SUMMARY
Sister chromatid cohesion is thought to involve entrapment of sister DNAs by a tripartite ring composed of the cohesin subunits Smc1, Smc3, and Scc1. Establishment of cohesion during S phase depends on acetylation of Smc3’s nucleotide-binding domain (NBD) by the Eco1 acetyl transferase. It is destroyed at the onset of anaphase due to Scc1 cleavage by separase. In yeast, Smc3 acetylation is reversed at anaphase by the Hos1 deacetylase as a consequence of Scc1 cleavage. Smc3 molecules that remain acetylated after mitosis due to Hos1 inactivation cannot generate cohesion during the subsequent S phase, implying that cohesion establishment depends on de novo acetylation during DNA replication. By inducing Smc3 deacetylation in postreplicative cells due to Hos1 overexpression, we provide evidence that Smc3 acetylation contributes to the maintenance of sister chromatid cohesion. A cycle of Smc3 NBD acetylation is therefore an essential aspect of the chromosome cycle in eukaryotic cells.
INTRODUCTION
Faithful segregation of chromosomes during mitosis requires physical interconnection of sister chromatids by a multi-subunit complex called cohesin (Nasmyth and Haering, 2009) from the time of their replication until their eventual disjunction at the onset of anaphase. The cohesin subunits Smc1 and Smc3 are rod-shaped molecules that dimerize to form V-shaped heterodimers by means of “hinge” domains situated at the ends of intramolecular antiparallel coiled coils. ABC-like nucleotide-binding domains (NBDs) at the other ends of each Smc molecule are connected by a kleisin subunit (Scc1 in yeast), thereby forming a tripartite ring with a 35-nm diameter.
Cohesion is dissolved at metaphase-to-anaphase transition by a site-specific protease called separase that is kept inactive for most of the cell cycle by an inhibitory chaperone called securin. Separase is activated only after all chromosomes have bi-oriented on mitotic spindles through securin’s sudden destruction at the hands of a ubiquitin protein ligase called the anaphase-promoting complex or cyclosome together with an accessory protein called Cdc20 (Peters, 2006). Thereupon, separase cleaves Scc1, which severs the cohesin ring and triggers the segregation of sisters to opposite poles (Oliveira et al., 2010; Uhlmann et al., 2000).
The fact that cohesin forms a ring whose severance releases it from chromatin has led to the suggestion that it holds sister DNAs by entrapping them. As predicted by this model, site-specific cross-linking of cohesin’s Smc-Smc and Smc-kleisin interfaces traps circular sister minichromosome DNAs inside a covalently circularized cohesin ring (Haering et al., 2008). Nevertheless, how this takes place remains enigmatic. Crucially, the process normally only occurs during S phase. Thus, cohesin produced in G2 or M phase associates with chromatin, possibly even using a topological mechanism, but fails to entrap sister DNAs even when these are held together by pre-existing cohesion (Haering et al., 2004). An exception to this rule are cells engaged in repairing double-strand DNA breaks (Strom et al., 2007; Strom and Sjogren, 2005; Unal et al., 2007).
In yeast, cohesin associates with chromatin during late G1 phase in a process requiring a separate complex composed of Scc2 and Scc4 proteins. It is presumed, but not yet proven, that this association is also topological in nature. A key question concerns the fate of cohesin rings during S phase. Do rings remain associated with chromatin during replication fork passage, in which case forks might actually pass through them, or are they displaced by the fork and replaced by rings that entrap nascent DNAs de novo shortly after fork passage? Cohesion establishment is facilitated by the RFCCtf18/Dcc1/Ctf8 complex thought to recruit proliferating cell nuclear antigen (PCNA) to replication forks and by Ctf4, a protein that connects the MCM/GINS helicase complex with DNA polymerase α responsible for lagging strand synthesis (Gambus et al., 2009; Tanaka et al., 2009).
More crucial for cohesion establishment is an acetyl-transferase known as Eco1 (Ivanov et al., 2002; Skibbens et al., 1999; Toth et al., 1999), whose essential function is to acetylate a pair of adjacent lysine residues (K112 and K113) on the upper surface of Smc3’s NBD (Rolef Ben-Shahar et al., 2008; Rowland et al., 2009; Unal et al., 2008; Zhang et al., 2008). Acetylation of Smc3 by Eco1 occurs during S phase and persists until its abrupt disappearance sometime during mitosis, raising the possibility that it acts as a switch that must be turned on and off during the cell cycle. However, the finding that substitution of K113 by threonine (Rowland et al., 2009) or asparagine (Rolef Ben-Shahar et al., 2008) permits yeast cells to build cohesion, albeit inefficiently, in the complete absence of Eco1 activity raises an alternative explanation: acetylation is merely necessary to alter Smc3 NBD activity in a constitutive manner. To address the importance of deacetylation at mitosis, de novo acetylation during S phase, and maintenance of acetylation during G2 and M phases, we set out to identify Smc3’s deacetylase.
RESULTS
Smc3 Acetylation Occurs during S Phase and Is Partly Dependent on Ctf4
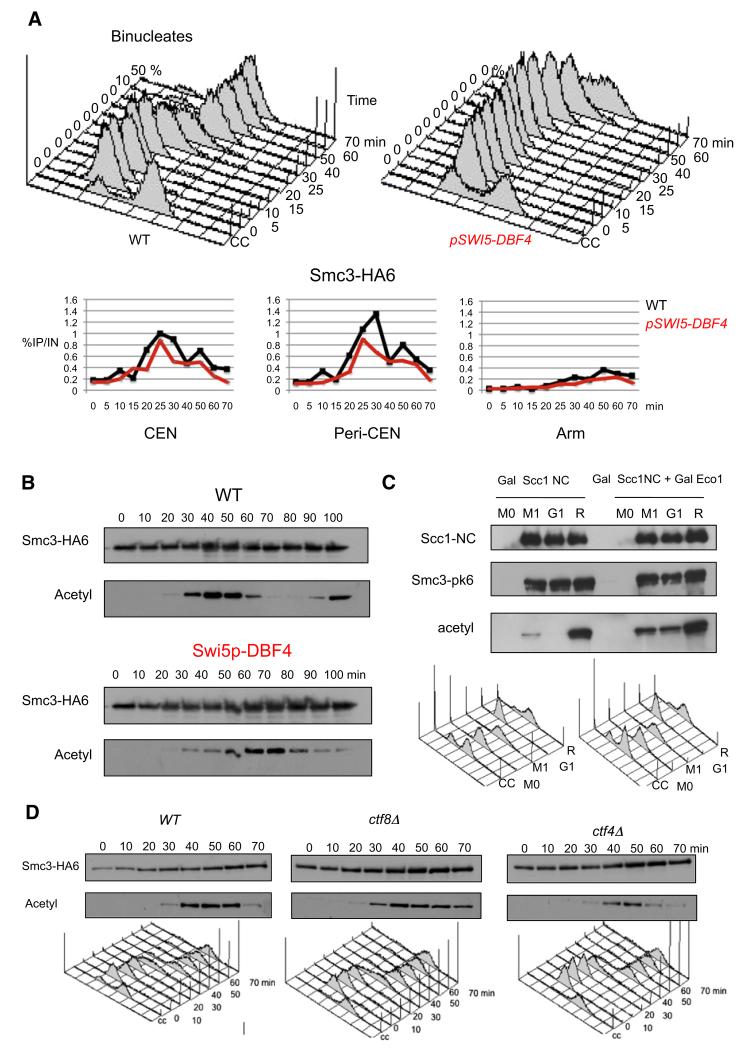
Few if any Smc3 molecules are acetylated in yeast cells arrested in G1 phase by mating pheromone. Entry into S phase upon release is accompanied by Smc3 acetylation, which persists until mitosis. Acetylation occurs shortly after cohesin’s association with chromatin (Figure 1B) and fails to take place in scc2 mutants (Unal et al., 2008) defective in loading cohesin onto chromosomes (Ciosk et al., 2000). To investigate whether acetylation is triggered by cohesin’s association with chromatin or by DNA replication, which occurs shortly afterward, we analyzed the effect of delayed activation of the Cdc7 kinase by expressing its Dbf4 regulatory factor from the SWI5 promoter (pSWI5-DBF4). This delayed both the onset of DNA replication and Smc3 acetylation by 25 min, but not cohesin’s association with centromeric, peri-centromeric, or arm sequences as measured by chromatin immunoprecipitation-quantitative polymerase chain reaction (ChIP-qPCR) (Figures 1A and 1B). Thus, in pSWI5-DBF4 cells, most Smc3 acetylation accompanied the delayed replication and occurred at least 30 min after cohesin’s association with chromatin. This observation, along with the finding that Smc3 acetylation is reduced in cells lacking the prereplication protein Cdc6 (Rolef Ben-Shahar et al., 2008), suggests that acetylation is stimulated by DNA replication. Because modest amounts of acetylated Smc3 accumulate prior to S phase in pSWI5-DBF4 cells, it is possible that some acetylation also occurs upon cohesin’s association with chromatin in late G1 phase.
Figure 1. Efficient Smc3 Acetylation by Eco1 at the Time of S Phase Depends on DNA Replication.
(A) HA6-tagged Smc3 associated with three chromosomal loci (as measured by ChIP and qPCR) in K16655 (MATa SMC3-HA6) and K16653 (MATa SMC3-HA6, dbf4::SWI5p-DBF4) cells that are released from α factor-induced G1 arrest into fresh medium at 25°C. Cell cycle progression was monitored by flow cytometry of DNA content, and the time of anaphase was microscopically determined by the occurrence of binucleated cells (upper panel). An untagged strain (K699) yielded no significant ChIP-qPCR signal.
(B) Smc3-HA6 was immunoprecipitated from wild-type (K16655) or SWI5p-DBF4 (K16653) cells that were released from α factor-induced G1 arrest into fresh medium at 25°C. The acetylation status of Smc3 was analyzed by western blotting using the α-acetyl lysine antibody (ST1027, Calbiochem).
(C). Strains K17957 (MATa SMC3-PK6::KanMX4, TRP1::PMET3CDC20, pGAL1-10SCC1(R180D, R268D)-HA3::LEU2, his3::tetR-GFP::HIS3, YEplac195) K17955 (MATa SMC3-PK6::KanMX4, TRP1:: PMET3CDC20, pGAL1-10SCC1(R180D,R268D)-HA3:: LEU2, YEplac195-Gal-Eco1) were grown to log phase in synthetic medium lacking methionine and arrested in metaphase for 2 hr in YEP medium supplemented with 2 mM methionine and 2% raffinose. After 2 hr, the sample was divided into three subsamples, one being maintained in metaphase arrest, the second being released to G1 arrest, and the third being released to the next S phase. In all cases 2% galactose was added to the media for 80 min to induce expression of Scc1-NC-HA3. Samples were taken at the indicated times for preparation of whole cell extract. Scc1-HA3 was immunoprecipitated using α-HA antibody, and the acetylation level of co-immunoprecipitated Smc3 was subsequently measured by α-Smc3-acetyl-K113 (Figure S1) anti-body via western blotting.
(D) Smc3-HA6 was immunoprecipitated from K16655 (MATa SMC3-HA6), K16842 (MATa SMC3-HA6 ctf8Δ), or K16845 (MATa SMC3-HA6 ctf4Δ) cells that were released from α factor-induced G1 arrest into fresh medium at 25°C. Western blots were performed using either HA-specific or acetyl-lysine-specific antibodies (ST1027, Calbiochem). Cell cycle progression was monitored by flow cytometry of DNA content, and the time of anaphase was microscopically determined by the occurrence of binucleated cells.
The fact that acetylation is maintained until the metaphase-to-anaphase transition (Figure 1B) raises the possibility that Eco1 continues to acetylate Smc3 during G2 or M phases. Cohesin expressed in postreplicative cells associates with chromosomes in an Scc2/4-dependent manner and occupies similar loci to those loaded during G1 or S phase (Lengronne et al., 2006). To test whether this cohesin population is acetylated, a modified version of Scc1 (Scc1-NC) that cannot be cleaved by separase due to mutations in both of its cleavage sites (Uhlmann et al., 1999) was induced from the GAL promoter in cells arrested in M phase by Cdc20 depletion. Western blotting revealed that Smc3 co-immunoprecipitated with Scc1-NC-HA3 in cdc20-arrested cells is only weakly acetylated in comparison with cells allowed to enter S phase (Figure 1C). Smc3 associated with Scc1-NC expressed in G1-arrested cells was not acetylated at all. Overexpression of Eco1 together with Scc1-NC increased the extent of Smc3 acetylation during both G1 and M phases, but not to the levels found in cells allowed to undergo S phase (Figure 1C). The low level of de novo Eco1-mediated acetylation accompanying cohesin’s association with chromosomes in G2/M could contribute to the lack of cohesion establishment under these circumstances (Haering et al., 2004). Indeed, the modest increase in Eco1-mediated cohesin acetylation brought about by Eco1 overexpression could be responsible for the finding that the latter permits cells to build cohesion during G2/M phase (Unal et al., 2007); however, under these circumstances, acetylation of Scc1, not Smc3, is thought to be involved (Heidinger-Pauli et al., 2008; Heidinger-Pauli et al., 2009).
How might the DNA replication machinery facilitate Smc3 acetylation? A report that Eco1 contains an essential N-terminal motif that binds PCNA (Moldovan et al., 2006) suggests that the acetylase might be recruited to replication forks by PCNA clamps. Inactivation of the RFCCtf18/Dcc1/Ctf8 clamp loading complex supposedly reduces the amount of PCNA associated with replication forks (Lengronne et al., 2006). However, inactivation of Ctf8 caused only a modest reduction of Smc3 acetylation during S phase (Figure 1D), an effect that is probably insufficient to explain the reduced sister chromatid cohesion of ctf8 Δ mutants (Mayer et al., 2001). Inactivation of Ctf4, which connects Mcm helicases to DNA polymerases on the lagging strand, had a greater effect (Figure 1D), which is consistent with the notion that Smc3 acetylation does indeed take place at replication forks.
Smc3 Deacetylation during Mitosis Depends on Hos1
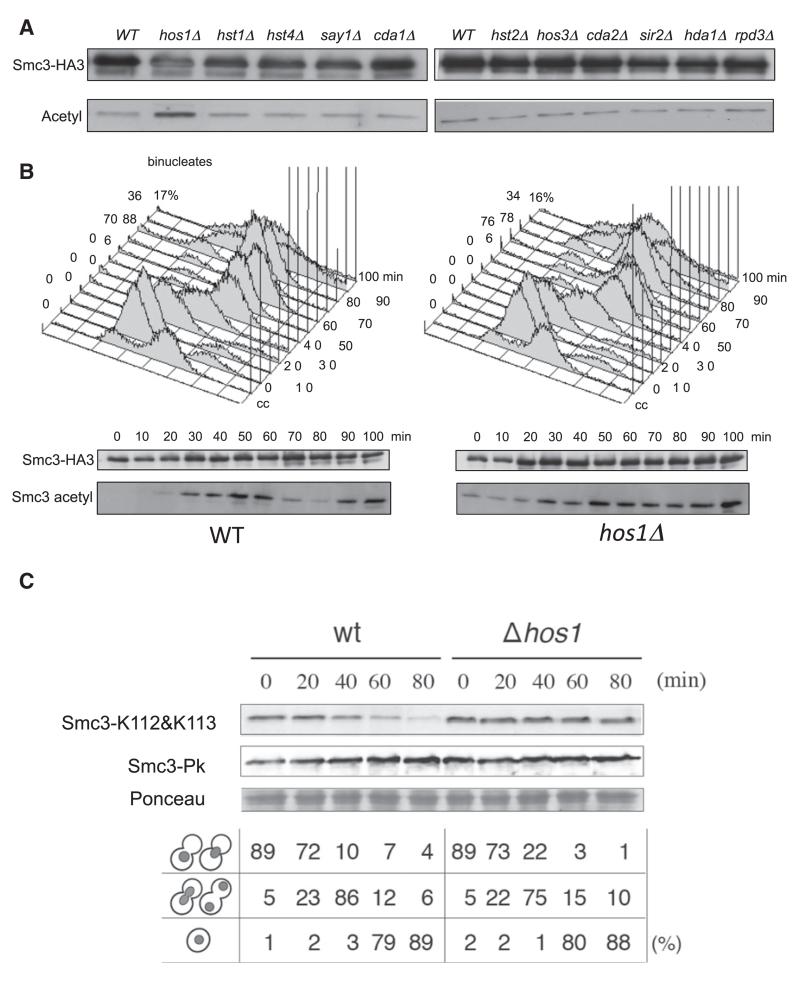
To identify the Smc3 deacetylase, we used an acetyl lysine-specific antibody to probe western blots of Smc3-HA3 immunoprecipitates prepared from cycling cultures of wild-type and known deacetylase deletion mutant strains (Figure 2A). Deleting the gene encoding the type I deacetylase Hos1 (Davie et al., 2003; Robyr et al., 2002; Rundlett et al., 1996) caused a modest increase in Smc3 acetylation. None of the other deletions had any effect (Figure 2A), including Hos1’s nearest relative Hos2 (Figure S2 available online). We also checked whether other enzymes contribute to Smc3 deacetylation by analyzing the phenotype of double mutants. The level of Smc3 acetylation in double mutant cells was similar to that of hos1 Δ single mutants, whether measured in cycling or G1 arrested cells (Figure S2).
Figure 2. The Histone Deacetylase Hos1 Acts on Smc3.
(A) Smc3-HA3 was immunopurified from 13 deacetylase deletion mutants, and the level of acetylated Smc3 was evaluated by western blotting using α-acetyl lysine antibody (ST1027, Calbiochem).
(B) Smc3 acetylation pattern in K15544 (WT) and K16351 (hos1 Δ) cells that are released from α factor-induced G1 arrest (160 min) into fresh medium at 25°C. Smc3-HA3 was immunopurified from aliquots of the culture at the indicated time points, and Smc3 acetylation was analyzed by western blotting using a specific α-acetylated K113 antibody. Cell cycle progression was monitored by flow cytometry of DNA content, and the time of anaphase was microscopically determined by the occurrence of binucleated cells.
(C) Strains ST231 (HOS1) and ST432 (hos1 Δ) growing in YPD medium were arrested at 23°C in G2/M phases for 2.5 hr after adding benomyl (80 μg/ml). Cells were then washed with YPD, re-suspended in YPD containing α factor, and cultured for 80 min at 23°C. The acetylation status and the level of Smc3-pk6 at the indicated time points were monitored by western blotting of crude extracts using α-pk- and α-Smc3-acetylated antibodies.
To test whether Hos1 is responsible for Smc3’s deacetylation during mitosis, we compared the cell cycle profile of Smc3 acetylation after release of wild-type and congenic hos1 Δ cells from pheromone-induced G1 arrest (Figure 2B). This revealed that Hos1 inactivation largely eliminated the drop in Smc3 acetylation that normally accompanies anaphase. A consequence is that Smc3 acetylation can be detected in hos1 Δ cells arrested in G1 by pheromone. To address whether deacetylation mediated by Hos1 takes place only as cells exit mitosis and not during mitosis itself, we compared Smc3 acetylation in wild-type and hos1 Δ cells released from a benomyl-induced mitotic arrest into fresh medium containing pheromone to block entry into S phase. Smc3 acetylation declined during the transition from M phase to G1 in wild-type but not in hos1 Δ cells (Figure 2C).
Scc1 Cleavage Is Necessary and Sufficient to Trigger Smc3 Deacetylation by Hos1
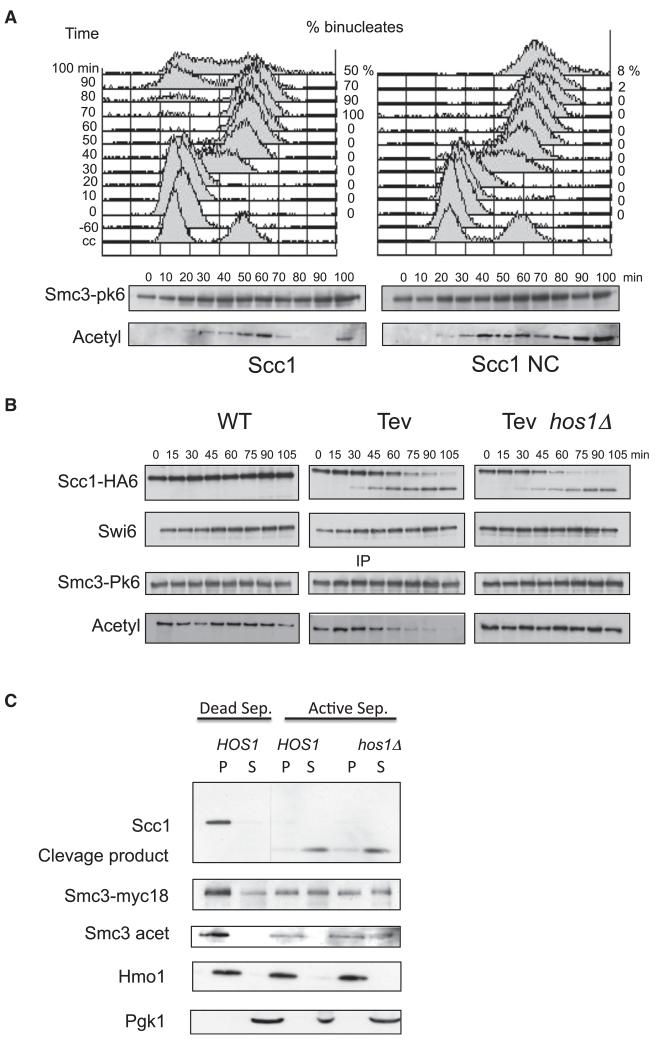
To test whether Smc3 deacetylation depends on Scc1 cleavage, we investigated the effect of expressing a version of Scc1 (Scc1-NC) that cannot be cleaved by separase due to mutations in both of its cleavage sites (Uhlmann et al., 1999). Cells arrested by pheromone in G1 were induced to express either wild-type or noncleavable Scc1 from the GAL promoter 1 hour before pheromone release. Western blotting of Smc3 immunoprecipitates using an acetyl-lysine specific antibody revealed that ectopic expression of Scc1-NC (but not Scc1) blocked Smc3 deacetylation as well as nuclear division (Figure 3A). Scc1 cleavage is therefore necessary for deacetylation. To test whether cleavage is sufficient, we analyzed the effect of expressing Tobacco Etch Virus (TEV) protease in cells whose Scc1 contains three tandem TEV recognition sites (Gruber et al., 2003; Uhlmann et al., 2000). Cells were first arrested in metaphase by depleting Cdc20 and then TEV expression from the GAL promoter induced by the addition of galactose. This triggered cleavage of most Scc1 molecules within 75 min, which was accompanied by Smc3 deacetylation (Figure 3B). Crucially, this deacetylation failed to take place in hos1 Δ cells despite equally efficient TEV-induced Scc1 cleavage (Figure 3B). These data imply that Hos1 is present and potentially active in metaphase as well as in anaphase cells. The event that triggers Smc3 deacetylation during anaphase is not a change in Hos1 activity but cleavage of the cohesin ring, which releases it from chromosomes.
Figure 3. Hos1 Acts on Smc3 Released from Chromatin after Scc1 Cleavage.
(A) Strain K16548 (MATa, SMC3-PK6, pGAL1-10-SCC1-HA3::LEU2) expresses Scc1-HA3 epitope from the GAL1-10 promoter. Strain K16546 (MATa, SMC3-PK6, pGAL1-10-SCC1(R180D, R268D)-HA3::LEU2) expresses a separase non-cleavable version of Scc1-HA3 from the GAL1-10 promoter. Both strains were arrested at 30°C in G1 by α factor in YEP medium containing 2% raffinose. After 1 hr in G1 phase, the GAL1-10 promoter was induced by addition of galactose. One hour after induction, cells were released from G1 arrest and samples were taken every 10 min for preparation of whole cell extract. Cell cycle progression was monitored by flow cytometry of DNA content and microscopic analysis of binucleate (anaphase) cells (upper panel). Cell extracts were prepared from aliquots of the culture at the indicated times and Smc3-PK6 immunoprecipitated using an antibody raised against the pk epitope tag. The acetylation level of Smc3 was monitored by western blotting using the α-acetyl lysine antibody (ST1027, Calbiochem). FACScan analysis shows that cells were arrested and released efficiently (upper panel).
(B) Yeast strains K16394 (MATα PMET3CDC20, SCC1-HA6, SMC3-PK6, YEp-PGAL1TEV), K16397 (MATα PMET3CDC20, SCC1(TEV220)-HA6, SMC3-PK6, YEp-PGAL1TEV) and K16465 (MATα PMET3CDC20, SCC1(TEV220)-HA6 SMC3-PK6 hos1Δ, YEp-PGAL1TEV) were grown to log phase in synthetic medium lacking methionine and arrested in metaphase for 2 hr in YEP medium supplemented with 2 mM methionine and 2% raffinose (0 min). Expression of TEV protease was induced by addition of 2% galactose (0-105 min). Smc3-PK6 was immunoprecipitated and the level of Smc3 acetylation was monitored using lysine-specific antibodies (ST1027, Calbiochem). Cleavage of Scc1-HA6 was monitored by probing whole cell extracts against the HA epitopes. Swi6 serves as a loading control.
(C) Chromatin isolated from the strain K17304 (MATa SCC1-HA6, SMC3-PK6, hos1Δ) arrested in nocodazole was incubated for 15 min at 25°C with a mixture of extracts from the mitotic strains K8965 (MATa esp1-1, pGAL1-10-ESP1), K17305 (MATa esp1-1, hos1 Δ, pGAL1-10-ESP1) and K8967 (MATa esp1-1, pGAL1-10-esp1-C1531A) overproducing wild-type (ESP1) or catalytically dead (esp1-C1531A) separase. Reactions were analyzed by western blotting using α-Myc, α-HA and α-Smc3 acetylated antibodies. Hmo1 and cytosolic pGK1 served as a loading control.
Hos1-Dependent Smc3 Deacetylation In Vitro
To address whether Smc3 deacetylation can be reproduced in vitro, crude preparations of yeast chromatin isolated from nocodazole treated hos1 Δ cells were incubated with soluble extracts prepared from esp1-1 HOS1 or esp1-1 hos1 Δ cells overexpressing from the GAL1-10 promoter either wild-type or catalytically dead separase (ESP1). Extracts containing active separase induced efficient Scc1 cleavage and release from the chromatin pellet of half of the Smc3 molecules (Figure 3C). Crucially, their degree of acetylation was unaltered when released by hos1 Δ extracts but greatly reduced when released by HOS1 extracts (Figure 3C). These data imply that cohesin cleavage not only releases it from chromatin but also triggers deacetylation of Smc3 by a soluble form of Hos1. It is also noticeable that Hos1 in the extracts induced little or no deacetylation of those Smc3 molecules that remained associated with chromatin pellets.
Hos1 Facilitates Sister Chromatid Cohesion
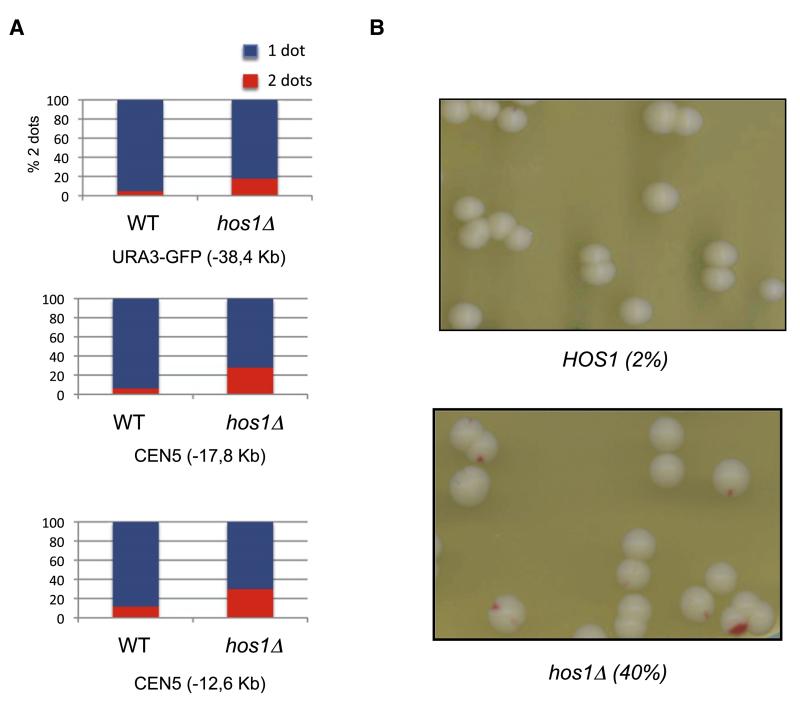
Though HOS1 is not an essential gene, it might nevertheless regulate sister chromatid cohesion. To test this, we used yeast strains with specific loci marked by Tet repressor-green fluorescent protein (GFP) bound to multiple tandem Tet operators (Michaelis et al., 1997) and with the anaphase-promoting complex or cyclosome activator Cdc20 expressed from the methionine-repressible MET3 promoter. Cells were arrested in metaphase by transferring them to rich medium containing methionine and scored as to whether they contained one (cohesion) or two (loss of cohesion) GFP dots. Deletion of HOS1 caused a modest increase in the fraction of cells with split GFP dots at loci 12.6, 17.8, or 38 kb from CEN5 (Figure 4A). To address whether hos1 Δ causes chromosome missegregation, we used an assay that measures loss of a supernumerary chromosome carrying the TRP1 gene and the nonsense suppressor SUP11, which suppresses the red colony color of ade2-1 mutants. Wild-type (HOS1) cells growing in rich medium rarely give rise to red-sectored colonies, but hos1 Δ increased their incidence from 2% to 40%, suggesting that Hos1 is necessary for high-fidelity chromosome transmission (Figure 4B). The viability of hos1 Δ mutants is not due to any single other deacetylase having an overlapping function, because no other deacetylase gene deletion is lethal when combined with hos1 Δ.
Figure 4. Cohesion Defect in hos1Δ.
(A) Cells carrying MET-CDC20 and GFP labels at various loci grown to log phase in synthetic medium lacking methionine were transferred to YPD plus methionine. After 150 min, the percentages of separated GFP foci were scored. The tet operators are integrated at: URA3-GFP (38.4 kb to left of CEN5) in wild-type (K16472) and hos1 Δ (K16470) (upper panel); 17.8CEN6-GFP (17.8 kb to left of CEN5) in wild-type (K17508) and hos1 Δ (K17506) (middle panel); and −12.6CEN5-GFP (12.6 kb to left of CEN5) in wild-type (K17512) and hos1 Δ (K17510) (lower panel).
(B) Sectoring assay for the chromosome loss using the SUP11-marked in HOS1 strain (K7040) and its hos1 Δ derivative (K16466).
Is De Novo Smc3 Acetylation Critical for Cohesion Establishment?
The ability of hos1 Δ cells to proliferate—albeit with increased rates of chromosome missegregation—in the absence of any apparent Smc3 deacetylation at anaphase raises the possibility that an acetylation/deacetylation cycle may not be strictly necessary for the establishment of sister chromatid cohesion. However, deacetylation during anaphase is not the only mechanism for providing Eco1 targets. Only about half of Smc1/3 heterodimers associate with Scc1 each cell cycle, and those that do not are presumably not acetylated. Moreover, it is likely that not all Smc1/3 heterodimers that form tripartite rings with Scc1 are acetylated during S phase, and heterodimers synthesized during G1 are presumably not acetylated during this period of the cell cycle. Therefore, hos1 Δ cells almost certainly enter S phase with a sizeable pool of unacetylated cohesin molecules that could serve as Eco1 targets.
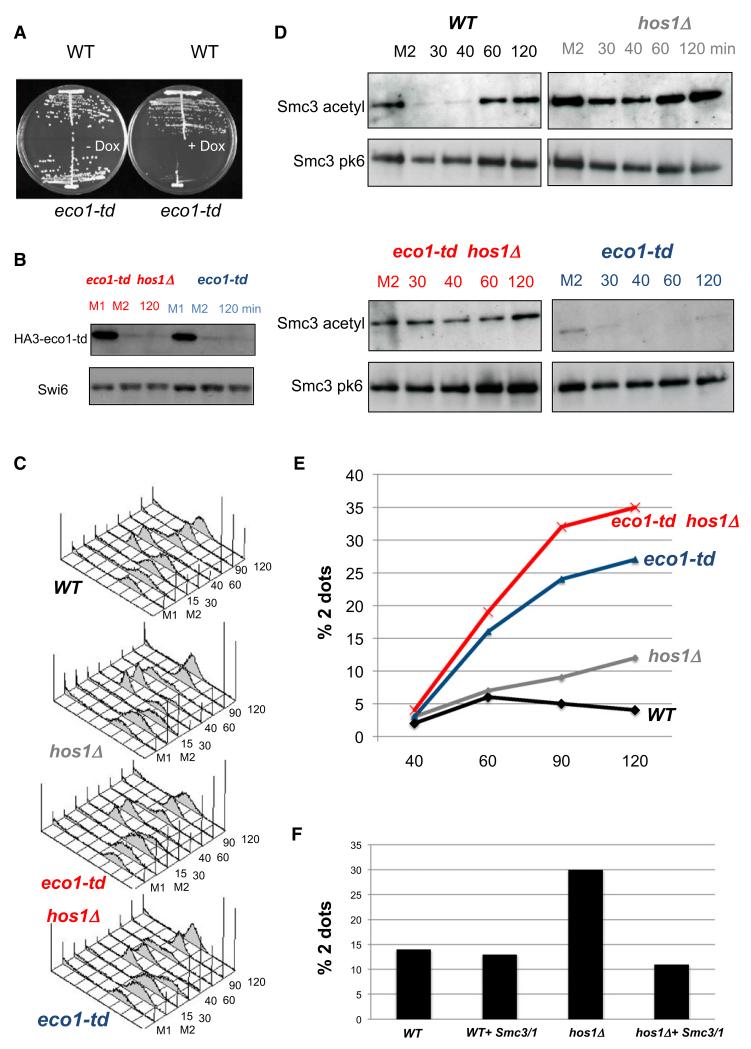
To address whether Smc3 must be acetylated anew each cell cycle, we asked whether hos1 Δ daughter cells that inherit a pool of acetylated Smc3 molecules can establish sister chromatid cohesion without Eco1 activity during the subsequent S phase. To do this, it was first necessary to create a strain with conditional Eco1 activity. Due to their low level of activity even at the permissive temperature, existing ts eco1 alleles were inappropriate. We therefore replaced ECO1 with a heat-inducible degradation allele (eco1-td) expressed from a Tet-repressible promoter in strains that express the degron-specific E3 ubiquitin ligase Ubr1 from the GAL1-10 promoter. Although eco1-td cells proliferate almost normally at 25°C in the absence of galactose or doxycline, they fail to do so at 37°C when galactose or doxycline are present (Figure 5A). To synchronize the cell cycle, we used strains that express Cdc20 from the MET3 promoter. To measure sister chromatid cohesion, we used strains whose TRP1 locus contains tandem arrays of Lac operators bound by Lac repressor fused to GFP.
Figure 5. Smc3 Needs to be Acetylated Specifically in S Phase to be Competent for Establishment of Cohesion.
Exponential phase cells of strains K17515 (WT, PMET3CDC20, SMC3-PK6), K17516 (hos1 Δ, PMET3CDC20, SMC3-PK6), and K17376 (td-Eco1, PMET3CDC20, SMC3-PK6) and K17382 (td-Eco1 hos1 Δ, PMET3CDC20, SMC3-PK6) were grown to log phase in synthetic medium lacking methionine and arrested in metaphase for 2 hr in YEP medium supplemented with 2 mM methionine, 2% raffinose, and 2% galactose (M1). Eco1 was degraded by shifting cultures to degron-mediated proteolysis conditions (YEP raff/gal, 20 μg/ml doxycycline, at 37°C) for 90 min (M2). Subsequently, the cells were released from G2/M arrest to raff/gal media (lacking methionine) containing 20 μg/ml doxycycline. Thirty minutes after release at 37°C, nocodazole was added to arrest the cells again in G2/M phase.
(A) Strains 16354 (WT) and 16353 (Eco1-deg) were grown at 25°C for 2 days on YP plates containing 2% raffinose and 2% galactose, then transferred to 37°C on plates containing 2% raffinose, 2% galactose, and doxycycline (5 μg/ml).
(B) Western blots show degradation of endogenous HA-tagged td-Eco1 proteins
(C) Fluorescence-activated cell sorting analysis shows that the release from Cdc20 arrest was complete and that all strains had completed DNA replication after 90 min.
(D) Smc3-PK6 was immunopurified, and the level of acetylated Smc3 was evaluated by western blotting using α-acetyl lysine antibody (ST1027, Calbiochem).
(E) Cohesion was measured by scoring the percentages of separated GFP dots at the TRP1 locus (12.6 kb from CEN4).
(F) K17987 (hos1 Δ, 256LacO::TRP1, PMET3CDC20, YEp-SMC1/SMC3) K17988 (hos1 Δ, 256LacO::TRP1, PMET3CDC20, YEp), K17989 (256LacO::TRP1, PMET3CDC20, YEp-SMC1/SMC3), and K17990 (256LacO::TRP1, PMET3CDC20) were grown to log phase in synthetic medium lacking methionine and uracil and arrested in metaphase for 2 hr in YEP medium supplemented with 2 mM methionine. Cohesion defect was measured by scoring the percentages of separated GFP dots at the TRP1 locus (12.6 kb from CEN4).
ECO1 or eco-td strains growing at 25°C with and without HOS1 were first arrested in metaphase due to addition of methionine and then incubated at 37°C in medium containing galactose and doxycline (in addition to methionine), which induced degradation of most Eco1 in the eco-td strains (Figure 5B). After 90 min at 37°C, cells were transferred to medium lacking methionine, which induced Cdc20 and triggered anaphase, cell division, and a new round of DNA replication (Figure 5C). To block the subsequent mitosis, nocodazole was added 30 min after Cdc20 induction. As expected, Eco1 depletion in eco1-td cells compromised the establishment of cohesion in HOS1 cells, which inherited little or no acetylated Smc3 from the previous cell cycle (Figures 5D and 5E). Crucially, deletion of HOS1 ensured that Smc3 molecules acetylated by eco1-td under permissive conditions persisted in the absence of eco1 activity during the subsequent cell cycle (Figure 5D). However, this did not ameliorate but rather exacerbated the cohesion establishment defect caused by the lack of Eco1 activity, even though the degree of Smc3 acetylation by the time cells completed phase was comparable to that in wild-type ECO1 HOS1 cells (Figures 5D and 5E). These data suggest that Smc3 molecules that fail to be deacetylated by Hos1 during anaphase are incapable of building sister chromatid cohesion during the next round of DNA replication. In other words, de novo acetylation of Smc3 during DNA replication might therefore be essential for the establishment of cohesion during S phase.
Our finding that non-deacetylated Smc3 cannot establish sister chromatid cohesion provides a potential explanation for the cohesion defect of hos1 Δ cells: namely, that they possess suboptimal amounts of unmodified Smc3 molecules as cells enter S phase. Consistent with this hypothesis, we found that expression of both SMC1 and SMC3 from a multicopy vector largely if not completely suppressed the cohesion defect of hos1 Δ cells (Figure 5F).
Is Smc3 Acetylation Important for Maintaining Cohesion?
The observation that Smc3 acetylation generated during DNA replication is only removed when sister chromatid cohesion is dissolved at the metaphase to anaphase transition raises the possibility that acetylation might be important for maintaining cohesion in post-replicative cells. This notion is still consistent with the finding that Eco1 is unnecessary for maintaining cohesion (Skibbens et al., 1999; Toth et al., 1999), because acetylation of K112 and K113 appears to be very stable. For example, modification inherited from a previous cell cycle persists for several hours in the absence of Eco1 and Hos1 activity (Figure 5D), and deacetylation mediated by Hos1 is largely if not completely delayed until Scc1 cleavage.
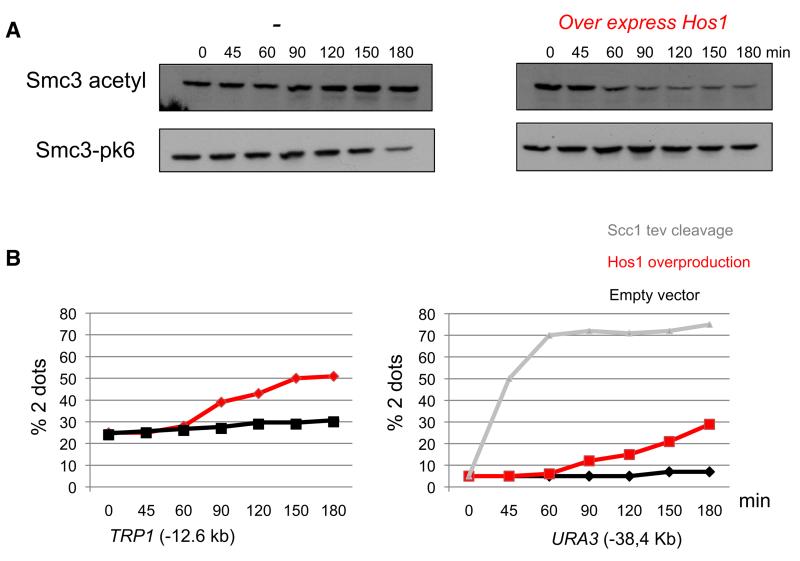
To address the role of Smc3 acetylation in maintaining cohesion, it is necessary to engineer a reduction in post-replicative cells. We have previously reported that shifting eco1-1 cells arrested in nocodazole to the restrictive temperature causes a modest decline in acetylation after two hours (Rowland et al., 2009), admittedly from already low levels, an effect possibly due to weak Hos1 activity even on intact cohesin rings associated with chromatin. This experiment did not however measure the effect on cohesion. The possibility that Hos1 might be partially active in mitotic cells led us to explore whether Hos1 overexpression can accelerate this process and induce more quantitative deacetylation. Figure 6A shows that overexpression of HOS1 from the GAL1-10 promoter induces a rapid albeit incomplete reduction in Smc3 acetylation in cells arrested in metaphase by depleting Cdc20. Deacetylation was accompanied by loss of cohesion not only at the TRP1 locus 12.6 kb from CEN4 but also at the URA3 locus 38 kb from CEN5 (Figure 6B). Loss of cohesion at URA3 was more modest than that caused by quantitative Scc1 cleavage triggered by expression of TEV protease in cells whose Scc1 contained three tandem TEV recognition sequences (Figure 6B). At present, we cannot be certain that deacetylation of Smc3 (as opposed to another cohesin subunit or protein) is responsible for the loss of cohesion or indeed whether the persistence of some cohesion is due to the incomplete deacetylation caused by Hos1 overexpression. Nevertheless, these data are consistent with the notion that Smc3 acetylation is important for maintaining cohesion in postreplicative cells.
Figure 6. Acetylation Stabilizes Cohesion.
Strains K17513 (256LacO::TRP1, PMET3CDC20, YEp-PGAL1HOS1, SMC3-PK6), K17514 (256La-cO::TRP1, PMET3CDC20, YEp-PGAL1, SMC3-PK6), K17383 (tetO112::URA3, PMET3CDC20, YEp-PGAL1HOS1), K17384 (tetO112::URA3, MET3-CDC20, YEp-PGAL1), and K9027 (MATα, te-tO112::URA3, PMET3CDC20, scc1Δ, SCC1-TEV268-HA3, GAL-TEV) were grown to log phase in synthetic medium lacking methionine and arrested in metaphase for 2 hr in YEP medium sup-plemented with 2 mM methionine and 2% raffinose (0 min). Expression of Hos1 was induced by addition of 2% galactose (0-180 min).
(A) Proteins from the strains K17513 and K17514 were extracted, and the levels of Smc3 acetylation and Smc3-PK6 at the indicated time points were monitored by western blotting of crude extracts using α-pk and α-acetylated Smc3 antibodies.
(B) Cohesion was measured by scoring the percentages of separated GFP dots at the TRP1 (12.6 kb from CEN4) and URA3 (38.4 kb from CEN5) loci.
DISCUSSION
Smc3 Is Acetylated during DNA Replication
Acetylation of cohesin’s Smc3 subunit is essential for sister chromatid cohesion. This modification is tightly cell cycle-regulated, increasing as cells enter S phase and declining as they undergo anaphase. Three lines of evidence indicate that the bulk of acetylation occurs not when cohesin associates with chromatin in late G1 (or in G2/M) but rather during the process of DNA replication itself. Smc3 acetylation is reduced in cells that enter the cell cycle without Cdc6 (Rolef Ben-Shahar et al., 2008), a factor required for prereplication complex formation, delayed in cells in which origin firing is retarded by expressing Dbf4 from the SWI5 promoter, and greatly reduced or absent when cohesin associates with DNA outside S phase, during either G2/M or G1 phase (Figures 1 and 2C).
It has been suggested that replication-specific acetylation is due to the association of Eco1 with PCNA polymerase clamps (Moldovan et al., 2006) placed at forks in large part by an “alternative” RFCCtf18 clamp loader (Lengronne et al., 2006; Mayer et al., 2001). It remains unclear whether this is indeed the mechanism, because the inactivation of RFCCtf18 by deleting CTF8 causes only a modest reduction in Smc3 acetylation. Furthermore, there is no convincing evidence that Eco1 is associated with forks in vivo (Lengronne et al., 2006).
A Type I Deacetylase Removes Smc3 Acetylation
We show here that Smc3 deacetylation is triggered by cleavage of cohesin’s Scc1 subunit at the onset of anaphase and that the enzyme responsible is a histone deacetylase called Hos1-Figure 7. Deacetylases belong to three main classes: the Rpd3, Hos1/Hos2-like (class I) enzymes, the Hda1-like (class II) enzymes and the Sir2-like (class III) enzymes. The main functions of type I enzymes such as Hos1 were thought to be in the control of transcription by deacetylating histones. However, members of this family are found in bacteria (Mycoplana bullata) and hyper-thermophilic bacteria (Aquifex aeolicus), which lack histones (Jackman et al., 2000; Leipe and Landsman, 1997). Moreover, it has been suggested that HDAC1 and SIRT1 deacetylate and thereby inactivate phosphatase and tensin homolog, a phosphatase that counteracts PI3 kinase (Ikenoue et al., 2008; Yao and Nyomba, 2008). Our finding that Hos1 is responsible for deacetylating Smc3 provides definitive evidence for a function for type I enzymes besides histone deacetylation.
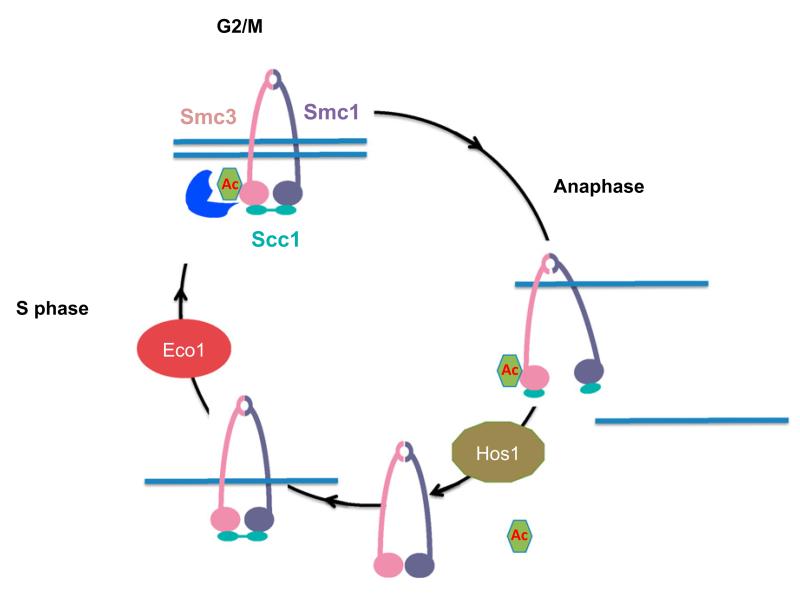
Figure 7. Smc3 Acetylation Cycle.
During S phase, Eco1 acetylates Smc3. This reaction is required for the establishment of sister chromatid cohesion. In G2/M phases, cohesion has been established, and we propose that Smc3 acetylation may act to stabilize it. At the metaphase-to-anaphase transition, separase cleavage of Scc1 allows Hos1 to deacetylate Smc3.
Regulation of Smc3 Deacetylation
Deacetylation of Smc3 is tightly cell cycle–regulated, taking place largely if not exclusively at the onset of anaphase in a manner dependent on cleavage of cohesin’s Scc1 subunit by separase. Our finding that deacetylation can also be triggered during metaphase by artificial Scc1 cleavage confirms that the process is regulated by the state of cohesin and not by cell cycle control of Hos1 activity. We envision two possible mechanisms. Cleavage may trigger a change in the conformation of cohesin that exposes acetylated K112 and K113 residues to Hos1. Alternatively, the crucial event may not be cleavage per se but rather cohesin’s release from chromatin, which is induced by cleavage. This raises the possibility that some protein or protein domain associated with acetylated Smc3 molecules inhibits deacetylation, while cohesin rings are tightly associated with chromatin. This putative Hos1 inhibitor could be a known cohesin subunit, like Scc3 or Pds5, which are required to stabilize cohesion in postreplicative cells and might do so in part by preventing Smc3 deacetylation. Another candidate is the N-terminal domain of Scc1, whose association with Smc3 NBDs might be destabilized by Scc1 cleavage.
A Cycle of Acetylation Is Essential
Our finding that hos1 Δ mutants have reduced sister chromatid cohesion indicates that Smc3 molecules that fail to be deacetylated might not be able to build cohesion during S phase. We tested this hypothesis by asking whether inactivation of Hos1, which ensures that daughter cells are born with a large pool of acetylated Smc3 molecules, enables cells to establish cohesion in the absence of Eco1 activity during the succeeding S phase. Instead of ameliorating the cohesion defect caused by the lack of de novo acetylation, Hos1 inactivation in fact exacerbates it. We cannot be certain that this effect is merely due to a failure to deacetylate Smc3, because Hos1 and Eco1 might have additional targets. However, the fact that mutations replacing K113 by threonine or asparagine permits yeast cells to build cohesion in the absence of Eco1 (Rolef Ben-Shahar et al., 2008; Rowland et al., 2009) is consistent with the notion that K112 and K113 might be the only crucial targets, as is our finding that overexpression of Smc1 and Smc3 suppresses the cohesion defect of hos1 Δ mutants. One explanation for our results is that deacetylation of Smc3 following Scc1 cleavage might be required to regenerate tripartite rings. For example, deacetylation might conceivably be required to release N-terminal Scc1 cleavage fragments from Smc3 NBDs, a process without which new rings would not be reformed in the next cell cycle. An alternative is that cohesin rings whose Smc3 subunit is acetylated may not be capable of building cohesion during S phase. In other words, the process of cohesion establishment may require unacetylated complexes as a crucial intermediate. If, for example, Smc3 acetylation helps to lock rings shut, then this locking could compromise the process of DNA entrapment inside rings, either before or during DNA replication. It is conceivable that a domain of some cohesin protein binding to unacetylated K112 and K113 has an important role in loading cohesin onto chromosomes in preparation for cohesion establishment. Future experiments should address whether the failure to deacetylate Smc3 compromises reformation of intact rings after anaphase, loading of cohesin onto chromatin in late G1 phase, or some event during establishment of cohesion during S phase.
Does Acetylation Stabilize Cohesion?
The mechanistic consequences of K112 and K113 acetylation are poorly understood. For example, it is unclear whether the modification facilitates DNA entrapment by cohesin rings during S phase or merely stabilizes this state immediately after it has been established. Our finding that Hos1 cannot deacetylate Smc3 until Scc1 is cleaved suggests that the acetyl marks at K112 and K113 are stable and are not turned over appreciably between DNA replication and anaphase. Thus, the acetylation mark could indeed have a postreplicative role. What might this be? The observation that overproduction of Hos1 in M phase cells induces rapid albeit incomplete Smc3 deacetylation that is accompanied by a reduction of sister chromatid cohesion is possibly an important clue that acetylation actively stabilizes cohesion. We cannot at this stage be certain that the loss of cohesion is due to the reduction in Smc3 acetylation as opposed to some other unknown target. Nevertheless, the notion that acetylation locks cohesin in a cohesive state (Unal et al., 2008) now warrants serious consideration. Indeed, locking cohesin into such a state might explain why acetylated Smc3 molecules cannot build cohesion in the first place. There is an interesting corollary of this idea. If K112 and K113 acetylation locks cohesin onto chromatin by preventing escape of DNAs trapped inside its ring, and if deacetylation depends on cohesin’s dissociation from chromatin, then the acetylated state will be naturally self sustaining, a perfect attribute for a lock. Evaluating whether such a locking function is an essential aspect of acetylation (possibly even its sole function) or merely an important one that improves the fidelity of chromosome segregation will require further experiments that achieve more complete deacetylation within a short time frame.
How might acetylation lock cohesive structures? One possibility is that it inhibits the binding and/or hydrolysis of adenosine triphosphate and thereby prevents reopening of cohesin rings. Another is that it provides a mark for the binding of other proteins required to keep cohesin rings firmly closed, possibly Scc3 or Pds5. It is curious in this regard that acetylation of Scc1 and not Smc3 is thought to be required to establish cohesion in G2 cells following DNA damage (Heidinger-Pauli et al., 2008). If acetylation does indeed lock cohesive structures, it would appear that locking can be achieved by multiple mechanisms. It is important to note that a function in locking cohesin rings does not preclude the possibility that acetylation also facilitates formation of cohesive structures.
EXPERIMENTAL PROCEDURES
ChIP-qPCR Analysis
ChIP-qPCR was performed with 3 μg of antibodies of α-pk antibodies (SV5-Pk1 from Serotec). qPCR was run using a Corbett Rotorgene cycler. The primers pairs used for chromosome VI were: -DEG1 (centromere), 5′-GCGGCCTTAAGTTCGTAGTG-3′ and 5′-AAGTGCCGGAAATTGTCTTG-3′; -SPB4 (pericentromere), 5′-GACGAAAGAACGGAAACTCG-3′ and 5′-CCTTGGATAGCTTTGCTGGA-3′; and CMK1 (inner arm), 5′-ACGGTTCAGTTCCTCCATTG-3′ and 5′-TGCAAAAGCTTTGCTGGTTA-3′.
Analysis of Smc3 Acetylation
A pellet from 109 cells was frozen in liquid nitrogen and stored at −80°C over-night. The cell pellet was resuspended in 400 μl extraction buffer (50 mM HEPES/KOH [pH 7.5], 5 mM EDTA, 150 mM NaCl, 0.25% NP40, 1 mM DTT, 10 mM sodium butyrate, protease inhibitors) and broken with glass beads. The soluble fraction was separated from the insoluble fraction by two consecutive centrifugations, each at 15,000 rpm and 4°C for 15 min.
In the case of Smc3-pk immunoprecipitation, extractions were clarified by centrifugation, and further cleared by incubation with immunoglobulin G sepharose beads (GE Healthcare) on a rotating wheel for 1 hr. A total of 3 μg antibody against the Smc3-Pk3 epitope was added for 1 hr (clone SV5-Pk1, Serotec), and immunocomplexes were isolated by adsorption to protein A sepharose beads (GE Healthcare) for 2 hr. Beads were washed extensively in extraction buffer, and immunocomplexes were recovered by incubation with elution buffer (50 mM Tris/HCl [pH 8.0], 10 mM EDTA, 1% SDS) for 15 min at 65°C. Eluates were analyzed by SDS-PAGE followed by western blotting with α-2 Pk and α-acetyl lysine antibodies (ST1027, Calbiochem) or with a mouse monoclonal α-Smc3 acetylated antibody.
To immunoprecipitate Smc3-HA or Scc1-HA, extractions were incubated with 60 μl anti-HA matrix (Sigma or Roche) for 90 min. To check the HA immunoprecipitation, western blotting was performed with α-12CA5 antibody.
GFP Dot Assay
Approximately 2 ×107 cells were pelleted at 10,000 rpm for 30 s. The cells were resuspended in 100 μl of 4% paraformaldehyde/3.4% sucrose and incubated for 15 min at room temperature. After fixation, cells were pelleted at 10,000 rpm for 30 s and resuspended in 100 μl KPO4/sorbitol with 1% Triton-X and incubated at room temperature for 4 min. Triton-X was subsequently washed away with 100 μl KPO4/sorbitol, and the cells were resuspended in 70 μl KPO4/sorbitol. GFP fluorescence was observed with a Zeiss Axio Imager.Z1 microscope (63× objective, NA = 1.40) equipped with a cool-SNAP HQ camera. For each experimental condition, at least 100 cells were scored for GFP dots.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to members of K.N.’s laboratory for useful discussions. Special thanks to S. Dixon and W. Upcher for critical reading of the manuscript. K.S. was supported by a grant from the Cell Innovation Program and Grant-in-Aid for Scientific Research (S) from the Ministry of Education, Culture, Sports, Science and Technology (Tokyo, Japan). K.N. was supported by Cancer Research UK and the Wellcome Trust.
Footnotes
Supplemental Information includes two figures and one table and can be found with this article online at doi:10.1016/j.molcel.2010.08.008.
REFERENCES
- Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- Davie JK, Edmondson DG, Coco CB, Dent SY. Tup1-Ssn6 interacts with multiple class I histone deacetylases in vivo. J. Biol. Chem. 2003;278:50158–50162. doi: 10.1074/jbc.M309753200. [DOI] [PubMed] [Google Scholar]
- Gambus A, van Deursen F, Polychronopoulos D, Foltman M, Jones RC, Edmondson RD, Calzada A, Labib K. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009;28:2992–3004. doi: 10.1038/emboj.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- Haering CH, Farcas A, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNAs. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Lowe J. Structure and stability of cohesin’s Smc1-kleisin interaction. Mol. Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Unal E, Koshland D. Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol. Cell. 2009;34:311–321. doi: 10.1016/j.molcel.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Unal E, Guacci V, Koshland D. The kleisin subunit of cohesin dictates damage-induced cohesion. Mol. Cell. 2008;31:47–56. doi: 10.1016/j.molcel.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Ikenoue T, Inoki K, Zhao B, Guan KL. PTEN acetylation modulates its interaction with PDZ domain. Cancer Res. 2008;68:6908–6912. doi: 10.1158/0008-5472.CAN-08-1107. [DOI] [PubMed] [Google Scholar]
- Ivanov D, Schleiffer A, Eisenhaber F, Mechtler K, Haering CH, Nasmyth K. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr. Biol. 2002;12:323–328. doi: 10.1016/s0960-9822(02)00681-4. [DOI] [PubMed] [Google Scholar]
- Jackman JE, Fierke CA, Tumey LN, Pirrung M, Uchiyama T, Tahir SH, Hindsgaul O, Raetz CR. Antibacterial agents that target lipid A biosynthesis in gram-negative bacteria. Inhibition of diverse UDP-3-O-(r-3-hydroxymyristoyl)-n-acetylglucosamine deacetylases by substrate analogs containing zinc binding motifs. J. Biol. Chem. 2000;275:11002–11009. doi: 10.1074/jbc.275.15.11002. [DOI] [PubMed] [Google Scholar]
- Leipe DD, Landsman D. Histone deacetylases, acetoin utilization proteins and acetylpolyamine amidohydrolases are members of an ancient protein superfamily. Nucleic Acids Res. 1997;25:3693–3697. doi: 10.1093/nar/25.18.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol. Cell. 2006;23:787–799. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Gygi SP, Aebersold R, Hieter P. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol. Cell. 2001;7:959–970. doi: 10.1016/s1097-2765(01)00254-4. [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell. 2006;23:723–732. doi: 10.1016/j.molcel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 2009;43:525–528. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- Oliveira RA, Hamilton RS, Pauli A, Davis I, Nasmyth K. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nat. Cell Biol. 2010;12:185–192. doi: 10.1038/ncb2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, Grunstein M. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell. 2002;109:437–446. doi: 10.1016/s0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouet F, Underwood P, Metson J, Imre R, et al. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol. Cell. 2009;33:763–774. doi: 10.1016/j.molcel.2009.02.028. [DOI] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom L, Sjogren C. DNA damage-induced cohesion. Cell Cycle. 2005;4:536–539. doi: 10.4161/cc.4.4.1613. [DOI] [PubMed] [Google Scholar]
- Strom L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjogren C. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science. 2007;317:242–245. doi: 10.1126/science.1140649. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Katou Y, Yagura M, Saitoh K, Itoh T, Araki H, Bando M, Shirahige K. Ctf4 coordinates the progression of helicase and DNA polymerase alpha. Genes Cells. 2009;14:807–820. doi: 10.1111/j.1365-2443.2009.01310.x. [DOI] [PubMed] [Google Scholar]
- Toth A, Ciosk R, Uhlmann F, Galova M, Schleifer A, Nasmyth K. Yeast Cohesin complex requires a conserved protein, Eco1p (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. Sister chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1p. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin E, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Koshland D. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7) Science. 2007;317:245–248. doi: 10.1126/science.1140637. [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- Yao XH, Nyomba BL. Hepatic insulin resistance induced by prenatal alcohol exposure is associated with reduced PTEN and TRB3 acetylation in adult rat offspring. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008;294:R1797–R1806. doi: 10.1152/ajpregu.00804.2007. [DOI] [PubMed] [Google Scholar]
- Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell. 2008;31:143–151. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.