Abstract
Pinpointing multi‐faceted, longitudinally‐changing factors that drive colorectal cancer is laborious and expensive, but doing so is necessary for more accurate CRC prognosis and therapy.

Subject Categories: Ageing, Cancer, Molecular Biology of Disease
More than 90% of colorectal cancer (CRC) cases occur in people 50 years or older. This fact accords with the well‐established notion that many cancers are diseases of old age that result from changes to or the deterioration of our cellular processes. So what specifically goes wrong?
“Of cancers that affect both men and women, colorectal cancer is the second leading cancer killer in the United States [and Europe], but it doesn't have to be.” (Centers for Disease Control and Prevention, http://www.cdc.gov/cancer/colorectal/).
Genes certainly play a role, but inherited genetic variation only partly explains the predisposition to sporadic CRC. More important, perhaps, are somatic mutations that accumulate in some cells and tumors over the years driving CRC. But such mutations build up at different rates in different people and do not impact all people the same way. Moreover, some somatic mutations are cell division‐driven, accumulating at a presumably constant rate, while others persist because they confer a cell proliferation and survival advantage, or because they provide that advantage in a context‐dependent manner, for example, upon intestinal inflammation. In fact, chronic inflammation establishes an ideal environment in which to nurture tumor development because it stimulates the recruitment of immune cells that produce free radicals, cytokines, and growth factors and couples tissue damage to adverse compensatory regeneration. A subclinical form of inflammatory signaling that contributes to heightened regeneration, as indicated by studies in flies and mice, might contribute to many cases of CRC 1. Thus, in addition to inherited and cell division‐driven mutations, our lifestyle‐shaped environment and our intestinal microbiota probably facilitate inflammation, metabolic deterioration, and epigenetic and genetic changes that accumulate as we age.
Environmental factors—such as diet, alcohol consumption, smoking, and lack of exercise—clearly predispose for CRC. Remarkably, however, the effects are partly reversible: CRC outcomes can significantly improve when we adopt healthier habits, either before or after diagnosis 2. This is likely due to reversible systemic and local (intestinal) metabolic and homeostatic changes that drive inflammatory signaling and facilitate tumorigenesis 1, 2. Thus, CRC incidence and outcome is not merely a result of “bad luck,” but is, to a certain degree, preventable, although the factors and mechanisms behind this are still unclear.
Intestinal microbiota are affected by environmental factors and are intensively studied in relation to inflammation and CRC. Nevertheless, proving that specific microbes are the causative agents of CRC has turned out to be difficult. For example, proving that Helicobacter pylori is a causative agent for gastric ulcers (and cancer) needed to satisfy most of Koch's postulates: that is, the bacteria had to be found in and isolated from ulcers, tested in a human, and tackled through antibiotic treatment for ulcer eradication.
To find environmental and/or microbial factors that contribute to CRC, combinatorial comparisons will need to be made in each person or animal model. For example, in Drosophila, genetic predisposition via K‐Ras/Ras1 oncogene expression synergizes with enterocyte‐damaging Pseudomonas aeruginosa to promote tumorigenesis, and P. aeruginosa becomes more virulent in the presence of peptidoglycan derived from other bacteria 3, 4. Moreover, intestinal P. aeruginosa synergizes with the K‐Ras/Ras1 oncogene to cause basal invasion and dissemination of Drosophila enterocytes 5. In mice, intestinal Citrobacter rodentium, Fusobacterium nucleatum, and Helicobacter hepaticus enhance tumorigenesis in animals that carry mutations in the tumor suppressor APC or lack the immunoregulatory cytokine IL‐10 and have been exposed to the mutagen azoxymethane 6, 7, 8.
To identify such synergisms in humans, we have to (i) assess personalized holo'omes in youth (disease‐free) versus old (disease‐prone) age—each holo'ome being the combination of the host and microbiota genome, transcriptome and proteome, the blood secretome and the intestinal metabolome (Fig 1); (ii) associate changes in holo'omes computationally, focusing on synergisms between host and microbe genes, blood factors and intestinal metabolites linked to disease; (iii) test the detrimental synergisms in model organisms, such as Drosophila and mice; (iv) provide preventive or therapeutic options that break those synergisms while following CRC occurrence.
Figure 1. Personalized holo'ome studied longitudinally in youth (disease‐free) versus old (disease‐prone) age.
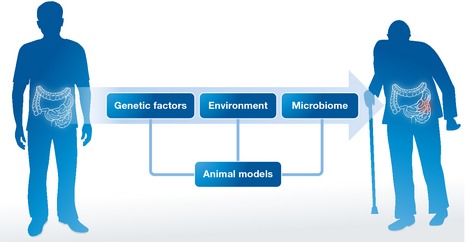
Holo'ome components include the host gene expression, the systemic and intestinal environment and the intestinal microbiome that can be modeled and studied in combination in animals.
Such a multi‐omic approach might also be applicable to other cancers influenced by microbiota and the environment. Unlike childhood cancers, such as retinoblastoma and neuroblastoma, which are usually dictated by the genetic background of neonates, most other malignancies, such as lung, liver, and pancreatic cancer, are heavily influenced by lifestyle. While the role of microbiota is likely more prominent and direct in CRC, many other organs are likely to be influenced by the intestinal, oropharyngeal, or skin flora.
Finally, whereas CRC research is focusing on tumor heterogeneity, microenvironment and mutation identification, other relevant research fields are making progress in assessing factors beyond genetics in disease. For example, research has linked inflammatory bowel and metabolic disease to specific host susceptibility mutations and dysbiosis (a shift in intestinal microbiota consistency) 9. Moreover, multi‐omic approaches have been used in microbiological, microbiota, and cancer studies 10. Despite significant progress, these are still in their infancy, because integrated analysis of raw multi‐omic data requires standardized methodologies in data acquisition, sophisticated, and specialized software and computational resources inaccessible to most biological research laboratories.
Pinpointing multifaceted, longitudinally changing factors that drive CRC is laborious and expensive, but doing so is necessary for more accurate CRC prognosis and therapy. The detrimental synergisms might be many and diverse, but to paraphrase Groucho Marx: These are the known principles of CRC, and if they do not suffice to explain reality, we will have to find others.
Contributor Information
Yiorgos Apidianakis, Email: apidiana@ucy.ac.cy.
Aristides G Eliopoulos, Email: eliopag@med.uoc.gr.
References
- 1. Panayidou S, Apidianakis Y (2013) Pathogens 2: 209–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee J, Jeon JY, Meyerhardt JA (2015) Hematol Oncol Clin North Am 29: 1–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Apidianakis Y, Pitsouli C, Perrimon N et al (2009) Proc Natl Acad Sci USA 106: 20883–20888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Korgaonkar A, Trivedi U, Rumbaugh KP et al (2013) Proc Natl Acad Sci USA 110: 1059–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bangi E, Pitsouli C, Rahme LG et al (2012) EMBO Rep 13: 569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Newman JV, Kosaka T, Sheppard BJ et al (2001) J Infect Dis 184: 227–230 [DOI] [PubMed] [Google Scholar]
- 7. Kostic AD, Chun E, Robertson L et al (2013) Cell Host Microbe 14: 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nagamine CM, Rogers AB, Fox JG et al (2008) Int J Cancer 122: 832–838 [DOI] [PubMed] [Google Scholar]
- 9. Kostic AD, Xavier RJ, Gevers D (2014) Gastroenterology 146: 1489–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Integrative HMP (iHMP) Research Network Consortium (2014) Cell Host Microbe 16: 276–289 [DOI] [PMC free article] [PubMed] [Google Scholar]


