Abstract
The pathological consequences of malaria infection are the result of parasite replication within red blood cells (RBCs). Invasion into RBCs is mediated by a large repertoire of parasite proteins that are distributed on the parasite surface and within specialised apical secretory organelles. As invasion is an essential step in the parasite life-cycle, targeting invasion-related molecules provides an avenue for therapeutic intervention. We have used genome and transcriptome data available for Plasmodium falciparum to identify proteins likely to be involved in RBC invasion. Of these candidates, we selected a protein which we have dubbed PfRON6 for detailed characterisation. PfRON6 contains a novel cysteine-rich domain that is conserved in other Apicomplexan parasites. We show that PfRON6 is localised in the rhoptry neck of merozoites and is transferred to the newly formed parasitophorous vacuole during invasion. Transfection experiments indicate that the gene which encodes PfRON6 is refractory to integration that disrupts the coding sequence, suggesting its absence is incompatible with the parasite life-cycle. Further, the cysteine-rich domain appears to be functionally important as it cannot be truncated. Taken together, these data identify PfRON6 as a novel and potentially important component of the Plasmodium invasion machinery.
Keywords: Malaria, Plasmodium falciparum, Invasion, Rhoptry neck protein
1. Introduction
Apicomplexan parasites are obligate intracellular organisms that invade target cells by an unusual mechanism referred to as gliding motility. Ligands on the parasite surface and within specialised secretory organelles are linked to an internal actin-myosin motor which provides the motile force. In the case of Plasmodium merozoites, more than 40 proteins have been identified that are believed to play a role in the invasion process (Cowman and Crabb, 2006). Antibodies against many of these proteins are able to block invasion and consequently proteins such as merozoite surface proteins (MSP) -1, -2, -4 and -5, apical membrane antigen 1 (AMA1) and rhoptry associated protein 2 (RAP2) are leading vaccine candidates (Ballou et al., 2004; Malkin et al., 2006; Matuschewski, 2006).
Proteins that are known to localise at the merozoite surface or within apical secretory organelles (rhoptries, micronemes and dense granules) share a number of features. Since trafficking of these proteins occurs via a classic eukaryotic secretory pathway, they typically possess an N-terminal hydrophobic signal sequence required for co-translational insertion into the endoplasmic reticulum (ER). They also lack additional signals (eg. apicoplast transit peptide or Plasmodium export element (PEXEL)) that would result in localisation within other membrane-bound organelles or traffic beyond the parasitophorous vacuole (PV) (Foth et al., 2003; Hiller et al., 2004; Marti et al., 2004; Tonkin et al., 2006). Consistent with the Plasmodium ‘just in time’ model of gene expression, most surface and apical secretory proteins are synthesised late during the asexual red blood cell (RBC) cycle in preparation for the next round of invasion (Ben Mamoun et al., 2001; Bozdech et al., 2003a, 2003b; Le Roch et al., 2003). A number of these proteins also contain cysteine-rich domains which are thought to play important roles in receptor-ligand interactions. While such domains share minimal similarity at the primary sequence level, they are thought to adopt related globular structures held together by a network of disulfide bonds (Tolia et al., 2005; Singh et al., 2006). The Cys6 family of proteins contains at least nine members, the best characterised of which is the gametocyte adhesin Pfs48/45 (van Dijk et al., 2001; Sanders et al., 2005). Other examples of such domains include those found in the reticulocyte-binding proteins of Plasmodium vivax and the related reticulocyte binding-like (RBL) proteins in Plasmodium falciparum, (Galinski et al., 1992; Rayner et al., 2000), the Duffy binding-like (DBL) domains of erythrocyte binding proteins (Mayor et al., 2005; Tolia et al., 2005; Singh et al., 2006), and the epidermal growth factor (EGF)-like domains of MSPs such as MSP1, MSP4, MSP5 and MSP10 (Blackman et al., 1991; Marshall et al., 1998; Black et al., 2001).
To identify novel proteins that are likely to be involved P. falciparum invasion we undertook a bioinformatic analysis using the resources available on the PlasmoDB (http://plasmodb.org) and ApiDB (http://apidb.org/apidb/) websites. Here we describe the detailed chracterisation of an unusual rhoptry neck protein which we have termed PfRON6. This protein is partially conserved across the Apicomplexa but is unrelated to any known proteins involved in invasion. The gene that encodes PfRON6 is refractory to genetic deletion suggesting that it has an indispensable role in the P. falciparum life-cycle.
2. Materials and methods
2.1. Plasmodium falciparum parasites
Plasmodium falciparum 3D7 parasites were cultured in vitro as previously described (Trager and Jensen, 1976; Cranmer et al., 1997). Human blood was obtained from the Australian Red Cross. Ethics approval for the use of human blood was obtained from the Monash University Standing Committee on Ethics in Research involving Humans (SCERH). Asynchronous parasite extracts were prepared as described (Black et al., 2001). Highly synchronised cultures (Lambros and Vanderberg, 1979) were sampled at various time points for stage-specific expression analysis.
2.2. Genomic sequences, cloning and analysis
Raw and annotated sequence data, transcriptomic and proteomic data was obtained from PlasmoDB (www.plasmodb.org and references therein) and ApiDB (www.apidb.org and references therein). Homology searches were carried out using the Basic Local Alignment Search Tool (BLAST) (Altschul et al., 1990, 1997; McGinnis and Madden, 2004; Ye et al., 2006). Potential motifs were identified using the Prosite, Pfam and Conserved Domain databases (www.expasy.org/tools and www.ncbi.nlm.nih.gov/BLAST/). Putative signal sequences were assigned based on results of the SignalP algorithm (Bendtsen et al., 2004). Apicoplast transit peptides and PEXEL motifs were identified using the tools on PlasmoDB (www.plasmodb.org and references therein).
Three non-overlapping fragments of PfRON6 and one fragment (encompassing amino acid residues 42 – 200) of the the rhoptry neck-located antigen PfRON4 (Alexander et al., 2006) were amplified by PCR from P. falciparum 3D7 cDNA. Fragments rPfRON6-A, -B and -C and PfRON4-1 were amplified using primers cattggatccGAACACGCCAATTTAATAA and cgcgaattcTTAATTTTCCTCTTCTTCCAA; catggatccAATGAAATAATTGAAAAGGAA and cgggaattcTTAATTATCATCATGTATATC; acgggattcTGTCCTATGGAATGTAATAAG and tcccaagcttTCCCGTTTTTTGTTTTTCATG; ccggaattctaAGCCATATAGAAGAACCTCAA and cccaagcttATGTGAATGATGATTTATATTATTAT, respectively (restriction sites underlined). Fragments PfRON-A and -B were ligated into the BamHI/EcoRI sites of expression vector pGEX-4T-1, (Guan and Dixon, 1991), fragment PfRON6-C was ligated into the BamHI/HindIII sites of pET24d (Novagen), and fragment PfRON4-1 was ligated into the EcoRI/HindIII sites of pGEX-KG (Guan and Dixon, 1991). Cloning and sequence analysis was performed as previously described (Black et al., 2001).
2.3. Plasmodium falciparum transfection and analysis
To disrupt PFB0680w, two ~1 kb sequences (F1 and F2) from the 5′ end of the gene were cloned into the SpeI/BglII (F1) and EcoRI/NcoI (F2) sites of the transfection plasmid pHHT-TK (Duraisingh et al., 2002) to generate pHTKΔRON6. PFB0680w F1 and F2 were amplified from P. falciparum 3D7 gDNA using primers ggactagtCCCTGGTATTTCTAGCTGTTTTAGC and gaagatctTCTCTGTTTCCTTTTTATCATCATC; and cggaattcCGAAGAAAAAGAATGGTAAAAATAAAG and gatgccatggcATGATTGTGAAGTATTGTATCCATGC, respectively (restriction sites underlined). To truncate PFB0680w and allow for expression of a shortened form of PfRON6, ~1 kb region from the centre of the gene was amplified using primers ggactagtGAGATTGAAAATGTAACAAATGC and ccgctcgagTTAGCTTCTTTTCATGATCTTTTCTTCC and cloned into SpeI/XhoI digested pHHT-TK to generate pHRON6Δt. To incorporate a C-terminal 3× heamagglutinin (HA) epitope tag either 1 kb or 500 bp of the 3′ end of the gene was amplified using the forward primer ggaagatctTTAGACCCATGACAATTG (BglII site) for the 500 bp fragment or ggaagatctCATCACTGTGCCATAGCAG (BglII site) for the 1 kb fragment and the reverse primer ggcctaggTCCCGTTTTTTGTTTTTCATG and cloned into BglII/AvrII digested pARL to generate pARON6-HA-500 and pARON6-HA-1000. Plasmodium falciparum 3D7 ring stage parasites were transfected as previously described (Wu et al., 1995; Fidock and Wellems, 1997) with either pHTKΔRON6 or pHRON6Δt. Parasites were initially cultured in the presence of 2.5 nM WR99210 (Jacobus Pharmaceuticals) for ~6 weeks, and then underwent five rounds of drug cycling. Parasites transfected with pHTKΔRON6 were then negatively selected with 4 μM Gancyclovir (Roche) for approximately 4 weeks to eliminate the episome. DNA from all parasite lines and the 3D7 parent line were purified using the Nucleon BACC2 kit (GE Healthcare) as per the manufacturer’s instructions. Parasite DNA and transfection plasmids were digested with XcmI and HincII (knockout), EcoRI (truncation) or SpeI and SacI (HA tag). The digested DNA was analysed by Southern blotting using standard protocols (Waller et al., 2003).
2.4. Recombinant protein expression and production of polyclonal antisera
Recombinant PfRON6-A (rPfRON6-A) and -B, and PfRON4-1 proteins were expressed in Escherichia coli BL21 DE3 and purified as GST fusions (Black et al., 2001). rPfRON6-C was expressed in E. coli BL21 DE3 and purified as a C-terminal His6 fusion (Wang et al., 1999). rPfRON6-C was subsequently solubilised and refolded using the Protein Refolding Kit (Novagen) as per the manufacturer’s instructions. Polyclonal antisera were raised in 8–10 week old female New Zealand White rabbits (Monash Animal Services, Clayton Vic, Australia). All animal experiments were performed in accordance with guidelines set by the Australian and New Zealand Council on Animal Care in Research and Teaching (ANZCART). Ethics approval was obtained from Monash University SOBSB Animal Ethics Committee. Immunisations were conducted by s.c. injection of 100 μg of purified rPfRON6 emulsified in FCA (Difco Laboratories), with monthly boosting of 100 μg protein emulsified in Incomplete Freund’s adjuvant (Difco Laboratories). The anti-PfRON6 sera was subsequently affinity purified using rPfRON6 linked to CNBr-activated Sepharose (Amersham Pharmacia).
2.5. SDS-PAGE and immunoblotting
rPfRON6 proteins and parasite lysate were resolved by SDS-PAGE using 12% (w/v) polyacrylamide gels and stained with coomassie blue or transferred to polyvinylidene fluoride (PVDF) membranes (NEN) for Western blot analysis as previously described (Black et al., 2001). All samples were resolved under denaturing conditions. Affinity purified rabbit anti-PfRON6 (1:500 dilution for fragments A and B and 1:100 dilution for fragment C) and polyclonal rabbit anti-RAMA (1:1,000 dilution) and anti-MSP4 (diluted 1:500) were used as the primary antibodies. Pooled human hyperimmune sera from malaria endemic regions of Vietnam and Papua New Guinea (PNG) and pooled sera from naïve individuals from Melbourne, Australia were also used to assess the reactivity of PfRON6. Anti-rabbit and anti-human immunoglobulin conjugated to horseradish peroxidise (Silenus) were used as the secondary antibodies.
2.6. Immunofluorescence assay (IFA)
Plasmodium falciparum parasites were cultured to 5% parasitemia and blood smears were used in IFA as previously described (Black et al., 2001). Affinity purified rabbit anti-PfRON6 (1:500 dilution), polyclonal mouse anti-RAMA, anti-AMA-1 (1:500 dilution), anti-Pf34 (1:250 dilution) and anti-HA (1:250 dilution; Invitrogen) were used as primary antibodies. Alexa Fluor 488-conjugated anti-rabbit immunoglobulin and Alexa Fluor 568-conjugated anti-mouse immunoglobulin (1:3,000 dilution; Molecular Probes Inc.) were used as the secondary antibodies. The smears were examined by wide-field fluorescence microscopy and selected samples were then examined using the Olympus SV1000 Confocal Laser Scanning Microscope. Co-localisation analysis was performed using WCIF ImageJ software (http://www.uhnresearch.ca/facilities/wcif/imagej/).
2.7. Immunoelectron microscopy
Parasitised RBCs were fixed in 0.1 M cacodylate buffer pH 7.4 containing 2% paraformaldehyde and 0.0075% glutaraldehyde for 20 min on ice. The cells were rinsed three times with PBS and partially serially dehydrated with 50%, 70% and 80% ethanol then embedded and polymerized in LR white resin medium grade (London Resin Company, UK) in accordance with the manufacturer’s instructions. Thin sections (80 nm) were cut and collected on formvard coated nickel grids. The sections were rehydrated in PBS for 5 min then blocked with PBS containing 5% nonfat dry milk and 0.01% Tween20 for 30 min. After rinsing in PBS containing 1% BSA and 0.01% Tween20 (TBPBS), the grids were incubated with affinity purified anti-rPfRON6-C (1:30 dilution) for 2 h, then rinsed five times with TBPBS and then incubated with anti-rabbit immunoglobulin antibodies conjugated to 15 nm gold beads (Aurion, NL) for 1 h. The grids were then washed three times with TBPBS and twice with PBS, fixed with 1% glutaraldehyde in water for 5 min and stained with uranyl acetate.
3. Results
3.1. Gene structure, transcription and identification of orthologues
We searched the P. falciparum transcriptome (http://plasmodb.org) for genes that are co-ordinately transcribed with known invasion related genes (maximum transcription at 42 ± 6 h and minimum transcription 12 ± 10 h, > 4-fold induction) (Supplementary Table S1). One of the genes identified was PFB0680w (Genbank accession XP_003149654) which is located on chromosome 2. PFB0680w is comprised of 15 exons, with the second exon containing most of the coding sequence. The PlasmoDB gene model predicts a protein with an apicoplast transit peptide. However, using manual annotation and reverse transcriptase-PCR analysis, Huestis and Fischer (2001) proposed an alternative gene structure which differs in the short first exon. Based on the experimentally confirmed model, PFB0680w encodes a protein of 950 amino acids which we termed PfRON6 based on protein localisation studies presented below. PfRON6 has a predicted N-terminal signal sequence, no apicoplast transit peptide or PEXEL motif, three blocks of degenerate repeats, and a C-terminal cysteine-rich region (Fig. 1A). The cysteine-rich region does not conform to any other family of cysteine-rich domains previously characterised in Plasmodium, nor is it similar to any domain in any other hypothetical protein in P. falciparum. Pattern and profile searches in the Genbank, Pfam and Prosite databases failed to identify any homologous sequences or motifs of known function.
Fig. 1.
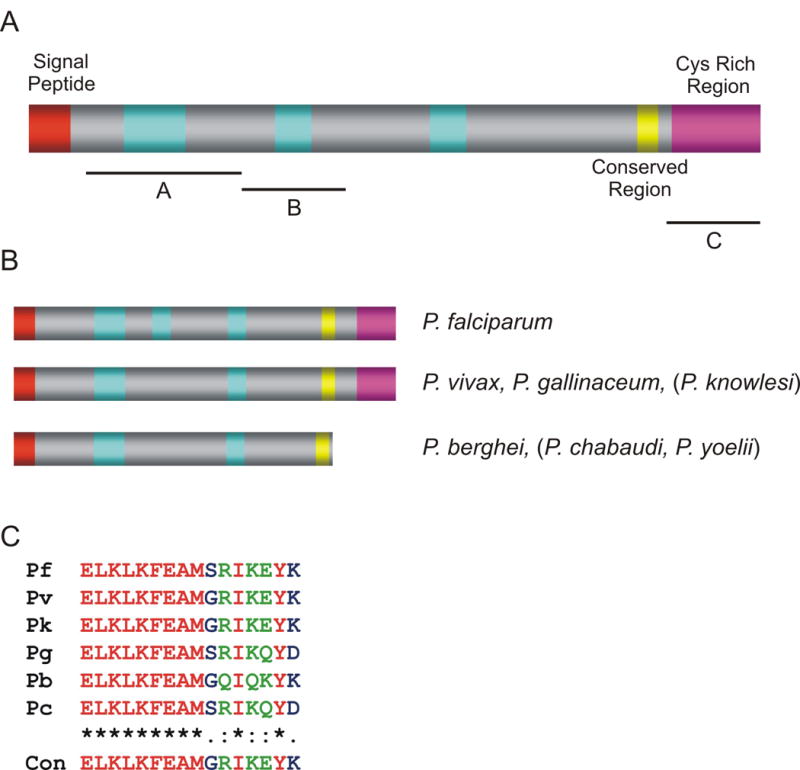
Predicted protein structure of PfRON6 and its orthologues. (A) Schematic representation of PfRON6. The signal peptide, repeat regions, the conserved region and the cysteine-rich region are indicated. The repeat regions consist of: R1 degenerate repeats of the sequence SDDHK[V/I]EE[N/V]KK, R2 tandem repeats of the sequence KDXXKEKX and R3 degenerate repeats containing the dipeptide EN. The numbers refer to the amino acid residues. Black bars indicate fragments corresponding to residues 21–308 (A), 308–441 (B) and 752–950 (C) that were produced as recombinant fusion proteins. (B) Schematic representation of PfRON6 orthologues from other Plasmodium species. Species where only partial sequence is available are indicated in brackets. (C) Amino acid alignments of the conserved regions of PfRON6 from different Plasmodium species. Pk – Plasmodium knowlesi; Pv – Plasmodium vivax; Pf – Plasmodium falciparum; Pb – Plasmodium. berghei; Pc – Plasmodium chabaudi; Pg – Plasmodium gallinaceum
Using a combination of BlastP and TBlastN searches, orthologous genes were identified in the genomes of P. vivax, Plasmodium berghei and Plasmodium gallinaceum. Partial sequences were also present in the unfinished genomes of Plasmodium yoelii, Plasmodium chabaudi and Plasmodium knowlesi (Fig. 1B). Where sufficient sequence data was available (P. vivax, P. knowlesi and P. berghei), synteny was established by comparing the surrounding genes. Alignment of protein sequences from different species suggests that PfRON6 is composed of three domains – an N-terminal repeat region, a sub-C-terminal conserved domain (Fig. 1C) and a C-terminal cysteine-rich domain. Interestingly P. berghei and P. chabaudi contain a truncated version of PfRON6 which is missing the C-terminal cysteine-rich region. In both cases, > 1 kb of sequence downstream of the putative stop codon is available, and for P. berghei this contains a gene that is orthologous to PFB0685c, the neighbouring gene to PFB0680w. Manual examination of this intergenic region confirmed that there is no potential exon that could encode the cysteine-rich region.
To determine whether individual domains were present in other Apicomplexans, regions of PfRON6 were used in BlastP searches against the genomes of Toxoplasma gondii, Cryptosporidium parvum, Cryptosporidium hominis, Theileria annulata, Theileria parva and Babesia bovis (www.apidb.org). Hypothetical proteins that contained a cysteine-rich region homologous to the cysteine-rich region of PfRON6 were identified in each of the genomes searched. Although the proteins showed low overall similarity (< 20%), each protein had a predicted signal sequence and the relative positions of all 10 cysteine residues in PfRON6 were conserved in all non-Plasmodium spp. PvRON6 and PgRON6 each had nine out of the 10 cysteines conserved (Supplementary Fig. S1). This is analogous to the dual EGF-like domains of MSP119, where 10 out of 12 cysteines in P. falciparum are conserved in P. knowlesi and Plasmodium cynomolgi (Garman et al., 2003).
3.2. PFB0680w is refractory to genetic deletion
To investigate the function of PfRON6 we attempted to disrupt PFB0680w by double cross-over homologous recombination using the pHHT-TK vector (Fig. 2A) (Duraisingh et al., 2002). We generated three independent tranfectant parasite lines and subjected them to five rounds of drug cycling and negative selection using ganciclovir. Southern blotting analysis demonstrated that no integration had occurred and that the plasmid was maintained as an episome (Fig. 2A). This result was confirmed by PCR (data not shown). Next we attempted to truncate PFB0680w to remove the cysteine-rich domain. The construct was designed so that integration of the plasmid by single cross-over homologous recombination would disrupt the coding region of PfRON6 at amino acid 738 (Fig. 2B). As with the knockout constructs, integration failed to occur and the plasmid was maintained episomally (Fig. 2B and PCR data not shown).
Fig. 2.
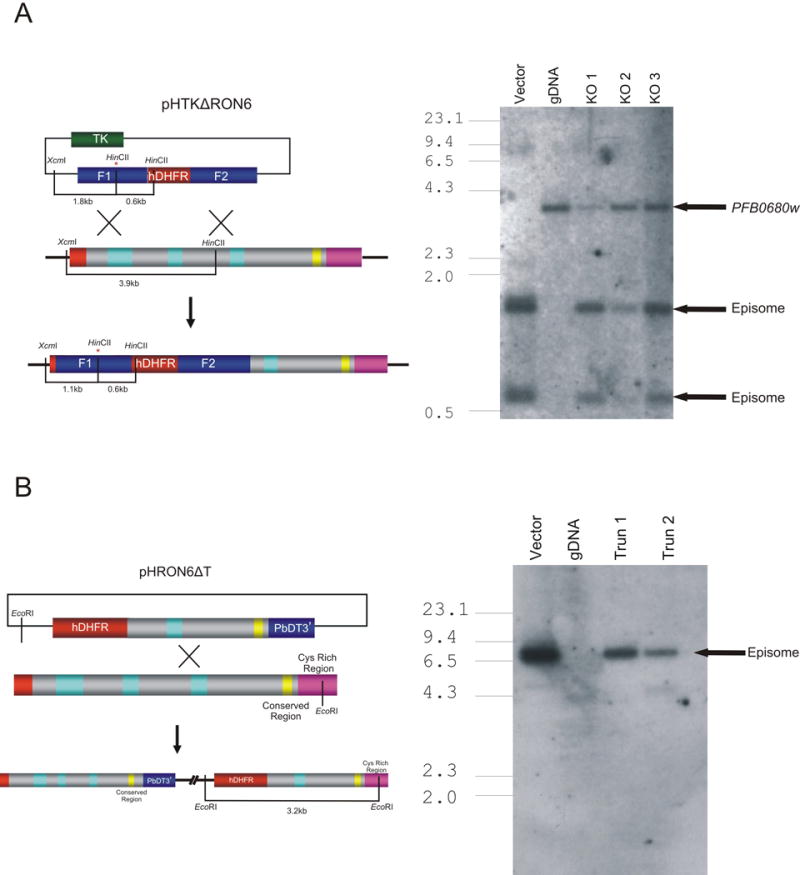
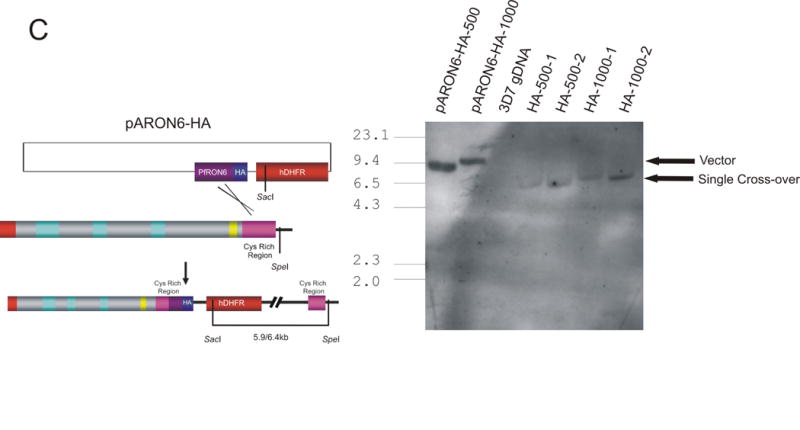
Genetic targeting of PFB0680w. (A) Schematic of integration vectors and Southern blot analysis of transgenic parasite lines transfected with pHTKΔRON6. Vector control, 3D7 genomic DNA (gDNA), and gDNA from three independent transfectant parasite lines (KO 1, KO 2 and KO 3) were digested with HincII and XcmI and probed with an F1-specific probe. The bars on the schematic show the location of the restriction sites with the sizes of the expected bands shown below. The asterix (*) indicates the introduction of a HincII site into pHTKΔRON6 during PCR amplification. Bands corresponding to undisrupted genomic copy of PFB0680w and plasmid maintained as an episome are indicated. (B) Southern blot analysis of transgenic parasite lines transfected with pHRON6Δt. Vector control, 3D7 gDNA and gDNA from two independent transfectant parasite lines (Trun 1 and Trun 2) were probed with a human dihydrofolate reductase (hDHFR) specific probe. Bands corresponding to plasmid maintained as an episome are indicated. (C) Southern blot analysis of transgenic parasite lines transfected with pARON6-HA-500 or pARON6-HA-1000. Vector controls, 3D7 gDNA and gDNA from two independent transfectant parasites lines from both vectors were probed with a hDHFR-specific probe. Bands corresponding to plasmid and single cross-over are indicated.
Failure to detect integration into PFB0680w indicates that either there is a strong selective pressure against parasites with significant disruptions to PFB0680w, or that the locus cannot be targeted by homologous recombination. To investigate these possibilities, we attempted to introduce a HA tag at the 3′ end of the gene (Fig. 2C). Following transfection and drug selection, Southern blot analysis (Fig. 2C) and PCR (results not shown) demonstrated that integration of this construct had occurred as expected. These results prove that PFB0680w is accessible to homologous recombination and strongly suggest that the gene is refractory to genetic manipulations that interfere with the production of a functional gene product.
3.3. PfRON6 is expressed in mature blood stages
To characterise expression of PfRON6 in blood stage parasites we produced three non-overlapping regions as recombinant proteins (termed rPfRON6-A, -B and -C) (Fig. 1A). Antisera were raised against the recombinant proteins and these were used in immunoblot experiments to identify one or more proteins in parasite lysate prepared from asynchronous in vitro culture (Fig. 3A). All three antisera consistently reacted with a parasite protein of 150 kDa. An additional parasite protein of 35 kDa was also recognised by anti-rPfRON6-A and -B, although reactivity to this was inconsistent. The predicted molecular mass of PfRON6 is 112 kDa. However, anomalously slow migration on SDS-PAGE is a common feature of many malarial proteins, presumably due to their low affinity for SDS (Anders et al., 1988). Therefore, it is likely that the 150 kDa band represents full-length PfRON6. Whether or not the 35 kDa protein is the result of physiologically relevant proteolytic processing or a cross-reactive distinct gene product requires further investigation.
Fig. 3.
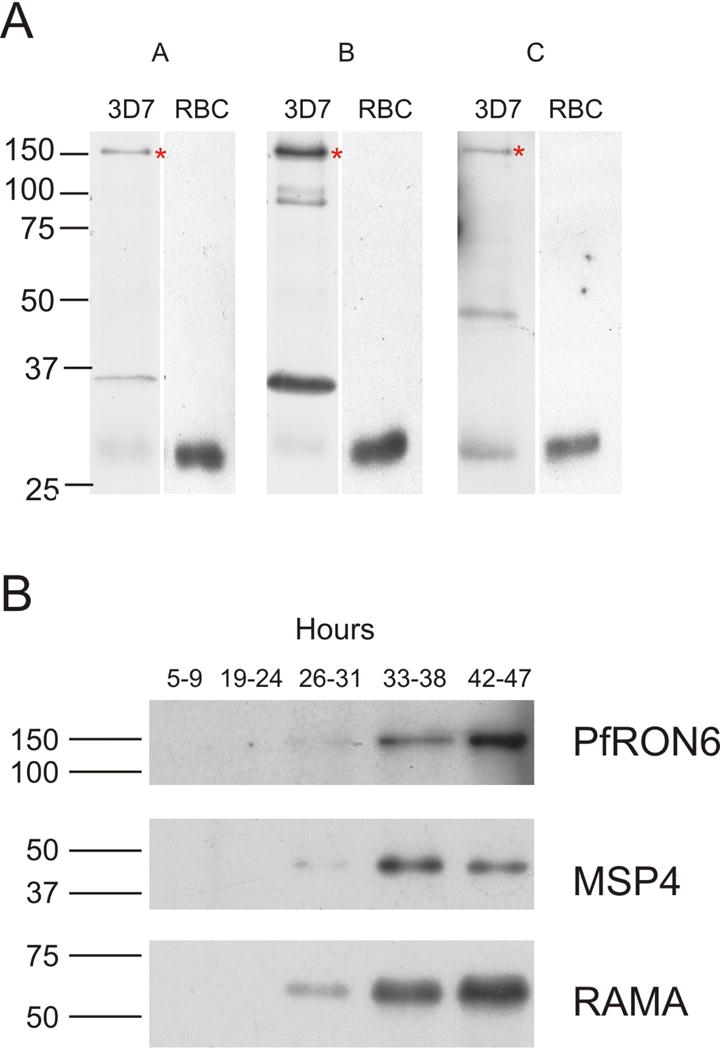
Expression of the PfRON6 protein in parasitised red blood cells (RBCs). (A) Reactivity of anti-PfRON6 antisera with parasite lysate. Immunoblots with anti-rPfRON6-A, -B and -C antisera on parasite extracts or uninfected RBCs. The asterix (*) indicates full-length PfRON6. (B) Time course of PfRON6 expression. Immunoblots with anti-rPfRON6-A, anti-RAMA, and anti-MSP4 antisera on parasite extracts from synchronised parasite samples at various time points post-invasion. Rings (5–9 h), early trophozoites (19–24 h), late trophozoites (26–31 h), early schizonts (33–38 h), late schizonts (42–47 h).
To study timing of PfRON6 expression, anti-rPfRON6-A was used to probe synchronised parasite samples (Fig. 3B and Supplementary Fig. S2). Full-length PfRON6 was detected in early and late schizont samples. Immunoblotting of the same samples with anti-RAMA and anti-MSP4 antisera demonstrates that PfRON6 appears later than RAMA, at approximately the same time as MSP4. These results are in agreement with microarray data which indicate that both MSP4 and PFB0680w have maximum transcription at 36–44 h post-invasion (Bozdech et al., 2003a; Le Roch et al., 2003).
3.4. PfRON6 is localised in the rhoptry neck
To localise PfRON6 within parasitised RBCs, anti-PfRON6 antisera were used in immunofluorescence experiments (Fig. 4). All three antisera produced a punctate pattern of fluorescence in segmented schizonts characteristic of localisation with the apical secretory organelles (Fig. 4A). The same pattern was also observed using an anti-HA antibody with the 3′ HA tagged transfectant parasite lines (Fig. 4B and Supplementary Fig. S3). To determine whether PfRON6 was localised within rhoptries or micronemes, double labelling experiments with anti-RAMA (a rhoptry marker), anti-Pf34 and anti-PfRON4 (rhoptry neck markers), and anti-AMA1 (a microneme marker) antibodies were performed (Fig. 4C) (Bannister et al., 2003; Topolska et al., 2004; Alexander et al., 2006; Proellocks et al., 2007). We analysed 10 images for each antibody pair and quantified co-localisation using Pearson’s coefficient (Supplementary Fig. S4) (Manders et al., 1992). Co-localisation was highest between PfRON6 and the two rhoptry neck markers PfRON4 and Pf34 (0.833 ± 0.054 and 0.794 ± 0.054, respectively). There was less overlap between PfRON6 and RAMA (0.719 ± 0.033) and less still between PfRON6 and AMA1 (0.627 ± 0.079). There was no statistically significant difference between co-localisation of PfRON6 with either of the rhoptry neck markers, but there was a significant difference between co-localisation with the rhoptry neck markers and either RAMA or AMA1. High resolution imaging of extracellular merozoites resolved two bulbous RAMA positive structures (corresponding to individual rhoptries) with partial overlap of PfRON6 at the anterior end (Fig. 4D). Similiarly, in electronmicrographs, PfRON6-specific immunogold labelling was predominantly confined to the apical end of pear-shaped organelles (Fig. 4E). These observations strongly suggest that PfRON6 is present within the rhoptry neck of merozoites.
Fig. 4.
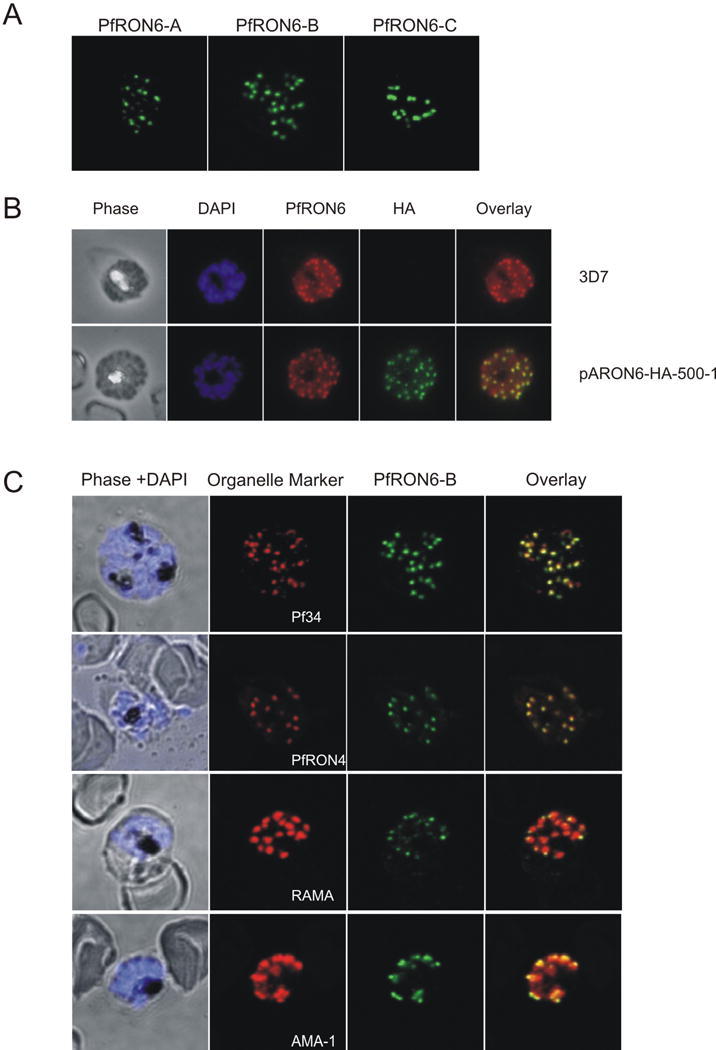
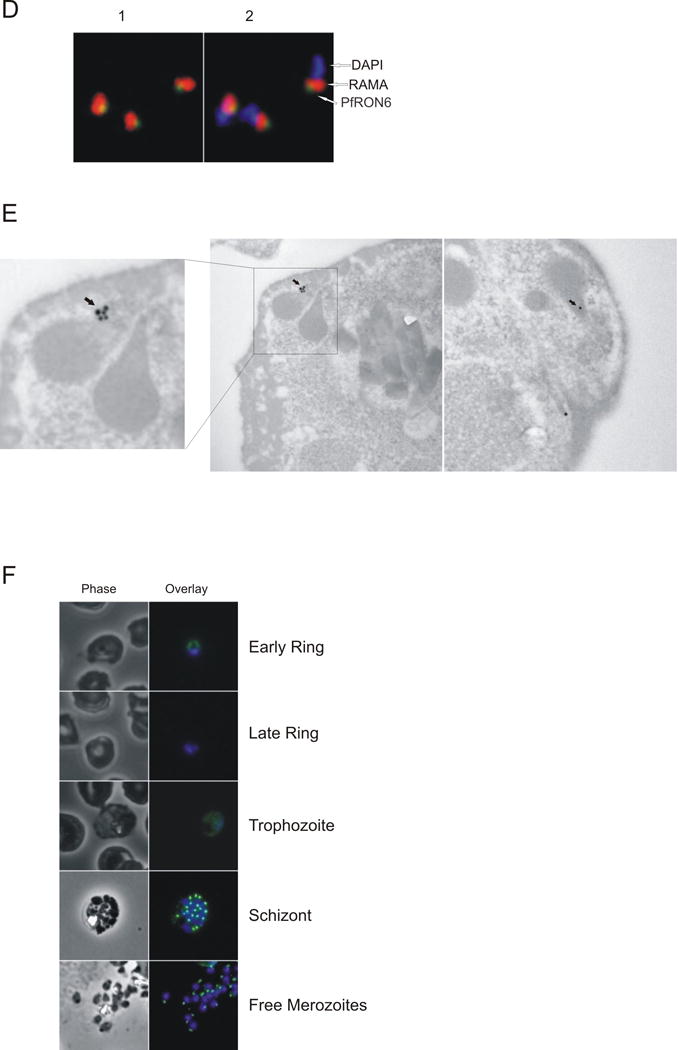
Localisation of the PfRON6 protein in parasitised red blood cells (RBCs). (A) Localisation of PfRON6 in segmented schizonts. Confocal microscopy using anti-rPfRON6-A, -B and -C antisera. (B) Double labelling using anti-rPfRON6-B and anti-haemagglutinin (HA) in HA-tagged transgenic parasites. Confocal microscopy using anti-rPfRON6-B with anti-HA. Corresponding overlay images are shown. (C) Double labelling of PfRON6 with Pf34, PfRON4, RAMA or AMA1 in segmented schizonts. Confocal microscopy using anti-rPfRON6-B with anti-Pf34 (rhoptry neck marker), anti-PfRON4 (rhoptry neck marker), anti-RAMA (rhoptry bulb marker), or anti-AMA1 (microneme marker). Corresponding overlay images are shown. (D) Localisation of PfRON6 in free merozoites. Overlay images of free merozoites labelled with anti-rPfRON6-B, anti-RAMA and DAPI. (E) Electron micrograph of immunogold staining using affinity purified anti-rRON6-C antibodies. Arrows indicate PfRON6. (F) Localisation of PfRON6 during the asexual red blood cell (RBC) cycle. Immunofluorescence microscopy using anti-rPfRON6-B antisera and DAPI on parasites at different stages of the asexual RBC cycle. The corresponding phase contrast images are shown.
Interesting differences in the distribution of PfRON6 were observed throughout the asexual RBC cycle (Fig. 4E). Consistent with the immunoblot results and microarray data (Bozdech et al., 2003a; Le Roch et al., 2003), PfRON6 was detectable in trophozoites, schizonts, free merozoites and young rings, but disappeared from older rings. In trophozoites, anti-PfRON6 antibodies produced a diffuse staining pattern, similar to that observed for other rhoptry proteins such as PfRhop148 and RAMA (Lobo et al., 2003; Topolska et al., 2004). Rhoptries are synthesised late in the erythrocytic cycle and the diffuse staining likely represents trafficking of the protein through the secretory pathway (Bannister et al., 2000). In schizonts and free merozoites PfRON6 was present at the apical end. In young rings, PfRON6 staining was present as a rim around the parasite, and co-localised with RAMA (Supplementary Fig. S3) indicating that during invasion the protein is transferred to the PV (Topolska et al., 2004).
3.5. Reactivity of PfRON6 recombinant proteins with human immune serum
To determine whether anti-PfRON6 antibodies are produced during natural malaria infection, rPfRON6-A, -B, -C and GST (control) were immunoblotted with sera from individuals living in malaria endemic regions of PNG and Vietnam or naïve individuals living in Melbourne, Australia (a non-endemic area) (Fig. 5). rPfRON6-A and -B recombinant proteins reacted with the PNG and Vietnam sera while rPfRON6-C reacted only with the Vietnam sera. None of the proteins reacted with the Melbourne sera. The GST control did not react with any of the sera. These results suggest that PfRON6 recombinant proteins are recognised specifically by sera from individuals exposed to malaria infection.
Fig. 5.
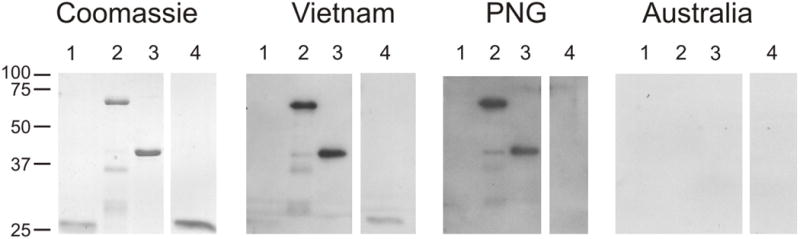
Reactivity of rPfRON6-A, -B and -C recombinant proteins with human immune sera. rPfRON6-A and -B were expressed and purified as GST fusion proteins. rPfRON6-C was expressed and purified as a hexa-histidine fusion. rPfRON6-A (lane 2), -B (lane 3), -C (lane 4) and a GST (lane 1) control were resolved by SDS-PAGE, and either stained with coomassie (loading control) or transferred to PVDF membrane and immunoblotted with human sera from individuals living in malaria endemic areas of Papua New Guinea (PNG) and Vietnam or with sera from naïve individuals living in Melbourne, Australia.
4. Discussion
The invasion machinery of Plasmodium is an attractive candidate for therapeutic intervention. Invasion is a necessary step in the parasite’s life-cycle and effective inhibition of sporozoite or merozoite invasion would prevent parasite replication and clinical manifestations. In this study, we used a bioinformatic approach to identify a novel malarial protein predicted to be involved in invasion. PfRON6 has a characteristic N-terminal signal sequence and appears to be composed of three domains – an N-terminal repetitive region, a sub-C-terminal conserved domain, and a C-terminal cysteine-rich domain.
Blocks of repeats or low complexity sequence are common amongst P. falciparum proteins (Anders et al., 1988). One possibility is that they function as a smoke screen to direct the immune response away from functionally important regions of the molecule (Cowman et al., 1985; Anders et al., 1988, 1993). More recently, repeats have been shown to function in protein-protein interactions (Waller et al., 1999; Magowan et al., 2000). Either of these are possible roles for the N-terminal repeats of PfRON6.
Domains based on a conserved arrangement of cysteine residues are also common in Plasmodium. They occur in numerous protein families and interact with a wide range of host receptors including the Duffy antigen receptor for chemokines (DARC), Glycophorin A and intracellular cell adhesion molecule I (ICAM-1) (Miller et al., 1976; Sim et al., 1994; Smith et al., 2000)}. The arrangement of cysteines within the C-terminal region of PfRON6 is unlike any previously described, suggesting that it is a novel domain, but one that is conserved across the Apicomplexa. It is unclear whether it interacts with a host receptor or with another parasite protein. However, the fact that rodent malaria species possess a truncated version of PfRON6 which lacks the cysteine-rich domain indicates that either those parasites lack the corresponding interacting protein or that mouse RBCs lack the corresponding receptor.
To investigate the function of PfRON6 we attempted to mutate PFB0680w. Only constructs that were designed to modify the C-terminal end of the protein by addition of a small epitope tag integrated into the genome. The expression and localisation of the tagged protein was indistinguishable from wild-type. In contrast, repeated attempts to disrupt the gene or to truncate the C-terminal cysteine-rich domain failed. The cysteine-rich domain may participate in essential ligand-receptor interactions, or may be required for correct trafficking of the protein. In either case, our findings suggest that PfRON6, including the cysteine-rich domain, is necessary for completion of the asexual RBC cycle.
Using protein-specific antibodies we have shown that PfRON6 is expressed during the asexual RBC cycle. Consistent with microarray data, PfRON6 synthesis begins during the late trophozoite stage (Bozdech et al., 2003a; Le Roch et al., 2003). The protein is trafficked through compartments of the secretory system and in schizonts and free merozoites it is present in the rhoptry neck. PfRON6 was also detected by IFA in newly invaded rings. However, PfRON6 could not be detected by immunoblotting in older rings (5–9 h) and could not be detected by IFA or immunoblotting in trophozoites. Taken together, this data indicates that PfRON6 is secreted during invasion and is transferred to the PV, but is then rapidly degraded, presumably by the action of a protease. The localisation of PfRON6 strongly suggests that it plays a role in invasion, rather than in subsequent intracellular growth. Recent studies in both Plasmodium and Toxoplasma have implicated rhoptry neck proteins in the formation of a specialised tight junction complex for exclusion of selected host and parasite proteins from the forming PV membrane (Alexander et al., 2005, 2006; Lebrun et al., 2005). Other rhoptry neck proteins, such as members of the RBL family are involved in host cell tropism and antigenic variation (Gruner et al., 2004; Stubbs et al., 2005; Triglia et al., 2005).
Recombinant PfRON6 protein fragments were specifically recognised by sera from people living in malaria endemic areas of PNG and Vietnam. This demonstrates that PfRON6 is expressed in field isolates and that it is capable of eliciting an immune response in a natural infection. Interestingly, PNG and Vietnam immune sera differed in their ability to recognise the cysteine-rich domain. This difference may reflect the variation in the temporal acquisition of antibodies against Plasmodium antigens in these two populations.
In summary, we have characterised a novel rhoptry protein. The primary amino acid sequence of PfRON6, its expression and localisation suggest that it is involved in merozoite invasion of RBCs. If the N-terminal and C-terminal regions are indeed protein-protein interaction domains, PfRON6 may be part of a larger protein complex, or may serve as a link between a host cell receptor and another parasite protein. In any case, these interactions appear to be necessary for the completion of the parasite life-cycle and further investigation is warranted to determine whether they can be targeted by rationally designed drugs or a subunit vaccine.
Supplementary Material
Supplementary Fig. S1. Amino acid alignment of the PfRON6-like cysteine-rich domains in different Apicomplexa. Cysteine residues are highlighted in yellow. Pk – Plasmodium knowlesi; Pv – Plasmodium vivax; Pf – Plasmodium falciparum; Pg – Plasmodium gallinaceum; Cp – Cryptosporidium parvum; Ch – Cryptosporidium hominis; Tg – Toxoplasma gondii; Ta – Theileria annulata; Bb – Babesia bovis; and Tp – Theileria parva.
Supplementary Fig. S2. Stage-specific expression of PfRON6, RAMA and MSP4 proteins in parasitised red blood cells (RBCs). Variation in protein expression over time as a percentage of the maximum level of expression of the particular protein quantified using ImageJ.
Supplementary Fig. S3. Localisation of the PfRON6 protein in transgenic and ring stage parasites. (A) Immunofluorescence microscopy on haemagglutinin (HA)-tagged transgenic parasite lines using anti-PfRON6 and anti-HA antibodies. Corresponding overlay images are shown. (B) Immunofluorescence microscopy on wild-type ring stage parasites using anti-PfRON6 and anti-RAMA antibodies. Corresponding overlay images are shown.
Supplementary Fig. S4. Co-localisation of the PfRON6 protein with RAMA, AMA1, Pf34 and PfRON4 in parasitised red blood cells. Each dot represents a Pearson’s coefficient calculated from an individual image and the horizontal lines represent the median for each group. Statistical comparison between the groups was calculated using Bonferroni’s multiple comparison test. NS – P > 0.05; * – P < 0.05; ** – P < 0.01; *** – P < 0.001.
Acknowledgments
This work was supported by the National Health and Medical Research Council (NHMRC). N.I.P. and L.M.K. are supported by the Australian Postgraduate Award. D.A.S. is supported by an NHMRC Medical and Dental Scholarship. The authors would like to thank Robert Huestis of the Victorian Bioinformatics Consortium for assistance with gene annotation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Note: Supplementary data associated with this article
References
- Alexander DL, Mital J, Ward GE, Bradley P, Boothroyd JC. Identification of the moving junction complex of Toxoplasma gondii: a collaboration between distinct secretory organelles. PLoS Pathog. 2005;1:e17. doi: 10.1371/journal.ppat.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DL, Arastu-Kapur S, Dubremetz JF, Boothroyd JC. Plasmodium falciparum AMA1 binds a rhoptry neck protein homologous to TgRON4, a component of the moving junction in Toxoplasma gondii. Eukaryot Cell. 2006;5:1169–1173. doi: 10.1128/EC.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders RF, Coppel RL, Brown GV, Kemp DJ. Antigens with repeated amino acid sequences from the asexual blood stages of Plasmodium falciparum. Prog Allergy. 1988;41:148–172. [PubMed] [Google Scholar]
- Anders RF, McColl DJ, Coppel RL. Molecular variation in Plasmodium falciparum: polymorphic antigens of asexual erythrocytic stages. Acta Trop. 1993;53:239–253. doi: 10.1016/0001-706x(93)90032-7. [DOI] [PubMed] [Google Scholar]
- Ballou WR, Arevalo-Herrera M, Carucci D, Richie TL, Corradin G, Diggs C, Druilhe P, Giersing BK, Saul A, Heppner DG, Kester KE, Lanar DE, Lyon J, Hill AV, Pan W, Cohen JD. Update on the clinical development of candidate malaria vaccines. Am J Trop Med Hyg. 2004;71:239–247. [PubMed] [Google Scholar]
- Bannister LH, Hopkins JM, Fowler RE, Krishna S, Mitchell GH. Ultrastructure of rhoptry development in Plasmodium falciparum erythrocytic schizonts. Parasitology. 2000;121(Pt 3):273–287. doi: 10.1017/s0031182099006320. [DOI] [PubMed] [Google Scholar]
- Bannister LH, Hopkins JM, Dluzewski AR, Margos G, Williams IT, Blackman MJ, Kocken CH, Thomas AW, Mitchell GH. Plasmodium falciparum apical membrane antigen 1 (PfAMA-1) is translocated within micronemes along subpellicular microtubules during merozoite development. J Cell Sci. 2003;116:3825–3834. doi: 10.1242/jcs.00665. [DOI] [PubMed] [Google Scholar]
- Ben Mamoun C, Gluzman IY, Hott C, MacMillan SK, Amarakone AS, Anderson DL, Carlton JM, Dame JB, Chakrabarti D, Martin RK, Brownstein BH, Goldberg DE. Co-ordinated programme of gene expression during asexual intraerythrocytic development of the human malaria parasite Plasmodium falciparum revealed by microarray analysis. Mol Microbiol. 2001;39:26–36. doi: 10.1046/j.1365-2958.2001.02222.x. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Black CG, Wu T, Wang L, Hibbs AR, Coppel RL. Merozoite surface protein 8 of Plasmodium falciparum contains two epidermal growth factor-like domains. Mol Biochem Parasitol. 2001;114:217–226. doi: 10.1016/s0166-6851(01)00265-1. [DOI] [PubMed] [Google Scholar]
- Blackman MJ, Ling IT, Nicholls SC, Holder AA. Proteolytic processing of the Plasmodium falciparum merozoite surface protein-1 produces a membrane-bound fragment containing two epidermal growth factor-like domains. Mol Biochem Parasitol. 1991;49:29–33. doi: 10.1016/0166-6851(91)90127-r. [DOI] [PubMed] [Google Scholar]
- Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003a;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdech Z, Zhu J, Joachimiak MP, Cohen FE, Pulliam B, DeRisi JL. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 2003b;4:R9. doi: 10.1186/gb-2003-4-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman AF, Saint RB, Coppel RL, Brown GV, Anders RF, Kemp DJ. Conserved sequences flank variable tandem repeats in two S-antigen genes of Plasmodium falciparum. Cell. 1985;40:775–783. doi: 10.1016/0092-8674(85)90337-x. [DOI] [PubMed] [Google Scholar]
- Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell. 2006;124:755–766. doi: 10.1016/j.cell.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Cranmer SL, Magowan C, Liang J, Coppel RL, Cooke BM. An alternative to serum for cultivation of Plasmodium falciparum in vitro. Trans R Soc Trop Med Hyg. 1997;91:363–365. doi: 10.1016/s0035-9203(97)90110-3. [DOI] [PubMed] [Google Scholar]
- Duraisingh MT, Triglia T, Cowman AF. Negative selection of Plasmodium falciparum reveals targeted gene deletion by double crossover recombination. Int J Parasitol. 2002;32:81–89. doi: 10.1016/s0020-7519(01)00345-9. [DOI] [PubMed] [Google Scholar]
- Fidock DA, Wellems TE. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc Natl Acad Sci U S A. 1997;94:10931–10936. doi: 10.1073/pnas.94.20.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foth BJ, Ralph SA, Tonkin CJ, Struck NS, Fraunholz M, Roos DS, Cowman AF, McFadden GI. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science. 2003;299:705–708. doi: 10.1126/science.1078599. [DOI] [PubMed] [Google Scholar]
- Galinski MR, Medina CC, Ingravallo P, Barnwell JW. A reticulocyte-binding protein complex of Plasmodium vivax merozoites. Cell. 1992;69:1213–1226. doi: 10.1016/0092-8674(92)90642-p. [DOI] [PubMed] [Google Scholar]
- Garman SC, Simcoke WN, Stowers AW, Garboczi DN. Structure of the C-terminal domains of merozoite surface protein-1 from Plasmodium knowlesi reveals a novel histidine binding site. J Biol Chem. 2003;278:7264–7269. doi: 10.1074/jbc.M210716200. [DOI] [PubMed] [Google Scholar]
- Gruner AC, Snounou G, Fuller K, Jarra W, Renia L, Preiser PR. The Py235 proteins: glimpses into the versatility of a malaria multigene family. Microbes Infect. 2004;6:864–873. doi: 10.1016/j.micinf.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Hiller NL, Bhattacharjee S, van Ooij C, Liolios K, Harrison T, Lopez-Estrano C, Haldar K. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306:1934–1937. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- Huestis R, Fischer K. Prediction of many new exons and introns in Plasmodium falciparum chromosome 2. Mol Biochem Parasitol. 2001;118:187–199. doi: 10.1016/s0166-6851(01)00376-0. [DOI] [PubMed] [Google Scholar]
- Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, De La Vega P, Holder AA, Batalov S, Carucci DJ, Winzeler EA. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- Lebrun M, Michelin A, El Hajj H, Poncet J, Bradley PJ, Vial H, Dubremetz JF. The rhoptry neck protein RON4 re-localizes at the moving junction during Toxoplasma gondii invasion. Cell Microbiol. 2005;7:1823–1833. doi: 10.1111/j.1462-5822.2005.00646.x. [DOI] [PubMed] [Google Scholar]
- Lobo CA, Rodriguez M, Hou G, Perkins M, Oskov Y, Lustigman S. Characterization of PfRhop148, a novel rhoptry protein of Plasmodium falciparum. Mol Biochem Parasitol. 2003;128:59–65. doi: 10.1016/s0166-6851(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Magowan C, Nunomura W, Waller KL, Yeung J, Liang J, Van Dort H, Low PS, Coppel RL, Mohandas N. Plasmodium falciparum histidine-rich protein 1 associates with the band 3 binding domain of ankyrin in the infected red cell membrane. Biochim Biophys Acta. 2000;1502:461–470. doi: 10.1016/s0925-4439(00)00069-7. [DOI] [PubMed] [Google Scholar]
- Malkin E, Dubovsky F, Moree M. Progress towards the development of malaria vaccines. Trends Parasitol. 2006;22:292–295. doi: 10.1016/j.pt.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Manders EM, Stap J, Brakenhoff GJ, van Driel R, Aten JA. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J Cell Sci. 1992;103(Pt 3):857–862. doi: 10.1242/jcs.103.3.857. [DOI] [PubMed] [Google Scholar]
- Marshall VM, Tieqiao W, Coppel RL. Close linkage of three merozoite surface protein genes on chromosome 2 of Plasmodium falciparum. Mol Biochem Parasitol. 1998;94:13–25. doi: 10.1016/s0166-6851(98)00045-0. [DOI] [PubMed] [Google Scholar]
- Marti M, Good RT, Rug M, Knuepfer E, Cowman AF. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science. 2004;306:1930–1933. doi: 10.1126/science.1102452. [DOI] [PubMed] [Google Scholar]
- Matuschewski K. Vaccine development against malaria. Curr Opin Immunol. 2006;18:449–457. doi: 10.1016/j.coi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Mayor A, Bir N, Sawhney R, Singh S, Pattnaik P, Singh SK, Sharma A, Chitnis CE. Receptor-binding residues lie in central regions of Duffy-binding-like domains involved in red cell invasion and cytoadherence by malaria parasites. Blood. 2005;105:2557–2563. doi: 10.1182/blood-2004-05-1722. [DOI] [PubMed] [Google Scholar]
- McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- Proellocks NI, Kovacevic S, Ferguson DJ, Kats LM, Morahan BJ, Black CG, Waller KL, Coppel RL. Plasmodium falciparum Pf34, a novel GPI-anchored rhoptry protein found in detergent-resistant microdomains. Int J Parasitol. 2007;37:1233–1241. doi: 10.1016/j.ijpara.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner JC, Galinski MR, Ingravallo P, Barnwell JW. Two Plasmodium falciparum genes express merozoite proteins that are related to Plasmodium vivax and Plasmodium yoelii adhesive proteins involved in host cell selection and invasion. Proc Natl Acad Sci U S A. 2000;97:9648–9653. doi: 10.1073/pnas.160469097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders PR, Gilson PR, Cantin GT, Greenbaum DC, Nebl T, Carucci DJ, McConville MJ, Schofield L, Hodder AN, Yates JR, 3rd, Crabb BS. Distinct protein classes including novel merozoite surface antigens in Raft-like membranes of Plasmodium falciparum. J Biol Chem. 2005;280:40169–40176. doi: 10.1074/jbc.M509631200. [DOI] [PubMed] [Google Scholar]
- Sim BK, Chitnis CE, Wasniowska K, Hadley TJ, Miller LH. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science. 1994;264:1941–1944. doi: 10.1126/science.8009226. [DOI] [PubMed] [Google Scholar]
- Singh SK, Hora R, Belrhali H, Chitnis CE, Sharma A. Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature. 2006;439:741–744. doi: 10.1038/nature04443. [DOI] [PubMed] [Google Scholar]
- Smith JD, Craig AG, Kriek N, Hudson-Taylor D, Kyes S, Fagan T, Pinches R, Baruch DI, Newbold CI, Miller LH. Identification of a Plasmodium falciparum intercellular adhesion molecule-1 binding domain: a parasite adhesion trait implicated in cerebral malaria. Proc Natl Acad Sci U S A. 2000;97:1766–1771. doi: 10.1073/pnas.040545897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs J, Simpson KM, Triglia T, Plouffe D, Tonkin CJ, Duraisingh MT, Maier AG, Winzeler EA, Cowman AF. Molecular mechanism for switching of P. falciparum invasion pathways into human erythrocytes. Science. 2005;309:1384–1387. doi: 10.1126/science.1115257. [DOI] [PubMed] [Google Scholar]
- Tolia NH, Enemark EJ, Sim BK, Joshua-Tor L. Structural basis for the EBA-175 erythrocyte invasion pathway of the malaria parasite Plasmodium falciparum. Cell. 2005;122:183–193. doi: 10.1016/j.cell.2005.05.033. [DOI] [PubMed] [Google Scholar]
- Tonkin CJ, Pearce JA, McFadden GI, Cowman AF. Protein targeting to destinations of the secretory pathway in the malaria parasite Plasmodium falciparum. Curr Opin Microbiol. 2006;9:381–387. doi: 10.1016/j.mib.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Topolska AE, Lidgett A, Truman D, Fujioka H, Coppel RL. Characterization of a membrane-associated rhoptry protein of Plasmodium falciparum. J Biol Chem. 2004;279:4648–4656. doi: 10.1074/jbc.M307859200. [DOI] [PubMed] [Google Scholar]
- Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- Triglia T, Duraisingh MT, Good RT, Cowman AF. Reticulocyte-binding protein homologue 1 is required for sialic acid-dependent invasion into human erythrocytes by Plasmodium falciparum. Mol Microbiol. 2005;55:162–174. doi: 10.1111/j.1365-2958.2004.04388.x. [DOI] [PubMed] [Google Scholar]
- van Dijk MR, Janse CJ, Thompson J, Waters AP, Braks JA, Dodemont HJ, Stunnenberg HG, van Gemert GJ, Sauerwein RW, Eling W. A central role for P48/45 in malaria parasite male gamete fertility. Cell. 2001;104:153–164. doi: 10.1016/s0092-8674(01)00199-4. [DOI] [PubMed] [Google Scholar]
- Waller KL, Cooke BM, Nunomura W, Mohandas N, Coppel RL. Mapping the binding domains involved in the interaction between the Plasmodium falciparum knob-associated histidine-rich protein (KAHRP) and the cytoadherence ligand P. falciparum erythrocyte membrane protein 1 (PfEMP1) J Biol Chem. 1999;274:23808–23813. doi: 10.1074/jbc.274.34.23808. [DOI] [PubMed] [Google Scholar]
- Waller KL, Muhle RA, Ursos LM, Horrocks P, Verdier-Pinard D, Sidhu AB, Fujioka H, Roepe PD, Fidock DA. Chloroquine resistance modulated in vitro by expression levels of the Plasmodium falciparum chloroquine resistance transporter. J Biol Chem. 2003;278:33593–33601. doi: 10.1074/jbc.M302215200. [DOI] [PubMed] [Google Scholar]
- Wang L, Black CG, Marshall VM, Coppel RL. Structural and antigenic properties of merozoite surface protein 4 of Plasmodium falciparum. Infect Immun. 1999;67:2193–2200. doi: 10.1128/iai.67.5.2193-2200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sifri CD, Lei HH, Su XZ, Wellems TE. Transfection of Plasmodium falciparum within human red blood cells. Proc Natl Acad Sci U S A. 1995;92:973–977. doi: 10.1073/pnas.92.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, McGinnis S, Madden TL. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 2006;34:W6–9. doi: 10.1093/nar/gkl164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Amino acid alignment of the PfRON6-like cysteine-rich domains in different Apicomplexa. Cysteine residues are highlighted in yellow. Pk – Plasmodium knowlesi; Pv – Plasmodium vivax; Pf – Plasmodium falciparum; Pg – Plasmodium gallinaceum; Cp – Cryptosporidium parvum; Ch – Cryptosporidium hominis; Tg – Toxoplasma gondii; Ta – Theileria annulata; Bb – Babesia bovis; and Tp – Theileria parva.
Supplementary Fig. S2. Stage-specific expression of PfRON6, RAMA and MSP4 proteins in parasitised red blood cells (RBCs). Variation in protein expression over time as a percentage of the maximum level of expression of the particular protein quantified using ImageJ.
Supplementary Fig. S3. Localisation of the PfRON6 protein in transgenic and ring stage parasites. (A) Immunofluorescence microscopy on haemagglutinin (HA)-tagged transgenic parasite lines using anti-PfRON6 and anti-HA antibodies. Corresponding overlay images are shown. (B) Immunofluorescence microscopy on wild-type ring stage parasites using anti-PfRON6 and anti-RAMA antibodies. Corresponding overlay images are shown.
Supplementary Fig. S4. Co-localisation of the PfRON6 protein with RAMA, AMA1, Pf34 and PfRON4 in parasitised red blood cells. Each dot represents a Pearson’s coefficient calculated from an individual image and the horizontal lines represent the median for each group. Statistical comparison between the groups was calculated using Bonferroni’s multiple comparison test. NS – P > 0.05; * – P < 0.05; ** – P < 0.01; *** – P < 0.001.


