Abstract
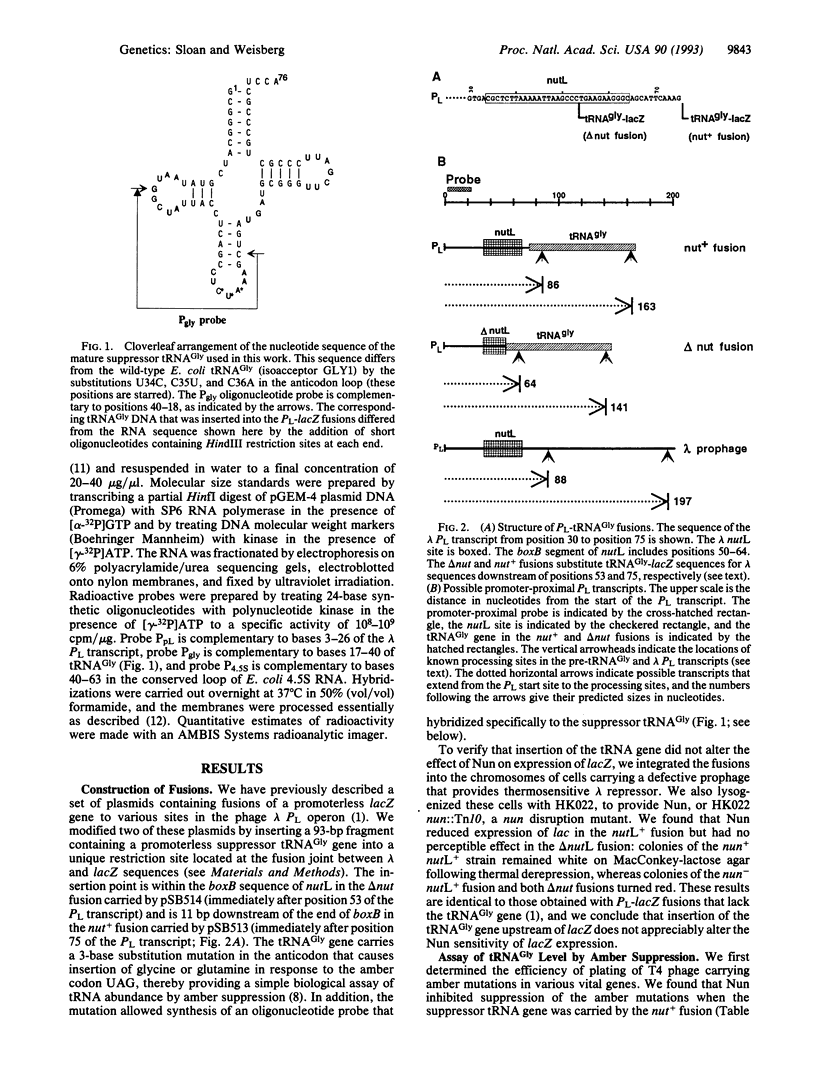
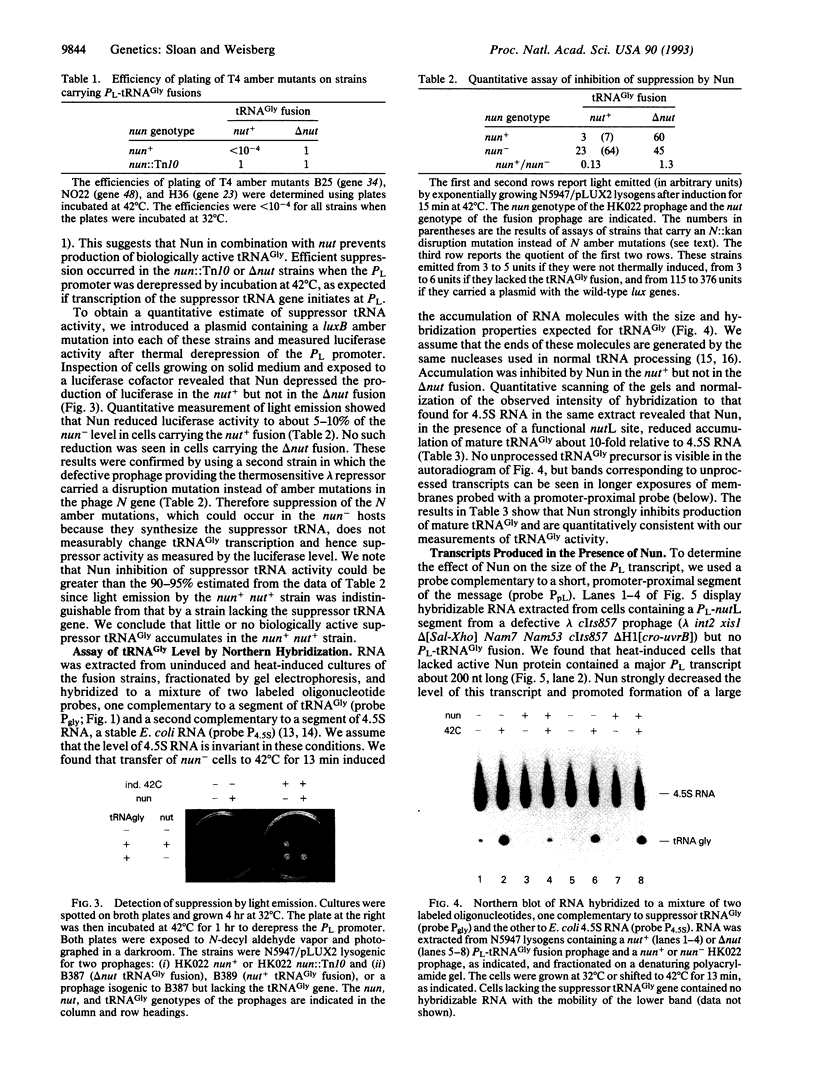
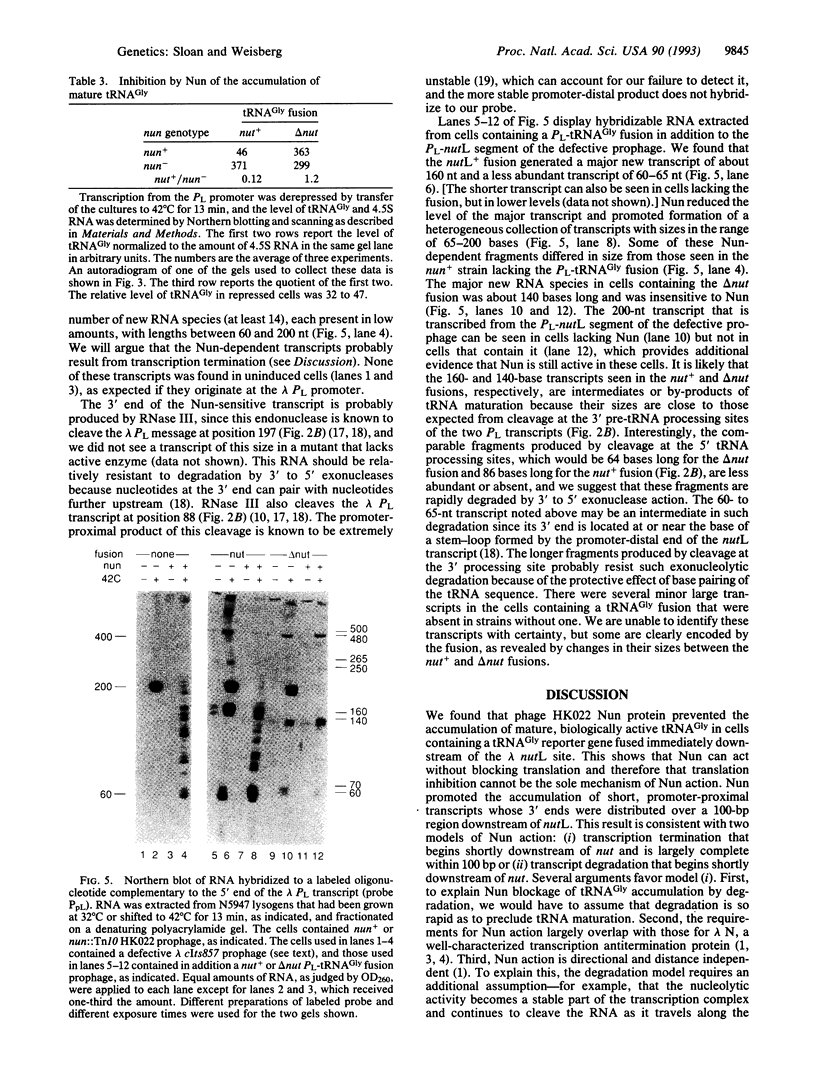
The Nun protein of phage HK022 blocks the expression of genes that lie downstream of the nut sites of phage lambda. Nun is believed to act by promoting premature termination of transcription at or near these sites. To test this hypothesis and to facilitate mapping the sites of termination, we inserted a gene encoding a suppressor tRNA immediately downstream of the lambda nutL site and determined the effect of Nun on tRNA level. We found that Nun severely reduced the accumulation of mature, biologically active tRNA and promoted the accumulation of short, promoter-proximal transcripts whose 3' ends were dispersed over a 100-nucleotide region downstream of nutL. These results are consistent with the hypothesis that Nun terminates transcription within the region immediately downstream of nutL and are inconsistent with the hypothesis that the only action of Nun is to prevent translation of genes located downstream of the nut site. The stability, small size, and easily assayable biological function of suppressor tRNA recommend it as a reporter of transcription in other systems.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Altman S., Kirsebom L., Talbot S. Recent studies of ribonuclease P. FASEB J. 1993 Jan;7(1):7–14. doi: 10.1096/fasebj.7.1.7916700. [DOI] [PubMed] [Google Scholar]
- Anevski P. J., Lozeron H. A. Multiple pathways of RNA processing and decay for the major leftward N- independent RNA transcript of coliphage lambda. Virology. 1981 Aug;113(1):39–53. doi: 10.1016/0042-6822(81)90134-3. [DOI] [PubMed] [Google Scholar]
- Bouvet P., Belasco J. G. Control of RNase E-mediated RNA degradation by 5'-terminal base pairing in E. coli. Nature. 1992 Dec 3;360(6403):488–491. doi: 10.1038/360488a0. [DOI] [PubMed] [Google Scholar]
- Brennan C. A., Dombroski A. J., Platt T. Transcription termination factor rho is an RNA-DNA helicase. Cell. 1987 Mar 27;48(6):945–952. doi: 10.1016/0092-8674(87)90703-3. [DOI] [PubMed] [Google Scholar]
- Brown S. 4.5S RNA: does form predict function? New Biol. 1991 May;3(5):430–438. [PubMed] [Google Scholar]
- Das A. How the phage lambda N gene product suppresses transcription termination: communication of RNA polymerase with regulatory proteins mediated by signals in nascent RNA. J Bacteriol. 1992 Nov;174(21):6711–6716. doi: 10.1128/jb.174.21.6711-6716.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehretsmann C. P., Carpousis A. J., Krisch H. M. mRNA degradation in procaryotes. FASEB J. 1992 Oct;6(13):3186–3192. doi: 10.1096/fasebj.6.13.1397840. [DOI] [PubMed] [Google Scholar]
- Emory S. A., Bouvet P., Belasco J. G. A 5'-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992 Jan;6(1):135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Peltz S. W., Jacobson A. Turnover of mRNA in prokaryotes and lower eukaryotes. Curr Opin Genet Dev. 1992 Oct;2(5):739–747. doi: 10.1016/s0959-437x(05)80134-0. [DOI] [PubMed] [Google Scholar]
- Hsu L. M., Zagorski J., Fournier M. J. Cloning and sequence analysis of the Escherichia coli 4.5 S RNA gene. J Mol Biol. 1984 Sep 25;178(3):509–531. doi: 10.1016/0022-2836(84)90236-5. [DOI] [PubMed] [Google Scholar]
- Jin D. J., Burgess R. R., Richardson J. P., Gross C. A. Termination efficiency at rho-dependent terminators depends on kinetic coupling between RNA polymerase and rho. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1453–1457. doi: 10.1073/pnas.89.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameyama L., Fernandez L., Court D. L., Guarneros G. RNaselll activation of bacteriophage lambda N synthesis. Mol Microbiol. 1991 Dec;5(12):2953–2963. doi: 10.1111/j.1365-2958.1991.tb01855.x. [DOI] [PubMed] [Google Scholar]
- Karn J., Graeble M. A. New insights into the mechanism of HIV-1 trans-activation. Trends Genet. 1992 Nov;8(11):365–368. doi: 10.1016/0168-9525(92)90284-b. [DOI] [PubMed] [Google Scholar]
- Lozeron H. A., Anevski P. J., Apirion D. Antitermination and absence of processing of the leftward transcript of coliphage lambda in the RNAase III-deficient host. J Mol Biol. 1977 Jan 15;109(2):359–365. doi: 10.1016/s0022-2836(77)80039-9. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Foss K., Jenkins R. A., Schneider J. Rapid determination of nucleotides that define tRNA(Gly) acceptor identity. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6147–6151. doi: 10.1073/pnas.88.14.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz S. W., Jacobson A. mRNA stability: in trans-it. Curr Opin Cell Biol. 1992 Dec;4(6):979–983. doi: 10.1016/0955-0674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- Robert J., Sloan S. B., Weisberg R. A., Gottesman M. E., Robledo R., Harbrecht D. The remarkable specificity of a new transcription termination factor suggests that the mechanisms of termination and antitermination are similar. Cell. 1987 Nov 6;51(3):483–492. doi: 10.1016/0092-8674(87)90644-1. [DOI] [PubMed] [Google Scholar]
- Robledo R., Atkinson B. L., Gottesman M. E. Escherichia coli mutations that block transcription termination by phage HK022 Nun protein. J Mol Biol. 1991 Aug 5;220(3):613–619. doi: 10.1016/0022-2836(91)90104-e. [DOI] [PubMed] [Google Scholar]
- Roland K. L., Powell F. E., Turnbough C. L., Jr Role of translation and attenuation in the control of pyrBI operon expression in Escherichia coli K-12. J Bacteriol. 1985 Sep;163(3):991–999. doi: 10.1128/jb.163.3.991-999.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmientos P., Sylvester J. E., Contente S., Cashel M. Differential stringent control of the tandem E. coli ribosomal RNA promoters from the rrnA operon expressed in vivo in multicopy plasmids. Cell. 1983 Apr;32(4):1337–1346. doi: 10.1016/0092-8674(83)90314-8. [DOI] [PubMed] [Google Scholar]
- Steege D. A., Cone K. C., Queen C., Rosenberg M. Bacteriophage lambda N gene leader RNA. RNA processing and translational initiation signals. J Biol Chem. 1987 Dec 25;262(36):17651–17658. [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981 Feb 26;289(5800):751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Yager T. D. The elongation-termination decision in transcription. Science. 1992 Feb 14;255(5046):809–812. doi: 10.1126/science.1536005. [DOI] [PubMed] [Google Scholar]