Summary
Autism spectrum disorders (ASD) are a group of devastating neurodevelopmental syndromes that affect up to 1 in 68 children. Despite advances in the identification of ASD risk genes, the mechanisms underlying ASD remain unknown. Homozygous loss-of-function mutations in Contactin Associated Protein-like 2 (CNTNAP2) are strongly linked to ASD. Here we investigate the function of Cntnap2 and undertake pharmacological screens to identify phenotypic suppressors. We find that zebrafish cntnap2 mutants display GABAergic deficits particularly in the forebrain and sensitivity to drug-induced seizures. High-throughput behavioral profiling identifies nighttime hyperactivity in cntnap2 mutants, while pharmacological testing reveals dysregulation of GABAergic and glutamatergic systems. Finally, we find that estrogen receptor agonists elicit a behavioral fingerprint anti-correlative to that of cntnap2 mutants and show that the phytoestrogen biochanin A specifically reverses the mutant behavioral phenotype. These results identify estrogenic compounds as phenotypic suppressors and illuminate novel pharmacological pathways with relevance to autism.
Introduction
Autism spectrum disorders (ASD) are a group of neurodevelopmental syndromes characterized by deficits in social interaction and communication as well as repetitive behaviors and restricted interests (American Psychiatric Association, 2013). Gene discovery in ASD has accelerated dramatically (De Rubeis et al., 2014; Iossifov et al., 2014; Sanders et al., 2015), providing a launching point for the illumination of relevant biological pathways (Parikshak et al., 2013; Willsey et al., 2013). Contactin Associated Protein-like 2 (CNTNAP2) is one of the first genes strongly linked to autism and epilepsy in consanguineous families (Strauss et al., 2006). This gene encodes a cell adhesion molecule of the neurexin family that localizes voltage-gated potassium channels at the juxtaparanodal region of myelinated axons (Poliak et al., 2003). Loss of CNTNAP2 function in mice leads to abnormal neuronal migration, reduced GABAergic neurons, spontaneous seizures, hyperactivity, social deficits, and increased repetitive behaviors (Penagarikano et al., 2011). However, the function of CNTNAP2 in the central nervous system and the consequences of its loss for ASD pathology are less well understood.
At present, our ability to advance rapidly from the identification of risk genes to the discovery of biological mechanisms and pharmacological suppressors remains limited. The zebrafish is a model vertebrate system well-suited for conducting small molecule screens to uncover modulators of signaling pathways (Ablain and Zon, 2013; Kokel and Peterson, 2011; Rihel et al., 2010). Quantitative behavioral profiling provides a high-throughput approach to characterize psychoactive molecules based on the behavioral readout of their effects in zebrafish larvae (Rihel et al., 2010). Here we investigate the consequences of the loss of Cntnap2 in zebrafish and utilize quantitative behavioral profiling as a platform to conduct rational pharmacological screens to identify phenotypic suppressors and novel pathways with relevance to autism.
Results
cntnap2 Mutants Display GABAergic Deficits
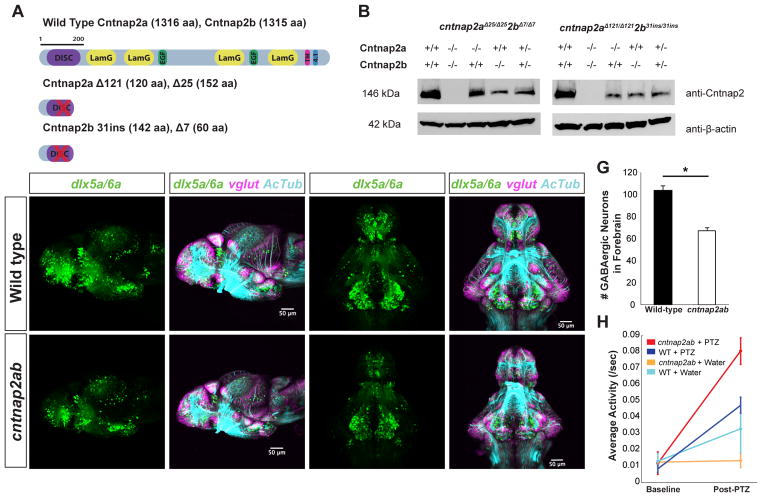
Zebrafish have two cntnap2 paralogs that are expressed broadly in the developing central nervous system (CNS), with higher expression in the telencephalon (Figure S1A–C). Cntnap2a and Cntnap2b proteins show 71% and 65% identity to the human protein, respectively, and contain the same functional domains (Figure 1A). To characterize their function, we generated two loss-of-function mutations in each paralog using zinc finger nucleases and crossed fish carrying these alleles to generate double mutant fish (cntnap2aΔ121/Δ121cntnap2b31i/31i and cntnap2aΔ25/Δ25cntnap2bΔ7/Δ7) (Figure S1D–F), referred to hereafter as cntnap2ab mutants. Each mutation causes a premature stop codon within the discoidin domain and results in loss of protein expression (Figures 1A and 1B), such that double mutants represent a loss of Cntnap2 function.
Figure 1. cntnap2ab mutants display GABAergic deficits.
A. Cntnap2a and Cntnap2b proteins showing functional domains. (DISC = Discoidin; LamG = Laminin G; EGF = Epidermal Growth Factor; TM = Transmembrane; 4.1 = 4.1 Binding Domain).
B. Western blot analysis of Cntnap2a and Cntnap2b in cntnap2aΔ25/Δ25cntnap2bΔ7/Δ7 and cntnap2aΔ121/Δ121cntnap2b31i/31i fish.
C–F, C′–F′. Reporter gene expression in Tg(dlx6a-1.4kbdlx5a/dlx6a:GFP) and Tg(vglut:DsRed) in wild-type (C–F) and cntnap2aΔ121/Δ121cntnap2b31i/31i (cntnap2ab) (C′–F′) larvae at 4 dpf. Note the deficit in dlx5a/6a:GFP+ cells in the telencephalon (arrowheads). (C–D, C′–D′): lateral views. (E–F, E′–F′): ventral views. (tel, telencephalon; hyp, hypothalamus; OT, optic tectum.)
G. Total number of dlx5a/6a:GFP+ cells in the forebrain of wild-type (n=13) and cntnap2ab mutants (n=14) (*p=3.08 × 10−7, one-way ANOVA).
H. Average activity in response to pentylenetetrazol (PTZ) (10 mM) in the progeny of incrossed cntnap2aΔ25/+cntnap2bΔ7/+ fish at 4 dpf (n=268; cntnap2ab + PTZ, n=6; wild-type + PTZ, n=11; cntnap2ab + water, n=11; wild-type + water, n=6; p=0.0013, two-way ANOVA, genotype x dose interaction).
It has been proposed that an imbalance in excitatory and inhibitory signaling in the CNS is a mechanism underlying ASD and epilepsy (Rubenstein and Merzenich, 2003). To test if this is occurring in cntnap2ab mutants, we analyzed inhibitory and excitatory neuronal populations in wild-type and cntnap2ab mutants during early brain development, using transgenic lines that label GABAergic neurons and precursors (Tg(dlx6a-1.4kbdlx5a/dlx6a:GFP)) and glutamatergic neurons (Tg(vglut:DsRed)) (Ghanem et al., 2003; Kinkhabwala et al., 2011; Zerucha et al., 2000). We observed a significant decrease in GABAergic cells in cntnap2ab mutants at 4 days post fertilization (dpf) (Figures 1C–F and 1C′–F′; and Figures S2A–H and S2A′–H′) in the forebrain and cerebellum (Figure S2I–J). In particular, there were an average of 34% fewer GABAergic neurons in the forebrain of cntnap2ab mutants compared to wild-type fish at 4 dpf (Figure 1G, p=3.08×10−7, one-way ANOVA). The decreased number of GABAergic cells in the dorsal telencephalon (pallium) at this stage is consistent with a failure in the migration of these cells from the ventral telencephalon (subpallium), similar to findings reported in Cntnap2 mutant mice (Penagarikano et al., 2011). In contrast, there were no significant regional deficits in glutamatergic neurons (Figure S2A–H, S2A′–H′ and S2K).
Although there were no gross morphological abnormalities in the structure of the axon scaffold in cntnap2ab mutants between 48 hpf and 5 dpf (Figures S1H–I and S1H′–I′), mutant head size is significantly smaller (Table S1, Figures S1N–R and S1N′–Q′). In addition, we did not observe gross differences in markers of apoptosis (TUNEL staining) at 28 hpf or proliferation (phospho-histone H3) at 48 hpf (Figures S2R–S and S2R′–S′). However, deficits in GABAergic neurons and transient delays in commissure formation are evident in the forebrain of mutants at 28 hpf (Figures S1J–M and S1J′–K′), indicating additional early roles for Cntnap2 in brain development. Together, these data indicate loss of Cntnap2 results in a deficit of inhibitory neurons, particularly in the forebrain.
Increased Seizure Susceptibility in cntnap2 Mutants
Loss of inhibitory neurons can increase susceptibility to seizures (Cobos et al., 2005). To determine the effect of loss of Cntnap2 on seizure susceptibility in zebrafish, we treated wild-type and mutant larvae with pentylenetetrazol (PTZ), a GABA-A receptor antagonist that induces seizures in rodents and zebrafish (Baraban et al., 2005; Watanabe et al., 2010). cntnap2ab mutants display increased sensitivity to PTZ-induced seizures (Figure 1H and Figure S3A–F). Drug-induced seizures appear as robust increases in activity and rapid, burst-like and circling movements, followed by periods of inactivity (Baraban et al., 2005). Homozygous double mutants display significantly more activity in response to PTZ (Figure S4B, p=0.0013, two-way ANOVA, genotype x dose interaction, n=268). These results are consistent with increased seizures associated with CNTNAP2 mutations in humans and mice (Penagarikano et al., 2011; Strauss et al., 2006). Consequently, increased sensitivity to PTZ provides further evidence for GABAergic deficits in cntnap2ab mutants.
cntnap2 Mutants Show Nighttime Hyperactivity
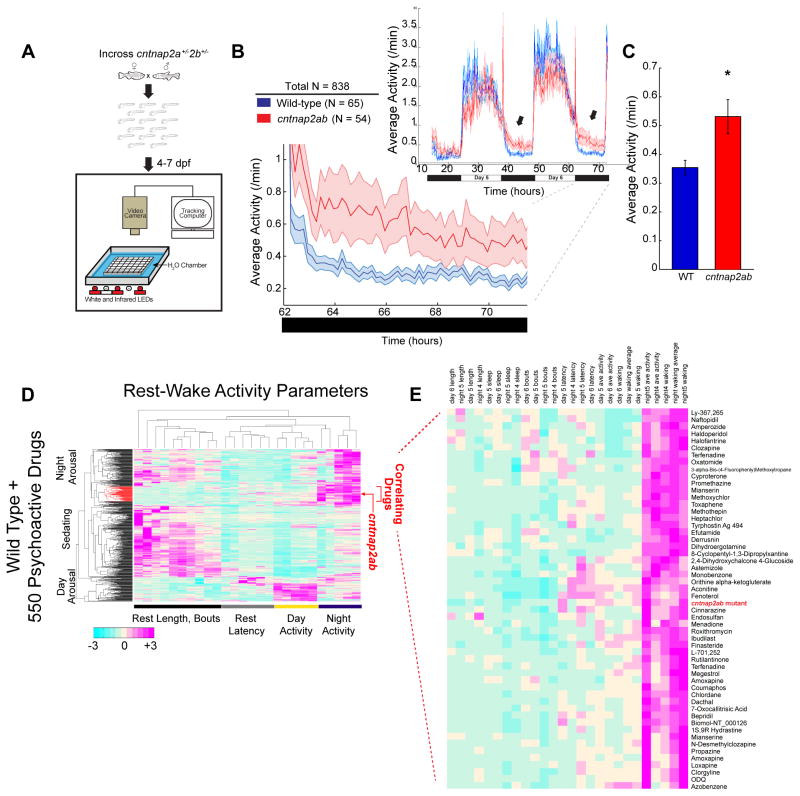
We next adapted a blinded, high-throughput assay to quantify a series of rest-wake cycle behavioral parameters over multiple days (Figure 2A) (Prober et al., 2006; Rihel et al., 2010). Quantitative profiling revealed that cntnap2ab mutants display significantly greater nighttime activity compared to wild-type siblings between 4 and 7 dpf (Figures 2B and 2C and Figure S3G, p=0.00012, one-way ANOVA, n=838). This phenotype was independently observed in cntnap2ab mutants harboring different alleles (Figure S3H, p=0.0198, n=187). In contrast, other rest-wake parameters (Table S2), as well as acoustic startle, habituation, and optokinetic response (Table S3) were not significantly affected, further highlighting the specificity of the nighttime hyperactivity phenotype. Combined, these results indicate that loss of Cntnap2 selectively causes nighttime hyperactivity, consistent with GABAergic deficits leading to an imbalance of excitatory and inhibitory signaling in mutant fish.
Figure 2. cntnap2ab mutants display nighttime hyperactivity.
A. Experimental setup (Prober et al., 2006; Rihel et al., 2010).
B. Locomotor activity of cntnap2aΔ25/Δ25cntnap2bΔ7/Δ7 (cntnap2ab, red) and wild-type (WT, blue) sibling-matched larvae over 72 h. Hyperactivity in mutants worsens on successive nights (arrows). The magnified activity profile on night 6 is shown.
C. Average locomotor activity of cntnap2ab vs. wild-type. (*p=0.00012, one-way ANOVA, comparing all genotypes on all nights; p=0.0193, 0.0236, 0.0073, nights 4, 5, and 6, respectively.)
D. Hierarchical clustering of the cntnap2ab behavioral fingerprint (red arrow) compared to the fingerprints of wild-type larvae exposed to a panel of 550 psychoactive agents from 4 to 6 dpf (Rihel et al., 2010). Each rectangle in the clustergram represents the Z score, or the average value in standard deviations relative to the behavioral profiles of wild-type exposed to DMSO alone (magenta, higher than DMSO; cyan, lower than DMSO). The cntnap2ab profile correlates with agents that induce nighttime arousal (“Correlating Drugs”).
E. A magnified section of the clustergram, highlighting compounds that correlate with the cntnap2ab mutant behavioral fingerprint.
Differential Responses to Psychoactive Agents in cntnap2 Mutants
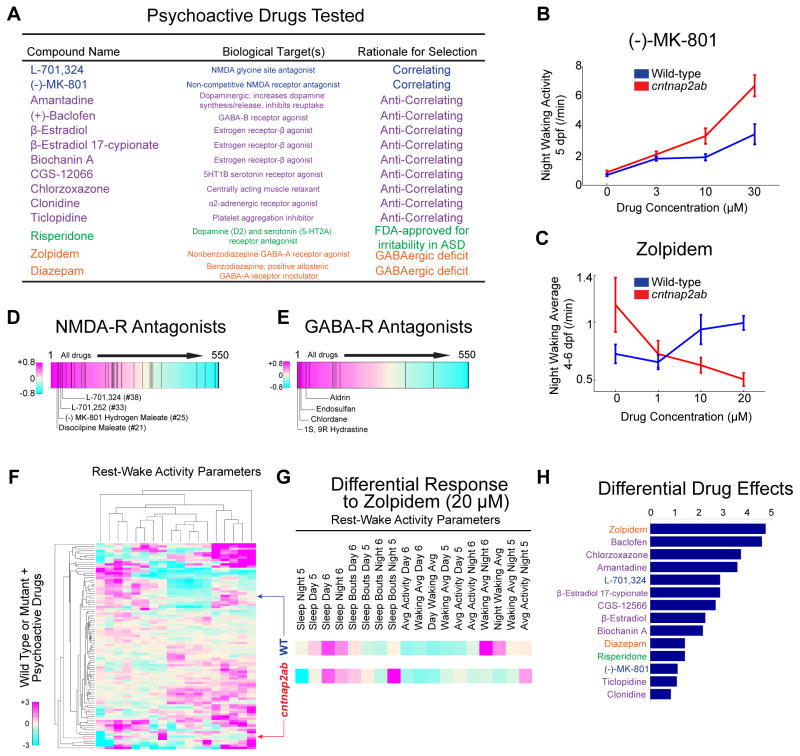
Collectively, the day-night, rest-wake cycle parameters of cntnap2ab mutants represent a genotypic behavioral fingerprint with a specific nighttime hyperactivity signature. To identify molecular pathways that are dysregulated in the absence of Cntnap2 function, we searched for drugs that phenocopy the mutant behavioral profile. To this end, we compared the cntnap2ab mutant behavioral fingerprint to a dataset of the behavioral profiles of wild-type larvae exposed to 550 psychoactive compounds (Rihel et al., 2010). This allows for an unbiased comparison of the different genetic and pharmacological conditions by cluster analysis (Rihel et al., 2010) (Figure 2D), and resulted in the identification of small molecules that strongly correlate with the mutant behavioral profile (Figure 2E). Next, we identified the top 14 drugs that anti-correlate, or generate the opposite phenotype of the mutant behavioral profile (Figure S4H). We reasoned that drugs that induce differential effects in wild-type and mutant larvae might indicate pathways that are dysregulated due to loss of Cntnap2. To test this, we exposed wild-type and cntnap2aΔ121/Δ121cntnap2b31i/31i larvae to a group of compounds from 4 to 7 dpf (Figure 3A) that were selected based on the following criteria: (1) correlating; (2) anti-correlating; (3) GABA-A receptor agonists, based on the structural GABAergic deficits; and (4) risperidone, because it is the first FDA-approved treatment for irritability and aggressive behavior in ASD (McCracken et al., 2002).
Figure 3. Differential behavioral responses of cntnap2ab mutants to psychoactive agents.
A. The 14 psychoactive drugs tested in cntnap2aΔ121/Δ121cntnap2b31i/31i and wild-type larvae, their biological targets, and the rationale for their selection. The following classes of drugs are shown: correlating (blue); anti-correlating (purple); drugs interacting with the GABA-A receptor (orange); and risperidone (green).
B, C. Dose-response effects of the NMDA receptor antagonist, (−)-MK-801, on night waking activity at 5 dpf (B) and the non-benzodiazepine GABA-A receptor agonist, zolpidem, on average night waking activity at 4–6 dpf (C) in wild-type (WT, blue) and cntnap2ab (red) larvae (p=0.002, (−)-MK-801; p=0.0003, zolpidem; two-way ANOVA, genotype x drug interaction).
D, E. Significant enrichment of NMDA receptor antagonists (D) and GABA receptor antagonists (E) in the top ranks of correlating drugs (p=0.031, NMDA-R antagonists and p=0.034, GABA-R antagonists; Kolmogorov-Smirnov).
F. Hierarchical clustering of the behavioral profiles of wild-type or cntnap2ab larvae exposed to 14 psychoactive agents at three doses each. Each rectangle in the clustergram represents the Z-score relative to the behavior of wild-type or mutant larvae exposed to DMSO alone (magenta, higher than DMSO; cyan, lower than DMSO).
G. Magnified sections highlight the behavioral fingerprints of wild-type and cntnap2ab larvae in response to zolpidem (20 μM).
H. Pairwise Euclidean distances between wild-type and cntnap2ab responses to psychoactive agents in the PCA. Note that zolpidem and (±)-baclofen produce the strongest differential responses.
Three lines of evidence indicate that both glutamatergic and GABAergic pathways are dysregulated in cntnap2ab mutants. First, we found that both GABA- and NMDA-receptor antagonists are significantly enriched among drugs that strongly correlate with the mutant phenotype (Figures 3D and 3E, p=0.031 and p=0.034, respectively, Kolmogorov-Smirnov). These findings are consistent with the behavioral profile of cntnap2ab mutants, given that NMDA receptor antagonists induce nighttime hyperactivity in wild-type larvae, and the increased sensitivity of mutants to the GABA-A receptor antagonist, PTZ. Second, cntnap2ab mutants are more sensitive to arousal by NMDA receptor antagonists across a range of doses (Figure 3B), suggesting attenuation of glutamatergic signaling.
Third, we found that GABA receptor agonists, such as zolpidem, induce differential behavioral effects in cntnap2ab mutants compared to wild-type larvae (Figures 3C and 3F–H). For this analysis, we performed hierarchical clustering and principal component analysis (PCA) to quantify differential mutant responses compared to wild-type across 18 rest-wake activity parameters (Rihel et al., 2010). Specifically, we calculated the Euclidean distance between the mutant + drug and wild-type + drug behavioral profiles, representing the difference between the mutant and wild-type responses to each drug (Figure 3H and Figure S4A–F; see Supplemental Experimental Procedures). Using this approach, we found that the GABA receptor agonists, zolpidem and (±)-baclofen, produce the strongest differential responses of the drugs tested (Figure 3H), suggesting perturbation of GABAergic signaling pathways in mutants. Taken together, our quantitative behavioral profiling reveals dysfunction in glutamatergic as well as GABAergic pathways due to loss of Cntnap2.
Estrogens Reverse the Behavioral Phenotype of cntnap2ab Mutants
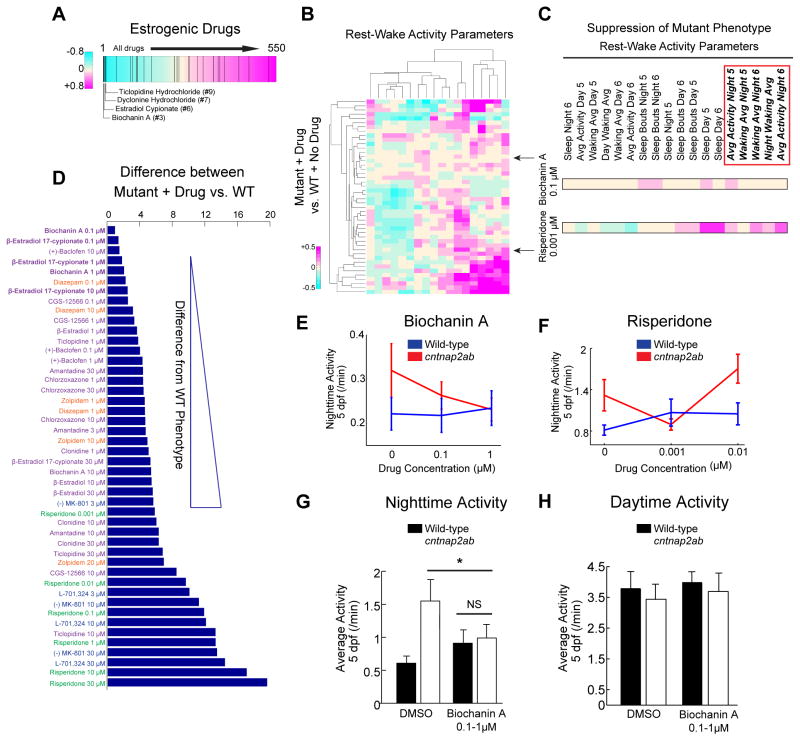
Next, we searched for small molecules that suppress the mutant phenotype of nighttime hyperactivity. We reasoned that such compounds might elicit a behavioral fingerprint in wild-type opposite to that of mutant larvae. We found that estrogenic compounds are significantly enriched in the top ranks of small molecules that anti-correlate with the cntnap2ab mutant behavioral fingerprint (Figure 4A, p=0.0003 by random permutation). Specifically, four of the top ten anti-correlating drugs have known estrogenic activity. Based on these results, we hypothesized that such drugs might be able to rescue the mutant phenotype. To test this, we analyzed the effects of estrogenic compounds and other selected molecules in wild-type and mutant larvae using quantitative behavioral profiling and PCA analysis (Figure 3A; see Supplemental Experimental Procedures). Behavioral rescue was defined as the shortest Euclidean distance between the mutant + drug and wild type + no drug behavioral profiles (Figure 4D).
Figure 4. Biochanin A reverses nighttime hyperactivity in cntnap2ab mutants.
A. Rank-sorting of the anti-correlating dataset with respect to estrogenic compounds shows significant enrichment of estrogenic agents in the top ranks (p=0.0003 by random permutation). Black lines indicate drugs defined as having estrogenic activity (25 compounds in total).
B. Hierarchical clustering of the behavioral fingerprints of cntnap2aΔ121/Δ121cntnap2b31i/31i larvae exposed to 14 psychoactive agents at three doses each relative to the wild-type + no drug fingerprint. Each rectangle in the clustergram represents the Z-score of drug-exposed mutants relative to untreated wild-type (magenta, higher than wild-type; cyan, lower than wild-type).
C. Magnified sections of the clustergram show relative suppression of the mutant fingerprint by biochanin A (0.1 μM) compared to risperidone (0.001 μM). The red box highlights parameters that measure nighttime activity.
D. Pairwise Euclidean distances (“Differential Drug Effects”) between the mutant responses to psychoactive agents compared to untreated wild-type in the PCA (Figure S4G). Biochanin A (0.1 μM) produces the strongest suppression of the mutant phenotype in this assay, with the fewest effects on other behavioral parameters.
E, F. Dose-response effects of biochanin A (E) and risperidone (F) on nighttime activity at 5 dpf (p=0.0001, biochanin A; p=0.0034, risperidone, two-way ANOVA, gene x drug interaction). While there is some experimental variability in the baseline activity of both wild-type and mutant larvae, nighttime hyperactivity is consistently observed.
G, H. Effect of the blind addition of biochanin A (0.1–1 μM) or DMSO on activity at night (G) and day (H) in the progeny of incrosses of cntnap2aΔ25/+cntnap2bΔ7/+ fish at 5 dpf (*p=0.045, two-way ANOVA, gene x dose interaction).
We found that biochanin A, a plant-derived estrogen, and β-estradiol 17-cypionate most strongly reverse the mutant behavioral phenotype (Figures 4B–D and Figure S4G). Indeed, the behavioral response of cntnap2ab larvae treated with biochanin A (0.1 μM) from 4 to 7 dpf most strongly correlates with the wild-type phenotype, decreasing nighttime activity with little effect on other measures of rest and activity (Figures 4B and 4C). In contrast, risperidone (0.001 μM) reverses nighttime hyperactivity, but alters other rest-wake cycle parameters, indicating that it elicits a less specific phenotypic rescue than biochanin A (Figures 4C and 4E–F). In addition, we found that early exposure to biochanin A does not reverse the GABAergic deficits (Figure S4M) or drug-induced seizures (Figure S4K), and that chronic exposure followed by washout does not rescue nighttime hyperactivity (Figure S4L), providing evidence for an acute mechanism of action. Consistent with estrogenic activity, we found that biochanin A (10 μM) significantly activates the expression of estrogen response genes in zebrafish larvae (Figure S4N). However, the rescue dose (1 μM) shows only a weak effect on target genes, suggesting that the behavioral rescue might occur independently of the robust transcriptional activation of estrogen target genes. Moreover, we found that biochanin A (0.1–1 μM) leads to selective suppression of nighttime activity at 5 dpf and no change in daytime activity in background-matched larvae (Figures 4G and 4H), indicating its effects are not due to background effects or generalized sedation. Furthermore, β-estradiol (1 μM) is also able to rescue nighttime hyperactivity in cntnap2ab mutants (Figure S4J). Together, these results provide evidence that biochanin A and β-estradiol acutely suppress a specific behavioral phenotype in a genetic loss-of-function model of an ASD risk gene.
Discussion
This study represents the first characterization of zebrafish mutants of an ASD risk gene using quantitative behavioral profiling to identify biological pathways with relevance to ASD. We demonstrate that zebrafish cntnap2ab mutants display reductions in GABAergic neurons, increased sensitivity to drug-induced seizures, and nighttime hyperactivity. Further, our pharmacological data support dysregulation of GABAergic and glutamatergic signaling in mutants. Altered NMDA signaling has been shown to cause GABAergic deficits, particularly in parvalbumin-positive (PV+) interneurons, and has been proposed as a mechanism underlying neuropsychiatric disorders (Keilhoff et al., 2004; Saunders et al., 2013). Moreover, mouse Cntnap2 knockouts display GABAergic deficits, most prominently in PV+ interneurons (Penagarikano et al., 2011). Further studies are required to determine if PV+ subpopulations are altered in cntnap2ab mutants, but our study highlights that GABAergic deficits are likely to occur in concert with alterations in NMDA circuits, not previously associated with Cntnap2.
Next, our results uncover the ability of estrogens to rescue the cntnap2 mutant behavioral phenotype, suggesting that these compounds serve as modifiers of neural circuits disrupted in mutants. Indeed, estrogens can signal to multiple downstream pathways, including transcriptional activation of estrogen response genes, regulation of other transcription factors, and rapid activation of intracellular signaling pathways (Marino et al., 2006). The extent to which estrogens alter one or more downstream intracellular signaling pathways remains to be determined, but our findings suggest that the rescue activity likely involves an acute mechanism other than robust activation of estrogen target genes. Moreover, estrogens have been shown to affect glutamatergic signaling. Estradiol increases dendritic spine density and long-term potentiation via an NR2B-dependent mechanism that can be blocked by exposure to NMDA antagonists, such as MK-801 (Smith et al., 2009). Therefore, estrogens may be acting upstream of the identified NMDA and GABA deficits. While there is growing interest in the effects of sex hormones on brain development and ASD risk, given the 4:1 male:female ratio of classical autism (Baron-Cohen et al., 2011; Schaafsma and Pfaff, 2014), further investigations are required to determine whether such a mechanism might contribute to the observed female protective effect suggested by recent human genetics studies of ASD (Jacquemont et al., 2014).
As the list of reliable ASD risk genes continues to expand, there is a growing need for biologically relevant systems to advance from gene discovery to pharmacological screens. We demonstrate that quantitative behavioral profiling of zebrafish cntnap2ab mutants can be used as a novel platform for the prediction and in vivo screening of compounds to identify suppressors of a behavioral phenotype resulting from loss of an ASD risk gene. Future testing in a mammalian system is a critical next step. Moreover, the study of zebrafish mutants of the ASD risk gene, CNTNAP2, and its differential responses to psychoactive agents reveals the strength of this approach to identify molecular mechanisms and potential pharmacological candidates for further evaluation.
Experimental Procedures
Zebrafish
Mutations in cntnap2a and cntnap2b were generated using zinc finger nucleases. The cntnap2aΔ121/Δ121cntnap2b31i/31i and cntnap2aΔ25/Δ25cntnap2bΔ7/Δ7 lines were generated by incrossing double homozygotes, which are viable and fertile. Tg(dlx6a-1.4kbdlx5a/dlx6a:GFP) and Tg(vglut:DsRed) were obtained from the laboratories of M. Ekker and J. Fetcho, respectively. cntnap2aΔ121/Δ121cntnap2b31i/31i mutants were crossed to these transgenic lines. Animal experiments were conducted in accordance with IACUC regulatory standards (Yale University) and the UK Animals (Scientific Procedures) Act 1986.
Pharmacological Screen
Larval activity was monitored from 4 to 7 dpf using a custom-modified Zebrabox and automated video tracking system (Viewpoint, LifeSciences) (Rihel et al., 2010). Correlation analysis was done in Matlab (R2014a; The Mathworks, Inc.) to identify compounds that correlate or anti-correlate with the cntnap2aΔ25/Δ25cntnap2bΔ7/Δ7 phenotype. Three doses of each compound (Figure 3A; Table S4) were tested in 10–12 cntnap2aΔ121/Δ121cntnap2b31i/31i or wild-type replicate larvae with 10–12 control larvae per 96-well plate (Rihel et al., 2010). Hierarchical clustering was conducted in Matlab with the statistics and bioinformatics toolboxes.
Supplementary Material
Acknowledgments
We thank J. Fetcho and M. Ekker for transgenic lines; B. Bill, D. Geschwind, P. Panula, and G. Wright for in situ probes; D. Geschwind, P. De Camilli, J. Leckman, M. Lee, V. Yartseva and N. Sestan for discussions and manuscript editing; K. Divito, J.M.F., and S. Lau, blinded raters. Supported by NIH grants K08MH096176, CTSA UL1TR000142, T32MH018268, AACAP junior investigator award (E.J.H.), R21 HD073768, R01 HD074078, R01 GM081602 (A.J.G.), RC2MH089956 (A.J.G., M.W.S.), Overlook International Foundation (M.W.S.), K99HD071968 (D.C.), MH092257 and MH109498 (M. Granato), ERC Grant and UCL Excellence Fellowship (J.R.), and BBSRC, EU and Wellcome Trust grants (S.W.W.).
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and four tables.
Author Contributions
E.J.H., M.W.S., and A.J.G. designed the project. E.J.H. generated and characterized cntnap2ab mutants. E.J.H. and K.J.T. performed immunohistochemistry. J.M.F. assisted in genotyping, zebrafish husbandry, and in situ hybridizations. D.C. adapted methods of mutagenesis and screening. R.A.J. and M. Granato analyzed startle and habituation. F.K. and H.B. analyzed the optokinetic response. B.R.B. provided in situ probes and discussions. E.J.H. and M.J.F.B. analyzed forebrain commissures. E.J.H., S.I., K.J.T. and S.W.W. analyzed CNS structure and GABAergic deficits. E.J.H., M. Ghosh, and J.R. performed behavioral tracking and pharmacological screens. J.R. performed correlation analyses and PCA. E.J.H. wrote the manuscript with input from S.W.W., J.R., M.W.S., and A.J.G.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ablain J, Zon LI. Of fish and men: using zebrafish to fight human diseases. Trends in Cell Biol. 2013;23:584–586. doi: 10.1016/j.tcb.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 5. Arlington, VA: American Psychiatric Publishing; 2013. [Google Scholar]
- Baraban SC, Taylor MR, Castro PA, Baier H. Pentylenetetrazole induced changes in zebrafish behavior, neural activity and c-fos expression. Neuroscience. 2005;131:759–768. doi: 10.1016/j.neuroscience.2004.11.031. [DOI] [PubMed] [Google Scholar]
- Baron-Cohen S, Lombardo MV, Auyeung B, Ashwin E, Chakrabarti B, Knickmeyer R. Why are autism spectrum conditions more prevalent in males? PLoS Biol. 2011;9:e1001081. doi: 10.1371/journal.pbio.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I, Calcagnotto ME, Vilaythong AJ, Thwin MT, Noebels JL, Baraban SC, Rubenstein JL. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci. 2005;8:1059–1068. doi: 10.1038/nn1499. [DOI] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem N, Jarinova O, Amores A, Long Q, Hatch G, Park BK, Rubenstein JL, Ekker M. Regulatory roles of conserved intergenic domains in vertebrate Dlx bigene clusters. Genome Res. 2003;13:533–543. doi: 10.1101/gr.716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont S, Coe BP, Hersch M, Duyzend MH, Krumm N, Bergmann S, Beckmann JS, Rosenfeld JA, Eichler EE. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. Am J Hum Genet. 2014;94:415–425. doi: 10.1016/j.ajhg.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilhoff G, Becker A, Grecksch G, Wolf G, Bernstein HG. Repeated application of ketamine to rats induces changes in the hippocampal expression of parvalbumin, neuronal nitric oxide synthase and cFOS similar to those found in human schizophrenia. Neuroscience. 2004;126:591–598. doi: 10.1016/j.neuroscience.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Kinkhabwala A, Riley M, Koyama M, Monen J, Satou C, Kimura Y, Higashijima S, Fetcho J. A structural and functional ground plan for neurons in the hindbrain of zebrafish. Proc Natl Acad Sci. 2011;108:1164–1169. doi: 10.1073/pnas.1012185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokel D, Peterson RT. Using the zebrafish photomotor response for psychotropic drug screening. Methods in Cell Biol. 2011;105:517–524. doi: 10.1016/B978-0-12-381320-6.00022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino M, Galluzzo P, Ascenzi P. Estrogen signaling multiple pathways to impact gene transcription. Curr Genomics. 2006;7:497–508. doi: 10.2174/138920206779315737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken JT, McGough J, Shah B, Cronin P, Hong D, Aman MG, Arnold LE, Lindsay R, Nash P, Hollway J, et al. Risperidone in children with autism and serious behavioral problems. N Engl J Med. 2002;347:314–321. doi: 10.1056/NEJMoa013171. [DOI] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, Sonnenblick LI, Gruver R, Almajano J, Bragin A, et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliak S, Salomon D, Elhanany H, Sabanay H, Kiernan B, Pevny L, Stewart CL, Xu X, Chiu SY, Shrager P, et al. Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J Cell Biol. 2003;162:1149–1160. doi: 10.1083/jcb.200305018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prober DA, Rihel J, Onah AA, Sung RJ, Schier AF. Hypocretin/orexin overexpression induces an insomnia-like phenotype in zebrafish. J Neurosci. 2006;26:13400–13410. doi: 10.1523/JNEUROSCI.4332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihel J, Prober DA, Arvanites A, Lam K, Zimmerman S, Jang S, Haggarty SJ, Kokel D, Rubin LL, Peterson RT, et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science. 2010;327:348–351. doi: 10.1126/science.1183090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes, Brain, and Behav. 2003;2:255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, Murtha MT, Bal VH, Bishop SL, Dong S, et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders JA, Tatard-Leitman VM, Suh J, Billingslea EN, Roberts TP, Siegel SJ. Knockout of NMDA receptors in parvalbumin interneurons recreates autism-like phenotypes. Autism Research. 2013;6:69–77. doi: 10.1002/aur.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaafsma SM, Pfaff DW. Etiologies underlying sex differences in Autism Spectrum Disorders. Front Neuroendocrinol. 2014;35:255–271. doi: 10.1016/j.yfrne.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Smith CC, Vedder LC, McMahon LL. Estradiol and the relationship between dendritic spines, NR2B containing NMDA receptors, and the magnitude of long-term potentiation at hippocampal CA3-CA1 synapses. Psychoneuroendocrinology. 2009;34(Suppl 1):S130–142. doi: 10.1016/j.psyneuen.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, Stephan DA, Morton DH. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Takechi K, Fujiwara A, Kamei C. Effects of antiepileptics on behavioral and electroencephalographic seizure induced by pentetrazol in mice. J Pharmacol Sci. 2010;112:282–289. doi: 10.1254/jphs.09225fp. [DOI] [PubMed] [Google Scholar]
- Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, Reilly SK, Lin L, Fertuzinhos S, Miller JA, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerucha T, Stuhmer T, Hatch G, Park BK, Long Q, Yu G, Gambarotta A, Schultz JR, Rubenstein JL, Ekker M. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci. 2000;20:709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.