R6/2 mice from 6 to 11 weeks ( Fig. 4 C and D). A significant difference in the latency to fall was measurable between R6/2 mice and CYP46A1-HA R6/2 mice from 8 to 11 weeks.
Abnormal hind limb clasping is reminiscent of dystonic movements in mouse models of Huntington’s disease ( Stack et al. , 2005 ; Bauer et al. , 2010 ; Paul et al. , 2014 ). The clasping test assessed the presence of dystonic movements that occur during 30 s of tail suspension (hind limb splay, limb clasping and sustained limb flexure). A gradual score was established to evaluate the progressive severity of this motor disturbance. While all wild-type mice showed normal hind limb splay, R6/2 mice exhibited progressive limb clasping by 6 weeks that became more pronounced with age, with sustained flexure of both forelimbs and hind limbs ( Fig. 4 E and F). CYP46A1-HA R6/2 mice showed markedly lower clasping scores until 10 weeks old ( Fig. 4 F).
Altogether, these data indicate that CYP46A1-HA restoration delays progressive deterioration of motor performance in R6/2 mice.
CYP46A1 restoration protects R6/2 mice from neuropathological alterations
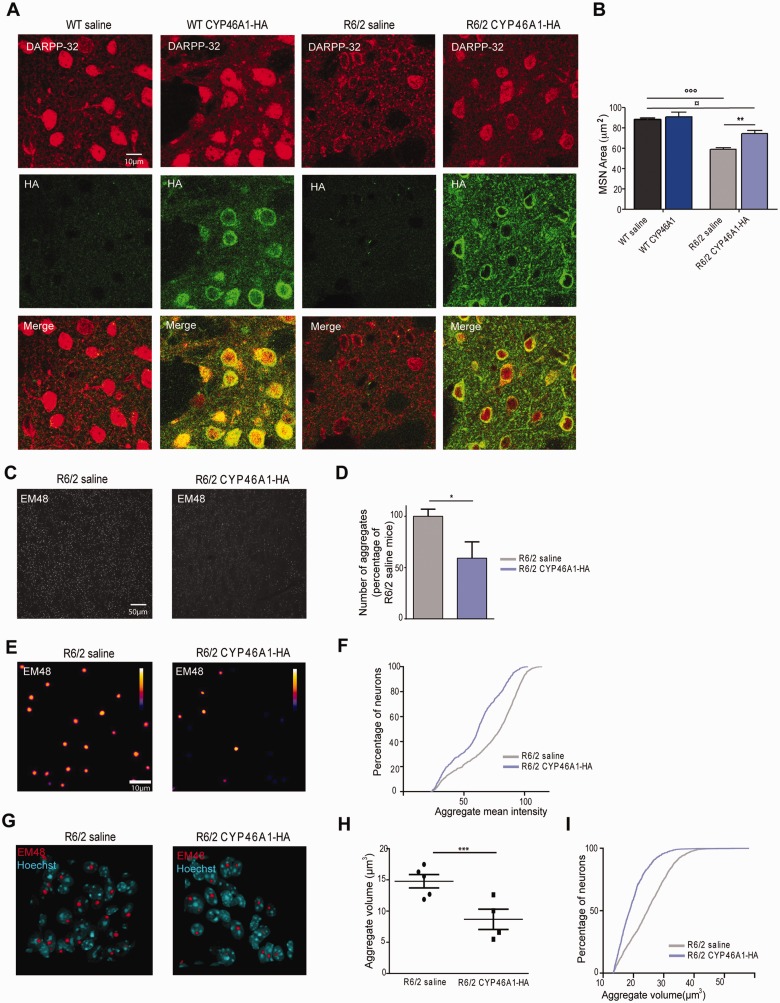
Twelve week old mice were sacrificed for neuropathological studies. Cresyl violet staining showed a global cellular atrophy in the striatum of R6/2 mice ( Stack et al. , 2005 ), which was positively impacted by CYP46A1-HA ( Supplementary Fig. 5 ). Medium spiny neuron atrophy can be detected owing to DARPP32 immunocytochemistry ( Bibb et al. , 2000 ). Double immunostaining with DARPP32 and haemagglutinin showed pronounced medium spiny neuron atrophy in control R6/2 when compared to wild-type mice, and CYP461-HA significantly improved medium spiny neuron area (59 µm 2 versus 74µm 2 , respectively; Fig. 5 B and C) and DARPP32 staining.
Figure 5.
CYP46A1-HA expression alleviates neuropathological hallmarks in R6/2 mice. ( A ) Dorsal striatal sections after double immunostaining with DARPP32 and haemagglutinin (HA) antibodies in 12-week-old wild-type and R6/2 mice injected with saline or AAVrh10-CYP46A1-HA. ( B ) Quantification of medium spiny neuron areas after DARPP32 staining. Histograms are represented as mean area ± SEM ( n = 5 independent mice). In each representative sections, 150 positive DARPP32 medium spiny neurons were quantified. Statistical analyses: one-way ANOVA followed by Bonferroni’s post hoc test. °°° P < 0.001, wild-type (WT) saline compared to R6/2 saline; ** P < 0.01, R6/2 saline compared to R6/2 CYP46A1-HA; ¤P < 0.05, wild-type saline compared to R6/2 CYP46A1-HA. ( C ) Immunohistochemical staining with the EM48 antibody in a dorsal striatal section of R6/2 mice (12 weeks old) injected with saline ( left ) or AAVrh10-CYP46A1-HA ( right ). ( D ) A threshold of aggregate intensity was applied to R6/2 mice and AAVrh10-CYP46A1-injected mice. Using this threshold, aggregate number was performed and quantified and normalized to R6/2 saline mice. Histograms are represented as mean ± SEM, n = 4–5 for AAVrh10-CYP46A1-HA and saline groups, respectively. Statistical analyses: unpaired Student's t -test * P < 0.05. ( E ) Two dimensional average projection from 3D images of aggregates in dorsal striatum. Analysis was performed in 3D but images are shown in 2D for easier visualization of intensities. Calibration bar shows the code of false colours representing the intensities. ( F ) Cumulative distribution frequency of mean intensities measured in 3D aggregates. ( G ) Three dimensional view of Hoechst stained nuclei with EM48 stained aggregates in the dorsal striatum. Note that aggregates are nuclear inclusions, and that their volumes are decreased in AAvrh10-CYP46A1-HA transduced cells. ( H ) Aggregate volume was quantified in R6/2 mice injected with either saline or AAVrh10-CYP46A1. Data are represented as mean ± SEM and each dot represents the variance of aggregate volume in each mouse ( n = 4–5 for AAVrh10-CYP46A1 and saline groups, respectively). Mann-Whitney test *** P < 0.001. ( I ) Cumulative distribution of aggregates volumes. 1400 aggregates were quantified from R6/2-saline mice (at least two striatal section per mouse, n = 5 independent mice) and 900 aggregates from AAVrh10-CYP46A1-HA R6/2 mice (at least two striatal section per mouse, n = 4 independent mice).
The formation of intracellular aggregates composed primarily of Exp-HTT is a common pathological hallmark of Huntington’s disease, and can be found in R6/2 mice ( Davies et al. , 1997 ). To determine whether CYP46A1 reduces aggregate load, striatal sections were stained with EM48 antibody, which specifically labels Exp-HTT aggregates. EM48-positive foci were counted in striatal areas that expressed CYP46A1-HA ( Fig. 5 C) and their quantification was normalized to control R6/2 mice ( Fig. 5 D). CYP46A1-HA R6/2 mice showed a significant decrease in the number of aggregates (−40%) when compared with control R6/2 mice. By contrast, non-transduced cortical area did not show any differences in aggregate number ( Supplementary Fig. 5 ). The mean intensity of Exp-HTT-induced aggregate formation decreased in CYP46A1-HA R6/2 mice ( Fig. 5 E and F). After Hoechst counterstaining, aggregates were found as nuclear inclusions in control R6/2 mice (97.3 ± 3.2%). CYP46A1-HA expression did not modify this nuclear localization of aggregates (97.8 ± 2.8%) ( Fig. 5 G). However, the mean volume of aggregates was significantly lower in CYP46A1-HA R6/2 as compared to control R6/2 mice ( Fig. 5 H and I). Collectively, these data reveal that CYP46A1 reduces the number, intensity level and size of aggregates.
Regulation of cholesterol homeostasis by CYP46A1
Quantitative measurements of sterols and oxysterols were performed by mass spectrometry from striatal extracts. We first compared cholesterol and 24S-OHC levels of wild-type and R6/2 mice injected or not with AAVrh10-GFP into the striatum ( Supplementary Fig. 6 ). No significant differences were found between wild-type or R6/2 mice injected with saline or AAV-GFP, indicating that the AAVrh10 vector by itself does not alter cholesterol homeostasis.
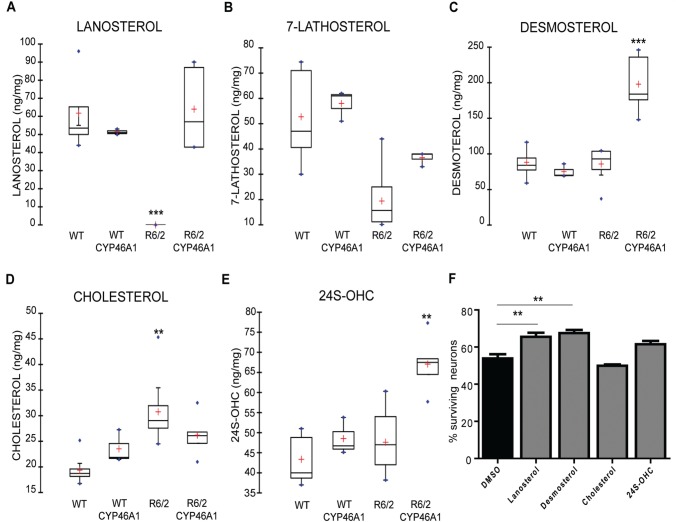
We then analysed the cholesterol metabolism of wild-type and R6/2 mice injected or not with CYP46A1-HA ( Fig. 6 ). Lanosterol, the first sterol of the metabolic pathway, along with cholesterol intermediates from the Kandutsch-Russell (lathosterol) and Bloch (desmosterol) pathways were analysed. As previously described, lanosterol levels were decreased in the striatum of R6/2 mice ( Valenza et al. , 2005 , 2010 ) ( Fig. 6 A), and 24S-OHC levels were unchanged ( Valenza et al. , 2007 b ), although reduced levels have also been observed ( Valenza et al. , 2010 ). The level of lathosterol, a common surrogate marker of cholesterol biosynthesis ( Kempen et al. , 1988 ), was significantly lowered in R6/2 mice ( Fig. 6 B), while the desmosterol content remained unchanged ( Fig. 6 C). In CYP46A1-HA R6/2 mice, normal levels of lanosterol were restored ( Fig. 6 A), with no change in lathosterol ( Fig. 6 B), and dramatical increase in desmosterol content ( Fig. 6 C). CYP46A1 restoration also resulted in normal cholesterol concentration ( Fig. 6 D) and increased 24S-OHC levels ( Fig. 6 E) in the striatum of R6/2 mice. None of these recoveries were linked to transcriptional regulation of the corresponding sterol synthesis enzymes ( Supplementary Fig. 7 ) . Together, these data indicate that restoration of CYP46A1 reinstates normal cholesterol levels by increasing cholesterol degradation and by increasing lanosterol and desmosterol levels.
Figure 6.
Regulation of cholesterol homeostasis by CYP46A1. At the end of the behavioural studies (12 weeks) groups of mice ( n = 5–7 per group) were rapidly sacrificed and the striatum from both hemispheres was dissected and flash frozen in liquid nitrogen. ( A – E ) Sterols and 24S-OHC levels were measured in the striatum of wild-type and R6/2 mice injected or not with AAVrh10-CYP46A1. Data are represented as mean ± SEM, n = 5–7 for all groups statistical analysis: Mann-Whitney test * P < 0.05, ** P < 0.01, *** P < 0.001 for wild-type versus R6/2 mice and ##P < 0.01 for wild-type AAVrh10-GFP versus wild-type AAVrh10CYP46A1-HA. ( F ) Striatal neurons were transfected with cDNA coding 103Q-HTT and treated with lanosterol, desmosterol, cholesterol, or 24S-OHC (1 µM) for 48 h. Quantification of surviving transfected neurons was based on Hoechst labelling. Data are expressed as mean ± SEM (at least three independent experiments; 100 transfected neurons per condition and per experiment). One-factor ANOVA followed by Bonferroni’s post hoc test was performed * P < 0.05; ** P < 0.01; *** P < 0.001.
Protection by lanosterol and desmosterol in Huntington’s disease striatal neurons
To study a potential neuroprotection of sterols and oxysterol regulated or produced by CYP46A1, i.e. cholesterol, lanosterol, desmosterol and 24S-OHC, we used striatal neurons in culture expressing Exp-HTT. The effect of these compounds was evaluated 48 h after exposure ( Fig. 6 F). Survival was not modified by either cholesterol or 24S-OHC treatments (49.95% and 61.49%, respectively) as compared to control (53.9%), but was significantly improved by lanosterol and desmosterol (+11.56% and +13.57%, respectively). These data indicate that lanosterol and desmosterol can be neuroprotective in striatal neurons expressing Exp-HTT.
Discussion
Cholesterol disturbance in Huntington’s disease has recently emerged as an important field of investigation. Both decreased sterol synthesis and increased neuronal accumulation of cholesterol have been observed, yet the cellular mechanisms accounting for this apparently opposing dysregulation in cholesterol homeostasis has not been well studied. We found that the expression of CYP46A1, the rate-limiting enzyme for cholesterol degradation in the brain ( Lund et al. , 1999 ; Bjorkhem and Meaney, 2004 ), is decreased in the putamen of patients with Huntington’s disease, in the striatum of R6/2 mice, and in Huntington’s disease striatal cell lines. In vivo , in a wild-type context, CYP46A1 knock-down induced motor deficits and spontaneous striatal neuron death. We further show that restoring CYP46A1 levels protected against Exp-HTT-induced striatal dysfunctions in vitro , including aggregate formation and neuronal death. In vivo , restoring CYP46A1 in the striatum of the Huntington’s disease R6/2 mouse model alleviated motor deficits, improved medium spiny neuron size, and restored cholesterol homeostasis. We conclude that this enzyme plays a neuroprotective role in Huntington’s disease.
Transcriptional dysregulation is an important event in the neuropathological process in Huntington’s disease ( Moumné et al. , 2013 ). A global microarray study from mouse models (R6/2 and knock-in mice) and samples from post-mortem human Huntington’s disease caudate previously showed downregulation of Cyp46A1 transcripts ( Kuhn et al. , 2007 ). Herein, we further show that Cyp46A1 transcripts are reduced in the striatum of R6/2 mice and with ST Hdh Q111 striatal cell lines. Altered histone acetylation levels and binding of the SP1 transcription factor to the promoter of Cyp46A1 ( Nunes et al. , 2010 ) could be responsible for Cyp46A1 dysregulation by Exp-HTT, as previously shown for other genes ( Dunah et al. , 2002 ; Sadri-Vakili et al. , 2007 ).
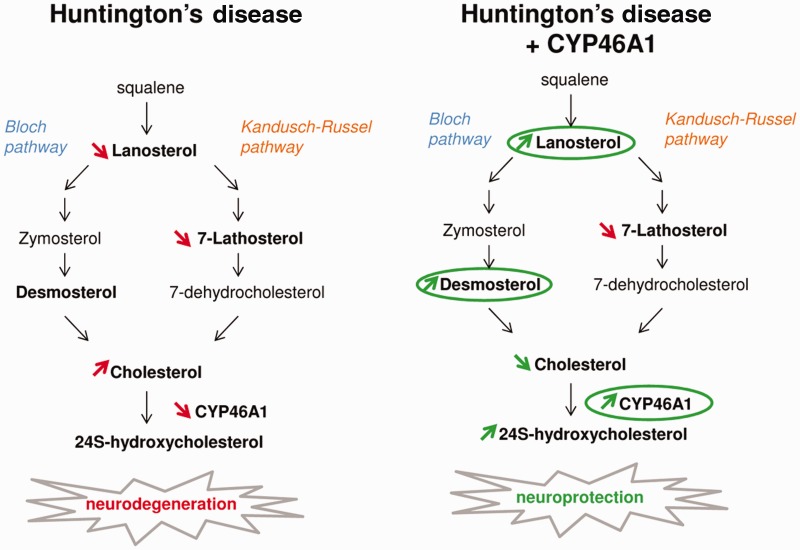
In agreement with previous studies ( Trushina et al. , 2006 ; Valenza et al. , 2007 a , b , 2010 ; del Toro et al. , 2010 ), we found decreased levels of lanosterol and lathosterol and a slight increase in cholesterol levels in the striatum of R6/2 mice. Although 24S-OHC plasma levels are reduced in patients with Huntington’s disease and parallels the degree of caudate atrophy ( Leoni et al. , 2013 ), we failed to detect any variation of 24S-OHC in the striatum, as previously described ( Valenza et al. , 2007 b ). Altogether our data suggest that altered cholesterol homeostasis in Huntington’s disease appears as a consequence of decreased sterol biosynthesis, and dysregulation of cholesterol clearance, occurring as a consequence of CYP46A1 deficiency. Importantly, increased expression of CYP46A1 restored normal cholesterol levels. The synthesis of new cholesterol and the production of 24S-OHC are closely related. Accordingly, a significant reduction of cholesterol synthesis was found in brain samples from CYP46A1 knockout mice, despite stable steady state levels of cholesterol ( Lund et al. , 2003 ). Although cholesterol is important for neuronal integrity in physiological context, its accumulation in membrane is deleterious. In the context of Huntington’s disease, Exp-HTT favours cholesterol accumulation in membranes, despite its decreased synthesis ( Trushina et al. , 2006 , 2014 ; del Toro et al. , 2010 ). This accumulation is thought to modify intracellular signalling of downstream neurotransmitter receptors distributed in lipid rafts. Within lipid rafts Exp-HTT accumulates with the active-phosphorylated form of glycogen synthase kinase-3β (GSK-3β, encoded by GSK3B ), a pro-apoptotic kinase whose activity can be attenuated by cholesterol depletion ( Valencia et al. , 2010 ). GSK-3β inhibitors reduce death of Exp-HTT-expressing neurons, suggesting that GSK-3β activity contributes to neuronal death in Huntington’s disease. In hippocampal neurons, CYP46A1 over-expression depletes membrane cholesterol ( Martin et al. , 2008 ) thereby increasing the pro-survival signalling pathway involving downstream BDNF receptors. In contrast, CYP46A1 loss of function decreases TrkB phosphorylation. Therefore the neuroprotective effect of CYP46A1 observed in the context of Huntington’s disease could rely on regulation of normal membrane cholesterol levels along with pro-survival intracellular pathways. Cholesterol content impacts the composition and organization of the synaptic membrane, and thus regulates the lateral diffusion of molecules and ultimately the stability and plasticity of neuronal synapses ( Renner et al. , 2009 ).
Two alternative pathways for cholesterol synthesis exist downstream of lanosterol ( Fig. 7 ): the Bloch (desmosterol) and the Kandutsch-Russel (lathosterol) pathways. In cell cultures, the Bloch pathway is preferentially used in astrocytes to efficiently produce cholesterol ( Nieweg et al. , 2009 ), while the Kandutsch-Russel pathway is used in neurons, but produces cholesterol with much less efficiency. The increase in desmosterol produced by CYP46A1 expression probably results from regulatory events between neuronal and glial cells. However, neuronal restoration of CYP46A1 failed to change lathosterol levels, suggesting that CYP46A1 regulates cholesterol homeostasis preferentially through the Bloch pathway.
Figure 7.
Regulation of cholesterol homeostasis by CYP46A1 is neuroprotective in Huntington’s disease. Left : Cholesterol homeostasis is dramatically dysregulated in Huntington’s disease (HD). Despite a decrease of lanosterol and 7-lathosterol, cholesterol is slightly increased, due to down regulation of CYP46A1 expression. Right : Restoration of CYP46A1 expression in Huntington’s disease increases cholesterol catabolism, thereby decreasing cholesterol content. It also increases levels of lanosterol and desmosterol, which protect striatal neurons from Exp-HTT-mediated toxicity. CYP46A1 displays a potent neuroprotective effect in Huntington’s disease likely by increasing cholesterol turnover and Bloch pathway activity.
Both lanosterol and desmosterol were protective in primary culture of striatal neurons expressing Exp-HTT. Neuroprotection by lanosterol was recently proposed in Parkinson’s disease. Similar to Huntington’s disease, lanosterol is decreased in a Parkinson’s disease mouse model and is neuroprotective in vitro for MPP+-treated dopaminergic neurons, likely by promoting mitochondria uncoupling and autophagy ( Lim et al. , 2012 ). Increasing autophagy, which is known to favour Exp-Htt clearance, is neuroprotective in Huntington’s disease ( Martin et al. , 2015 ). Neuroprotection by desmosterol has yet to be reported, although it is known to suppress proinflammatory gene expression in macrophage foam cells ( Spann et al. , 2012 ). Early inflammatory reactions, such as reactive microglia and elevated cytokine levels, are observed in the brains of patients with Huntington’s disease as well as mouse models ( Sapp et al. , 2001 ). Taken together, lanosterol and desmosterol could be neuroprotective by acting on mitochondria, autophagy and inflammatory response that are impaired in Huntington’s disease.
Central regulators of cholesterol metabolism in mammals include the nuclear liver X receptors (LXRs) that bind the promoters of cholesterogenic enzymes ( Karazinska et al. , 2011 ). 24S-OHC is classically described as an endogenous regulator of LXR-target genes in vitro ( Brown and Goldstein, 1997 ; Leoni and Caccia, 2015 ). However, LXR target genes were not upregulated in transgenic mice that overexpress brain CYP46A1 ( Shafaati et al. , 2011 ) or in the hippocampus of Alzheimer mice injected with AAV-CYP46A1 ( Hudry et al. , 2010 ). Accordingly, we failed to detect any regulation of these genes on restoration of CYP46A1 in vivo , likely owing to the diversity of cellular populations and the complexity of CYP46A1-mediated effects in neuronal versus glial cells. It is noteworthy that CYP46A1 strongly decreased Exp-HTT-induced aggregate formation both in vitro and in vivo . Interestingly, 24S-OHC regulates the expression of glucose-regulated protein 78 (GRP78, encoded by hspa5 ), an endoplasmic reticulum chaperone that counteracts aggregation of Exp-HTT ( Jiang et al. , 2012 ) and reduces amyloid-β production in a mouse model of Alzheimer disease ( Urano et al. , 2013 ). Therefore, these observations open a possible link between CYP46A1 neuroprotective effect and aggregate clearance.
In summary, our findings reveal a previously unknown and unexpected role for CYP46A1 in Huntington’s disease pathogenesis. CYP46A1 prevented cholesterol accumulation in R6/2 mice and displayed neuroprotective effect via regulation of sterols, more specifically lanosterol and desmosterol ( Fig. 7 ). We have revealed new cholesterol-targeting therapeutic strategies in Huntington’s disease, and presumably in other neurodegenerative diseases where dysregulation of cholesterol homeostasis has been clearly identified. This study paves the way to restore CYP46A1 expression in the striatum/putamen and caudate of Huntington’s disease patients by direct injection of viral vector (AAV or lentivirus), as in Parkinson’s disease ( Palfi et al. , 2014 ).
Supplementary Material
Acknowledgements
We acknowledge Philippe Mailly for his contribution to 3D reconstruction of microscope images of AAVrh10-GFP injected mice, Ariel Di Nardo for critical reading of the manuscript, Peter Vanhoutte for helpful scientific discussions and Emmanuel Brouillet for fruitful comments on transcriptomic data from the literature. We thank the vectorology platform (Inserm U1089, Nantes, France) for the production of all AAV vectors.
Glossary
Abbreviations
- AAV
adeno-associated virus
- Exp-HTT
expanded HTT
- GFP
green fluorescent protein
- 24S-OHC
24S-hydroxycholesterol
Funding
These studies were supported in part by Agence Nationale pour la Recherche (ANR) “13-BSV1-022-01” grant, DIM Biotherapy Region Ile de France grant, Centre National pour la Recherche Scientifique (CNRS); Institut National pour la Santé et la Recherche Médicale (INSERM) and Université Pierre et Marie Curie (UPMC). The IBPS Cellular Imaging and Rodent facilities are supported by the “Conseil Général de la Région Ile-de France”, Sorbonne Universités and CNRS.
Supplementary material
Supplementary material is available at Brain online.
References
- Aylward EH, Codori AM, Rosenblatt A, Sherr M, Brandt J, Stine OC, et al. . Rate of caudate atrophy in presymptomatic and symptomatic stages of Huntington's disease . Mov Disord 2000. ; 15 : 552 – 60 . [DOI] [PubMed] [Google Scholar]
- Bauer PO, Goswami A, Wong HK, Okuno M, Kurosawa M, Yamada M, et al. . Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein . Nat Biotechnol 2010. ; 28 : 256 – 63 . doi: 10.1038/nbt.1608. Epub 2010 Feb 28 . [DOI] [PubMed] [Google Scholar]
- Benchoua A, Trioulier Y, Zala D, Gaillard MC, Lefort N, Dufour N, et al. . Involvement of mitochondrial complex II defects in neuronal death produced by N-terminus fragment of mutated huntingtin . Mol Biol Cell 2006. ; 17 : 1652 – 63 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb JA, Yan Z, Svenningsson P, Snyder GL, Pieribone VA, Horiuchi A, et al. . Severe deficiencies in dopamine signaling in presymptomatic Huntington's disease mice . Proc Natl Acad Sci USA 2000. ; 97 : 6809 – 14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkhem I, Lutjohann D, Breuer O, Sakinis A, Wennmalm A . Importance of a novel oxidative mechanism for elimination of brain cholesterol. Turnover of cholesterol and 24(S)-hydroxycholesterol in rat brain as measured with 18O2 techniques in vivo and in vitro . J Biol Chem 1997. ; 272 : 30178 – 84 . [DOI] [PubMed] [Google Scholar]
- Bjorkhem I, Meaney S . Brain cholesterol: long secret life behind a barrier . Arterioscler Thromb Vasc Biol 2004. ; 24 : 806 – 15 . [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL . The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor . Cell 1997. ; 89 : 331 – 40 . [DOI] [PubMed] [Google Scholar]
- Charvin D, Vanhoutte P, Pages C, Borrelli E, Caboche J . Unraveling a role for dopamine in Huntington's disease: the dual role of reactive oxygen species and D2 receptor stimulation . Proc Natl Acad Sci USA 2005. ; 102 : 12218 – 23 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevy F, Humbert L, Wolf C . Sterol profiling of amniotic fluid: a routine method for the detection of distal cholesterol synthesis deficit . Prenat Diagn 2005. ; 25 : 1000 – 6 . [DOI] [PubMed] [Google Scholar]
- Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D . Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration . Cell 2006. ; 127 : 59 – 69 . [DOI] [PubMed] [Google Scholar]
- Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, et al. . Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation . Cell 1997. ; 90 : 537 – 48 . [DOI] [PubMed] [Google Scholar]
- del Toro D, Xifro X, Pol A, Humbert S, Saudou F, Canals JM, et al. . Altered cholesterol homeostasis contributes to enhanced excitotoxicity in Huntington's disease . J Neurochem 2010. ; 115 : 153 – 67 . [DOI] [PubMed] [Google Scholar]
- Deyts C, Galan-Rodriguez B, Martin E, Bouveyron N, Roze E, Charvin D, et al. . Dopamine D2 receptor stimulation potentiates PolyQ-Huntingtin-induced mouse striatal neuron dysfunctions via Rho/ROCK-II activation . PLoS One 2009. ; 4 : e8287 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietschy JM, Turley SD . Thematic review series: brain Lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal . J Lipid Res 2004. ; 45 : 1375 – 97 . [DOI] [PubMed] [Google Scholar]
- Djelti F, Braudeau J, Hudry E, Dhenain M, Varin J, Bièche I, et al. . ( 2015) CYP46A1 inhibition, brain cholesterol accumulation and neurodegeneration pave the way for Alzheimer's disease . Brain 2015. ; 13 : 2383 – 98 . [DOI] [PubMed] [Google Scholar]
- Dunah AW, Jeong H, Griffin A, Kim YM, Standaert DG, Hersch SM, et al. . Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease . Science 2002. ; 296 : 2238 – 43 . [DOI] [PubMed] [Google Scholar]
- Dzeletovic S, Babiker A, Lund E, Diczfalusy U . Time course of oxysterol formation during in vitro oxidation of low density lipoprotein . Chem Phys Lipids 1995. ; 78 : 119 – 28 [DOI] [PubMed] [Google Scholar]
- Gauthier LR, Charrin BC, Borrell-Pages M, Dompierre JP, Rangone H, Cordelieres FP, et al. . Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules . Cell 2004. ; 118 : 127 – 38 . [DOI] [PubMed] [Google Scholar]
- Gines S, Ivanova E, Seong IS, Saura CA, MacDonald ME . Enhanced Akt signaling is an early pro-survival response that reflects N-methyl-D-aspartate receptor activation in Huntington's disease knock-in striatal cells . J Biol Chem 2003. ; 278 : 50514 – 22 . [DOI] [PubMed] [Google Scholar]
- Hudry E, Van Dam D, Kulik W, De Deyn PP, Stet FS, Ahouansou O, et al. . Adeno-associated virus gene therapy with cholesterol 24-hydroxylase reduces the amyloid pathology before or after the onset of amyloid plaques in mouse models of Alzheimer's disease . Mol Ther 2010. ; 18 : 44 – 53 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Lv H, Liao M, Xu X, Huang S, Tan H, et al. . GRP78 counteracts cell death and protein aggregation caused by mutant huntingtin proteins . Neurosci Lett 2012. ; 516 : 182 – 7 . [DOI] [PubMed] [Google Scholar]
- Karasinska JM, Hayden MR . Cholesterol metabolism in Huntington disease . Nat Rev Neurol 2011. ; 7 : 561 – 72 . [DOI] [PubMed] [Google Scholar]
- Kempen HJ, Glatz JF, Gevers Leuven JA, van der Voort HA, Katan MB . Serum lathosterol concentration is an indicator of whole-body cholesterol synthesis in humans . J Lipid Res 1988. ; 29 : 1149 – 55 . [PubMed] [Google Scholar]
- Korade Z, Kenworthy AK . Lipid rafts, cholesterol, and the brain . Neuropharmacology 2008. ; 55 : 1265 – 73 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, Goldstein DR, Hodges A, Strand AD, Sengstag T, Kooperberg C, et al. . Mutant huntingtin's effects on striatal gene expression in mice recapitulate changes observed in human Huntington's disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage . Hum Mol Genet 2007. ; 16 : 1845 – 61 . [DOI] [PubMed] [Google Scholar]
- Leoni V, Mariotti C, Tabrizi SJ, Valenza M, Wild EJ, Henley SM, et al. . Plasma 24S-hydroxycholesterol and caudate MRI in pre-manifest and early Huntington's disease . Brain 2008. ; 131 : 2851 – 9 . [DOI] [PubMed] [Google Scholar]
- Leoni V, Long JD, Mills JA, Di Donato S, Paulsen JS . Plasma 24S-hydroxycholesterol correlation with markers of Huntington disease progression . Neurobiol Dis 2013. ; 55 : 37 – 43 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni V, Caccia C . The impairment of cholesterol metabolism in Huntington disease . Biochim Biophys Acta 2015. ; 1851 : 1095 – 105 . doi: 10.1016/j.bbalip.2014.12.018. Epub 2015 Jan 14 . [DOI] [PubMed] [Google Scholar]
- Li H, Li SH, Yu ZX, Shelbourne P, Li XJ . Huntingtin aggregate-associated axonal degeneration is an early pathological event in Huntington's disease mice . J Neurosci 2001. ; 21 : 8473 – 81 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L, Jackson-Lewis V, Wong LC, Shui GH, Goh AX, Kesavapany S, et al. . Lanosterol induces mitochondrial uncoupling and protects dopaminergic neurons from cell death in a model for Parkinson's disease . Cell Death Differ 2012. ; 19 : 416 – 27 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund EG, Guileyardo JM, Russell DW . cDNA cloning of cholesterol 24-hydroxylase, a mediator of cholesterol homeostasis in the brain . Proc Natl Acad Sci USA 1999. ; 96 : 7238 – 43 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund EG, Xie C, Kotti T, Turley SD, Dietschy JM, Russell DW . Knockout of the cholesterol 24-hydroxylase gene in mice reveals a brain-specific mechanism of cholesterol turnover . J Biol Chem 2003. ; 278 : 22980 – 8 . Epub 2003 Apr 9 . [DOI] [PubMed] [Google Scholar]
- Martin MG, Perga S, Trovo L, Rasola A, Holm P, Rantamaki T, et al. . Cholesterol loss enhances TrkB signaling in hippocampal neurons aging in vitro . Mol Biol Cell 2008. ; 19 : 2101 – 12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DD, Ladha S, Ehrnhoefer DE, Hayden MR . Autophagy in Huntington disease and huntingtin in autophagy . Trends Neurosci 2015. ; 38 : 26 – 35 . [DOI] [PubMed] [Google Scholar]
- Marullo M, Valenza M, Leoni V, Caccia C, Scarlatti C, De Mario A, et al. . Pitfalls in the detection of cholesterol in Huntington's disease models . PLoS Curr 2012. ; 4 : e505886e9a1968 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menalled L, El-Khodor BF, Patry M, Suárez-Fariñas M, Orenstein SJ, Zahasky B, et al. . Systematic behavioral evaluation of Huntington's disease transgenic and knock-in mouse models . Neurobiol Dis 2009. ; 35 : 319 – 36 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moumné L, Betuing S, Caboche J . Multiple aspects of gene dysregulation in Huntington's disease . Front Neurol 2013. ; 4 : 127 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milnerwood AJ, Raymond LA . Early synaptic pathophysiology in neurodegeneration: insights from Huntington's disease . Trends Neurosci 2010. ; 33 : 513 – 23 . [DOI] [PubMed] [Google Scholar]
- Nieweg K, Schaller H, Pfrieger FW . Marked differences in cholesterol synthesis between neurons and glial cells from postnatal rats . J Neurochem 2009. ; 109 : 125 – 34 . [DOI] [PubMed] [Google Scholar]
- Nunes MJ, Milagre I, Schnekenburger M, Gama MJ, Diederich M, Rodrigues E . Sp proteins play a critical role in histone deacetylase inhibitor-mediated derepression of CYP46A1 gene transcription . J Neurochem 2010. ; 113 : 418 – 31 . [DOI] [PubMed] [Google Scholar]
- Paul BD, Sbodio JI, Xu R, Vandiver MS, Cha JY, Snowman AM, Snyder SH . Cystathionine γ-lyase deficiency mediates neurodegeneration in Huntington's disease . Nature 2014. ; 509 : 96 – 100 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palfi S, Gurruchaga JM, Ralph GS, Lepetit H, Lavisse S, Buttery PC, et al. . Long-term safety and tolerability of ProSavin, a lentiviral vector-based gene therapy for Parkinson's disease: a dose escalation, open-label, phase 1/2 trial . Lancet 2014. ; 383 : 1138 – 46 . doi: 10.1016/S0140-673661939-X . [DOI] [PubMed] [Google Scholar]
- Pfrieger FW . Role of cholesterol in synapse formation and function . Biochim Biophys Acta 2003. ; 1610 : 271 – 80 . [DOI] [PubMed] [Google Scholar]
- Puglielli L, Tanzi RE, Kovacs DM . Alzheimer's disease: the cholesterol connection . Nat Neurosci 2003. ; 6 : 345 – 51 . [DOI] [PubMed] [Google Scholar]
- Renner M, Choquet D, Triller A . Control of the postsynaptic membrane viscosity . J Neurosci 2009. ; 29 : 2926 – 37 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Poirier MA . Protein aggregation and neurodegenerative disease . Nat Med 2004. ; 10 ( Suppl ): S10 – 7 . [DOI] [PubMed] [Google Scholar]
- Roze E, Saudou F, Caboche J . Pathophysiology of Huntington's disease: from huntingtin functions to potential treatments . Curr Opin Neurol 2008a. ; 21 : 497 – 503 . [DOI] [PubMed] [Google Scholar]
- Roze E, Betuing S, Deyts C, Marcon E, Brami-Cherrier K, Pages C, et al. . Mitogen- and stress-activated protein kinase-1 deficiency is involved in expanded-huntingtin-induced transcriptional dysregulation and striatal death . FASEB J 2008b. ; 22 : 1083 – 93 . [DOI] [PubMed] [Google Scholar]
- Sadri-Vakili G, Bouzou B, Benn CL, Kim MO, Chawla P, Overland RP, et al. . Histones associated with downregulated genes are hypo-acetylated in Huntington's disease models . Hum Mol Genet 2007. ; 16 : 1293 – 306 . [DOI] [PubMed] [Google Scholar]
- Sapp E, Kegel KB, Aronin N, Hashikawa T, Uchiyama Y, Tohyama K, et al. . Early and progressive accumulation of reactive microglia in the Huntington disease brain . J Neuropathol Exp Neurol 2001. ; 60 : 161 – 72 . [DOI] [PubMed] [Google Scholar]
- Shafaati M, Olin M, Bavner A, Pettersson H, Rozell B, Meaney S, et al. . Enhanced production of 24S-hydroxycholesterol is not sufficient to drive liver X receptor target genes in vivo . J Intern Med 2011. ; 270 : 377 – 87 . [DOI] [PubMed] [Google Scholar]
- Shobab LA, Hsiung GY, Feldman HH . Cholesterol in Alzheimer's disease . Lancet Neurol 2005. ; 4 : 841 – 52 . [DOI] [PubMed] [Google Scholar]
- Sipione S, Rigamonti D, Valenza M, Zuccato C, Conti L, Pritchard J, et al. . Early transcriptional profiles in huntingtin-inducible striatal cells by microarray analyses . Hum Mol Genet 2002. ; 11 : 1953 – 65 . [DOI] [PubMed] [Google Scholar]
- Sondhi D, Hackett NR, Peterson DA, Stratton J, Baad M, Travis KM, et al. . Enhanced survival of the LINCL mouse following CLN2 gene transfer using the rh.10 rhesus macaque-derived adeno-associated virus vector . Mol Ther 2007. ; 15 : 481 – 91 . [DOI] [PubMed] [Google Scholar]
- Spann NJ, Garmire LX, McDonald JG, Myers DS, Milne SB, Shibata N, et al. . Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses . Cell 2012. ; 151 : 138 – 52 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack EC, Kubilus JK, Smith K, Cormier K, Del Signore SJ, Guelin E, et al. . Chronology of behavioral symptoms and neuropathological sequela in R6/2 Huntington's disease transgenic mice . J Comp Neurol 2005. ; 490 : 354 – 70 . [DOI] [PubMed] [Google Scholar]
- Suzuki S, Kiyosue K, Hazama S, Ogura A, Kashihara M, Hara T, et al. . Brain-derived neurotrophic factor regulates cholesterol metabolism for synapse development . J Neurosci 2007. ; 27 : 6417 – 27 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Huntington's Disease Collaborative Research Group . A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes . Cell 1993. ; 72 : 971 – 83 [DOI] [PubMed] [Google Scholar]
- Tourette C, Farina F, Vazquez-Manrique RP, Orfila AM, Voisin J, Hernandez S, et al. . The Wnt receptor Ryk reduces neuronal and cell survival capacity by repressing FOXO activity during the early phases of mutant huntingtin pathogenicity . PLoS Biol 2014. ; 12 : e1001895 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F, et al. . Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells . Hum Mol Genet 2000. ; 9 : 2799 – 809 . [DOI] [PubMed] [Google Scholar]
- Trushina E, Singh RD, Dyer RB, Cao S, Shah VH, Parton RG, et al. . Mutant huntingtin inhibits clathrin-independent endocytosis and causes accumulation of cholesterol in vitro and in vivo . Hum Mol Genet 2006. ; 15 : 3578 – 91 . [DOI] [PubMed] [Google Scholar]
- Trushina E, Canaria CA, Lee DY, McMurray CT . Loss of caveolin-1 expression in knock-in mouse model of Huntington's disease suppresses pathophysiology in vivo . Hum Mol Genet 2014. ; 23 : 129 – 44 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, Roque RA, et al. . PGC-1alpha rescues Huntington's disease proteotoxicity by preventing oxidative stress and promoting TFEB function . Sci Transl Med 2012. ; 4 : 142ra97 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano Y, Ochiai S, Noguchi N . Suppression of amyloid-beta production by 24S-hydroxycholesterol via inhibition of intracellular amyloid precursor protein trafficking . FASEB J 2013. ; 27 : 4305 – 15 . [DOI] [PubMed] [Google Scholar]
- Valenza M, Rigamonti D, Goffredo D, Zuccato C, Fenu S, Jamot L, et al. . Dysfunction of the cholesterol biosynthetic pathway in Huntington's disease . J Neurosci 2005. ; 25 : 9932 – 9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenza M, Carroll JB, Leoni V, Bertram LN, Bjorkhem I, Singaraja RR, et al. . Cholesterol biosynthesis pathway is disturbed in YAC128 mice and is modulated by huntingtin mutation . Hum Mol Genet 2007a. ; 16 : 2187 – 98 . [DOI] [PubMed] [Google Scholar]
- Valenza M, Leoni V, Tarditi A, Mariotti C, Bjorkhem I, Di Donato S, et al. . Progressive dysfunction of the cholesterol biosynthesis pathway in the R6/2 mouse model of Huntington's disease . Neurobiol Dis 2007b. ; 28 : 133 – 42 . [DOI] [PubMed] [Google Scholar]
- Valenza M, Leoni V, Karasinska JM, Petricca L, Fan J, Carroll J, et al. . Cholesterol defect is marked across multiple rodent models of Huntington's disease and is manifest in astrocytes . J Neurosci 2010. ; 30 : 10844 – 50 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenza M, Cattaneo E . Emerging roles for cholesterol in Huntington's disease . Trends Neurosci 2011. ; 34 : 474 – 86 . [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP, Jr., et al. . Neuropathological classification of Huntington’s disease . J Neuropathol Exp Neurol 1985. ; 44 : 559 – 77 . [DOI] [PubMed] [Google Scholar]
- Xiang Z, Valenza M, Cui L, Leoni V, Jeong HK, Brilli E, et al. . Peroxisome-proliferator-activated receptor gamma coactivator 1 α contributes to dysmyelination in experimental models of Huntington's disease . J Neurosci 2011. ; 31 : 9544 – 53 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, et al. . Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease . Science 2001. ; 293 : 493 – 8 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.