Abstract
Earthworms belonging to the family Lumbricidae are extremely abundant in terrestrial temperate regions. They affect soil properties and nutrient cycling, thus shaping plant community composition and aboveground food webs. Some lumbricids are also model organisms in ecology and toxicology. Despite the intense research efforts dedicated to lumbricids over the last 130 years, the evolutionary relationships and taxonomic classification of these organisms are still subject to great debate. Resolution of their systematics is hampered by the structural simplicity of the earthworm body plan and the existence of cryptic species. We sampled 160 earthworm specimens belonging to 84 lumbricid species (28 genera) and 22 Lumbricoidea outgroups, sequenced two nuclear genes, four mitochondrial genes and seven mitochondrial tRNAs and examined 22 morphological characters. We then applied a combination of phylogenetic methods to generate the first robust genus-level phylogeny of the Lumbricidae. Our results show that the current Lumbricidae classification and the underlying hypotheses of character evolution must be revised. Our chronogram suggests that lumbricids emerged in the Lower Cretaceous in the holarctic region and that their diversification has been driven by tectonic processes (e.g. Laurasia split) and geographical isolation. Our chronogram and character reconstruction analysis reveal that spermathecae number does not follow a gradual pattern of reduction and that parthenogenesis arose from sexual relatives multiple times in the group; the same analysis also indicates that both epigeic and anecic earthworms evolved from endogeic ancestors. These findings emphasize the strong and multiple changes to which morphological and ecological characters are subjected, challenging the hypothesis of character stasis in Lumbricidae.
Keywords: Earthworms, Lumbricidae, Bayesian divergence time analysis, Ancestral state reconstruction, Spermathecae, Endogeic ancestors
1. Introduction
Earthworms represent the main animal biomass in most terrestrial temperate ecosystems (Lavelle and Spain, 2001). Where abundant, earthworms significantly impact soil physical, chemical and biological properties, hence modifying soil structure and accelerating organic matter decomposition and nutrient turnover, and ultimately shaping plant community composition and aboveground food webs (Lee, 1985; Edwards and Bohlen, 1996; Lavelle et al., 2001).
Earthworms belonging to the family Lumbricidae (Rafinesque-Schmaltz, 1815) are the most abundant invertebrates in soils of temperate regions, where they account for 90% of the invertebrate biomass (Edwards, 2004). Lumbricids are also important in animal food webs and serve as prey for a wide range of invertebrate and vertebrate predators. Several species are also model organisms in ecology, toxicology, physiology and reproductive biology (Lavelle and Spain, 2001; Domínguez and Velando, 2013). Bioresources resulting from cultivation of lumbricids (vermiculture and vermicomposting) are also of great economic value and provide important environmental benefits, especially in light of global concerns regarding sustainable land use, food security and climate change (Domínguez et al., 2010).
Despite the biological and economic importance of lumbricids and the great amount of research on these species over the last 130 years, very little is known about their evolutionary relationships (Pop et al., 2003). The Lumbricidae encompasses around 300 known species and is considered a monophyletic group included in the monophyletic Crassiclitellata (comprising all earthworms except Moniligastridae) (Jamieson, 1988, 2006; James and Davidson, 2012), which are united by the presence of a multilayered clitellum. However, there is no consensus on the classification of Lumbricidae, with proposals ranging from 6–14 genera in historical studies (Bouchè, 1972; Omodeo, 1956; Pop, 1941) to 31–45 genera in modern revisions (Blakemore, 2008; Csuzdi and Zicsi, 2003; Mrsic, 1991; Qiu and Bouché, 1998a). More importantly, all of the proposed classifications are somewhat “intuitive” rather than based on explicit phylogenetic analyses.
Lumbricidae taxonomy has always been hindered by the structural simplicity of the earthworm body plan that lacks complex appendages or highly specialized copulatory apparatuses (Pop et al., 2003; Pérez-Losada et al., 2009). Approximately 100 morphological characters have been routinely used for species identification, but they have never been included in a phylogenetic framework to infer lumbricid evolutionary relationships. Variation in the few morphological features used to identify lumbricids (e.g. prostomium, arrangement of the setae, position and form of the clitellum, tubercula pubertatis, and spermathecae) usually overlaps across both closely and distantly related taxa (Pérez-Losada et al., 2009; Pérez-Losada et al., 2011; Briones et al., 2009; Pop et al., 2003). This has led to the proposition of multiple morphotypes for the same species, as well as to the creation of invalid species complexes (Bouchè, 1972; Sims, 1983; Gates, 1972) and “catchall” genera (Briones et al., 2009; Pop et al., 2005). Although earthworm morphological systematics seems unreliable, no morphological matrix has been developed for the Lumbricidae and tested in a phylogenetic framework. Hence, the usefulness of earthworm morphology for inferring Lumbricidae evolutionary relationships and validating Lumbricidae taxonomy remains undetermined.
Over the last ten years several molecular phylogenies including some Lumbricidae taxa have been published (Pérez-Losada et al., 2005; Pérez-Losada et al., 2009; Pérez-Losada et al., 2011; Novo et al., 2011; Fernández et al., 2012) and some systematic issues (e.g., systematics of Eisenia fetida and Eisenia andrei, systematics of the Aporrectodea caliginosa species complex and Postandrilus radiation) have been solved. However, so far no comprehensive, robust phylogeny of the Lumbricidae has been produced and the few attempts published have been limited by their restricted taxonomic sampling and/or the low phylogenetic signal of the chosen genes (Pop et al., 2003; Briones et al., 2009; Pérez-Losada et al., 2012). The aforementioned studies have unanimously revealed systematic instability within the family (e.g. many genera do not form monophyletic assemblages) and have emphasized the urgent need for a classification reflecting evolutionary relationships.
The family Lumbricidae has a Holarctic distribution, naturally occurring in eastern and central USA, temperate Europe, and western and central Asia (Gates, 1972; Mrsic, 1991). Like other Crassiclitellata families, lumbricids are believed to be an old lineage (Buckley et al., 2011; James and Davidson, 2012) which distribution has been at least partly determined by paleogeography (e.g. Lee, 1994; Omodeo, 2000; James, 2004); although the patterns of diversification and times of divergence of the main Lumbricidae clades remain unknown. This raises general questions as to whether Lumbricidae lineages are correlated with their current geographic distributions and whether current natural distributions are good indicators of vicariant events and/or of past land area relationships (e.g. colonization of North America). An integrative approach combining molecular phylogenies and external information in the form of calibrations (e.g. fossils) and species distribution may help to address these questions. Unfortunately, the earthworm fossil record is very poor and other sources of evidence must be used as calibrations trees for time divergence estimation.
Although most lumbricids are hermaphrodites, some are parthenogenic and lack functional testes. This is often correlated with the presence or absence of spermathecae – a female reproductive organ in the hermaphroditic earthworms that stores the spermatozoa from the partner during copulation for later fertilization of the eggs (Velando et al., 2008; Domínguez and Velando, 2013). It has been hypothesized that following the evolution of parthenogenesis, the spermathecae and male reproductive organs tend to decrease in size and number (Gates, 1972). Hence, a robust lumbricid phylogeny is essential for understanding the evolution of spermathecal loss and reproductive strategies (hermaphroditism or parthenogenesis) in the Lumbricidae.
Ecologically, lumbricids are classified on the basis of their feeding and burrowing habits into three general ecotypes (Bouché, 1977; Lee, 1985): (i) epigeic earthworms, which live above mineral soil, rarely form burrows and feed preferentially on plant litter; (ii) endogeic earthworms, which forage below the surface soil, ingest large quantities of mineral soils and humified material, and build ramified, predominantly horizontal, burrows; and (iii) anecic earthworms, which build permanent vertical burrows deep into the mineral soil layer and come to surface to feed on decomposed plant litter, manure or other organic residues. A robust phylogeny is instrumental for testing whether feeding and burrowing habits have driven Lumbricidae evolution or whether adaptation to these three ecotypes has evolved multiple times.
The main aim of the present study was to develop a robust genus-level phylogeny of the family Lumbricidae using multiple gene regions and morphological characters. Then we will use that phylogeny to validate Lumbricidae taxonomy, time its radiation, and study both its geographical distribution and the evolution of its reproductive and ecological strategies.
2. Material and methods
2.1. Earthworm sampling
Sampling of the family Lumbricidae included 160 specimens, representing 84 species and 28 genera (Table S1 in supplementary material). Earthworms were collected in Spain, Andorra, France, Italy, Germany, United Kingdom, Finland, Denmark, Poland, Romania, Hungary, Serbia, Israel, Austria, Turkey, South Africa, USA, Brazil, China and Vietnam. Nearly all currently recognized genera are represented in the study, with the exception of five monospecific genera and four small genera (<10 species), with very restricted distributions. To root the Lumbricidae tree we used representatives of five other Crassiclitellata families including: Criodrilidae (1 species), Hormogastridae (2 species), Glossoscolecidae (7 species), Megascolecidae (5 species) and Microchaetidae (7 species).
All earthworm specimens were identified following descriptions in Bouchè (1972), Mrsic (1991), Qiu and Bouché (1998a, 1998b, 1998c), Sims and Gerard (1999), Csuzdi and Zicsi (2003) and Blakemore (2006). Specimens or tissues collected for DNA extraction were preserved in absolute ethanol and stored at −20 °C.
2.2. DNA isolation and sequencing
Total genomic DNA was extracted using the DNAeasy Tissue kit (Qiagen). Recent phylogenetic analyses (Pérez-Losada et al., 2009, 2011; Novo et al., 2011) have demonstrated the importance of using multiple genes for inferring earthworm evolutionary relationships. Hence, regions of the nuclear 28S rDNA and 18S rDNA and mitochondrial 16S rDNA, 12S rDNA, NADH dehydrogenase (ND1), cytochrome oxidase subunit II (COII) and tRNA Asn, Asp, Val, Leu, Ala, Ser, and Leu genes were amplified using the polymerase chain reaction (PCR), as described in Pérez-Losada et al. (2009). PCR products were purified using a MultiScreen PCRl96 (Millipore) kit and sequenced bidirectionally using an Applied Biosystems (ABI) 377XL automated sequencer. The ABI Big-dye Ready-Reaction kit was used following the standard cycle sequencing protocol, but with a 16th of the suggested reaction size. DNA sequences were deposited in GenBank under the Accession Numbers KJ911919–KJ912618.
2.3. Morphological data
A total of 22 morphological characters, all commonly used in earthworm alpha-taxonomy, were scored for 82 lumbricids and 20 representatives of the other Crasiclitellata families (Table S2 in supplementary material) using available species descriptions in Bouchè (1972), Mrsic (1991), Qiu and Bouché (1998a, 1998b, 1998c), Sims and Gerard (1999), Csuzdi and Zicsi (2003) and Blakemore (2006). Only one specimen per species was chosen to represent the species diversity. We considered the following external characters (Table S2 in supplementary material): number of body segments, body pigmentation, type of prostomium (a lobe overhanging the mouth), arrangement of the setae, presence of dorsal pores (small openings situated in the intersegmental grooves on the mid-dorsal line) and position of the first dorsal pore, arrangement of the nephridiopores, location of the male pores and presence of associated tumescences; type, start and end, and number of segments of the clitellum (glandular swelling involved in cocoon formation), and presence and length of the tubercula pubertatis (skin fold associated with sperm transfer to partners). We considered the following internal characters (Table S2 in supplementary material): number of seminal vesicles (sperm maturation and storage sacs), number of spermathecae (allosperm storage sacs), presence of prostate glands (add secretions to the seminal fluid), presence, location and extension of the gizzard and presence of extramural calciferous glands. Twelve characters were binary and ten were unordered multistate with up to eight states (Table S2 in supplementary material). The symbol ‘?’ was assigned in all cases where the character state was unknown.
2.4. Phylogenetic analysis
Nucleotide sequences from tRNAs were combined and all sequences from each gene region were aligned in MAFFT v6 (Katoh et al., 2005; Katoh, 2008) under the global (G-INS-i) algorithm and default settings. Alignment quality was assessed in GBlocks v0.91b (Castresana, 2000). Full and conserved (after GBlocks analysis) alignments generated similar maximum likelihood trees for all genes, hence full alignments were used in all subsequent phylogenetic analyses. Phylogenetic congruence among mitochondrial (COII: 689 bp, 12S: 351 bp, 16S: 1409 bp, ND1: 930 bp, tRNAs: 713 bp; bp of aligned DNA) genes and the two nuclear genes 18S (809 aligned bp) and 28S (965 aligned bp) was assessed using Wiens’ (1998) protocol. No areas of strongly supported incongruence were observed among gene trees. Three different datasets were assembled and analyzed: (i) DNA dataset including all seven gene regions (seven data partitions), (ii) DNA + morphological dataset (eight data partitions), and (iii) morphological dataset (one data partition). JModelTest v1.0.1 (Posada, 2009) was used to select the appropriate model of evolution for each DNA partition under the Akaike Information Criterion AIC (Posada and Buckley, 2004). The general time reversible model of evolution (Tavaré, 1986), with proportion of invariable sites and gamma distribution, was selected for each partition (GTR + G + I). Morphological data were analyzed using a simple model analogous to the JC model (Jukes and Cantor, 1969) (equal substitution rates), except that it has a variable number of states (two to eight in our case).
Both maximum likelihood (ML) and Bayesian methods of phylogenetic inference were applied to the three datasets. ML analysis was performed in GARLI under default settings for the genetic algorithm, except that searchreps = 10. Clade support was assessed using the non-parametric bootstrap procedure (Felsenstein, 1985) with 1000 bootstrap replicates. Bayesian analysis coupled with Markov chain Monte Carlo (BMCMC) inference was performed in MrBayes v3.1.2 (Ronquist and Huelsenbeck, 2003). Four independent BMCMC analyses were run in the CIPRES Science Gateway portal (Miller et al., 2010), each consisting of four chains. Each Markov chain was started from a random tree and run for 2 × 107 cycles, with sampling every 1000th generation. Sequence evolution model parameters were estimated independently for each data partition starting as unknown variables with uniform default priors. Convergence and mixing were monitored using Tracer v1.5 (Rambaut and Drummond, 2009). All sample points prior to reaching stationary levels were discarded as burn-in. The posterior probabilities for individual clades obtained from separate analyses were compared for congruence and then combined and summarized on a 50% majority-rule consensus tree.
Confidence in our best hypotheses of phylogenetic relationships was tested by first creating alternative hypotheses (e.g. Dendrobaena octaedra is monophyletic) in MacClade, as indicated by Pérez-Losada et al. (2004), and then comparing them under both likelihood and Bayesian frameworks. Likelihood topological tests were conducted using the Shimodaira and Hasegawa (1999) test as implemented in RAxML v7.2.0 (Stamatakis et al., 2008). Bayesian topological tests were performed as described in Huelsenbeck et al. (2002).
2.5. Divergence time estimation
Divergence times were estimated in BEAST under a relaxed lognormal clock (Drummond et al., 2006) for each gene partition and a Yule speciation prior. Since earthworm fossil record is unknown, other sources of evidence must be used as calibrations to estimate chronograms. To calibrate the Lumbricidae molecular tree, we used two calibrations. Thus, for example, some lumbricids of the genus Postandrilus are restricted to the island of Majorca in the western Mediterranean, of known geological history (Rosenbaum et al., 2002). Hence, as in Pérez-Losada et al. (2011), we used the cladogenic event separating the Baleares from the proto-Iberian Peninsula (Late Oligocene; 30–28 Mya) (Alvarez et al., 1974; Rosenbaum et al., 2002) to time the split of Aporrectodea morenoe and Postandrilus bertae from Postandrilus majorcanus and Postandrilus sapkarevi. This calibration was integrated into the analysis as a normal prior (mean = 29 Myr; SD = 3; 95% interval = 24.1–33.9 Myr). We also calibrated the root of the tree using a lognormal prior (soft bounds) of log (mean) = 0.0, minimum age (offset) = 200 Myr and log (SD) = 2.5, which gives the root a 95% interval of 200–261.1 Myr. We used this calibration to represent the minimum age of the main earthworm lineages in our tree, which are considered of Gondwanan origin (James and Davidson, 2012). We used the gene evolutionary rates estimated in Pérez-Losada et al. (2011) as initial values for the uniform priors chosen for the lognormal relaxed clock rates in our BEAST analyses. Two runs 2 × 107 generations long were completed and combined using LogCombiner. All the output generated by BEAST was analyzed in Tracer v1.5 and a chronogram was depicted in FigTree. One-hundred and twenty-one of the 127 estimated parameters presented effective sample size (ESS) values >200.
2.6. Evolution of morphological and ecological characters
We assessed the evolutionary history of the lumbricid spermathecae and ecotypes by using the Bayesian approach implemented in BEAST v1.7.4 (Drummond and Rambaut, 2007). The spermathecae included eight states (0, 1, 2, 3, 5, 6, 7 and ≥8 pairs), whereas the ecotypes included three states (epigeic, endogeic and anecic). The error associated with the characters under study (mapping uncertainty) was taken into account by estimating posterior probabilities for the ancestral states. DNA partitions were analyzed separately using the GTR + G + I model of nucleotide substitution, a Yule speciation prior – a traditional speciation model for species-level data (Drummond et al., 2006), and the relaxed lognormal molecular clock rate variation model. We used a symmetric model of trait substitution with an approximate continuous time Markov chain rate reference for the trait.clock.rate (Ferreira and Suchard, 2008). Such prior estimation is recommended when explicit prior information is unavailable. An exponential prior (mean = 1 change/Myr) was also tested, but no significant differences in character state posterior probabilities were observed. Two independent runs 2 × 107 generations long were performed and then combined in LogCombiner v1.7.4 (part of the BEAST package). Convergence and mixing was evaluated in Tracer v1.5. Phenotypic characters were annotated in TreeAnnotator v1.7.4 (part of the BEAST package) and visualized in FigTree v1.3.1.
3. Results
The DNA and DNA + morphology ML and Bayesian trees, including all of the 182 taxa used in the analysis, are presented in Figs. S3–S6 in supplementary material. Summary versions of these trees are shown in Figs. 1–4 to facilitate the visualization and comparison of Lumbricidae evolutionary relationships within and among datasets. The summary trees included only the three closest outgroup species (Criodrilidae and Hormogastridae; see below) to the ingroup and 86 Lumbricidae taxa representing conspecific monophyletic assemblages in the full trees. The chronogram in Fig. 4 included all outgroups, so the ages of the deeper Lumbricina nodes (root included) could be compared.
Fig. 1.
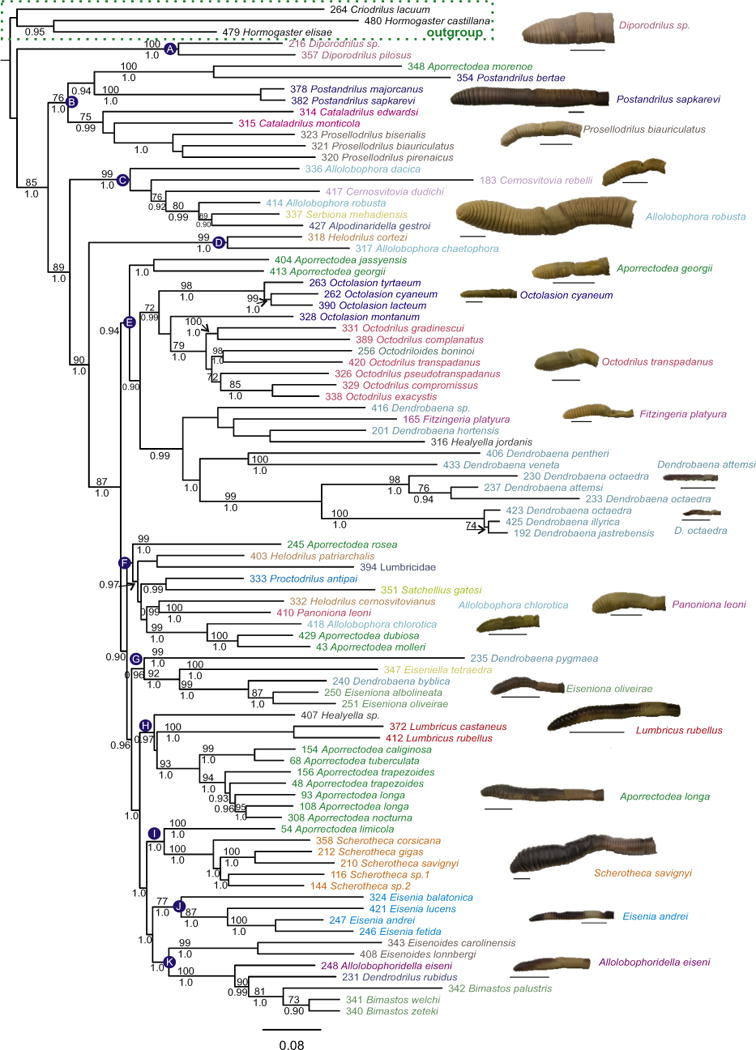
Lumbricidae maximum likelihood molecular tree. Bootstrap proportions (if ≥70%) and Bayesian posterior probabilities (if ≥95%) are shown above and below the branches, respectively. Selected taxa were pruned to facilitate visualisation of lumbricid relationships. The full tree is shown in Fig. S3 in supplementary material. Letters A–K refer to clades discussed in the text. Specimen photographs for selected lineages are also shown (bar = 1 cm). Code numbers are provided for each specimen.
Fig. 4.
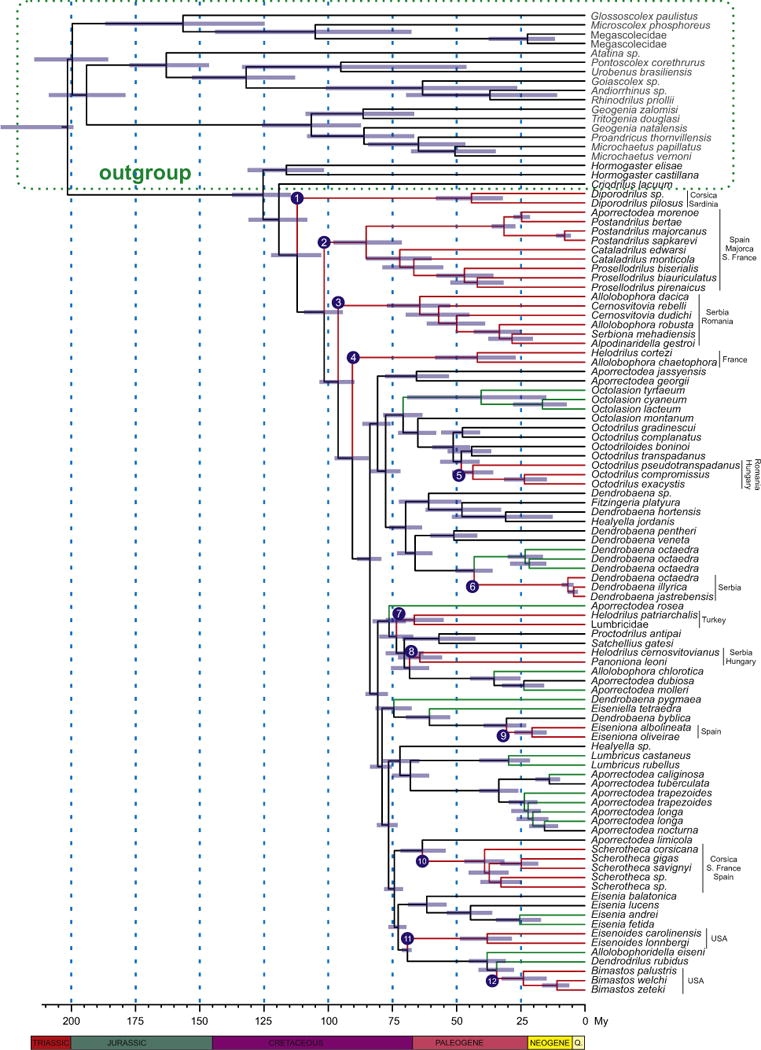
Lumbricidae chronogram. Node bars represent 95% confidence intervals for the divergence time estimates. Red and green branches represent species with restricted distributions and peregrine earthworms, respectively. Node numbers refer to the geographical distributions of the clades discussed in the text. Code numbers are provided for each specimen. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.1. Lumbricidae systematics
The 28 Lumbricidae genera were distributed in 11 clades (A to K in Fig. 1). Our ML and Bayesian phylogenetic trees were very similar to each other for each of the three datasets (Figs. 1–3 and Figs. S3–S6 in supplementary material), with only a few shallow nodes with low support and short branch lengths varying between methods; therefore both ML and Bayesian trees will be considered together unless otherwise stated. Criodrilidae and Hormogastridae were the closest relatives to Lumbricidae, while the other Lumbricina families in the outgroup were significantly separated (Bootstrap proportions [BP] > 70% and Bayesian posterior probability [PP] ≥ 0.95) (Figs. S5 and S6 in supplementary material). All the trees depicted Lumbricidae as a monophyletic group, but with low support (BP < 70% and PP < 0.95). However, high clade support (BP = 85% and PP = 1) for the family was obtained in the DNA tree (Fig. 1) and DNA + morphology tree (Fig. 2) below the Diporodrilus clade – the next node in the Lumbricidae tree.
Fig. 3.
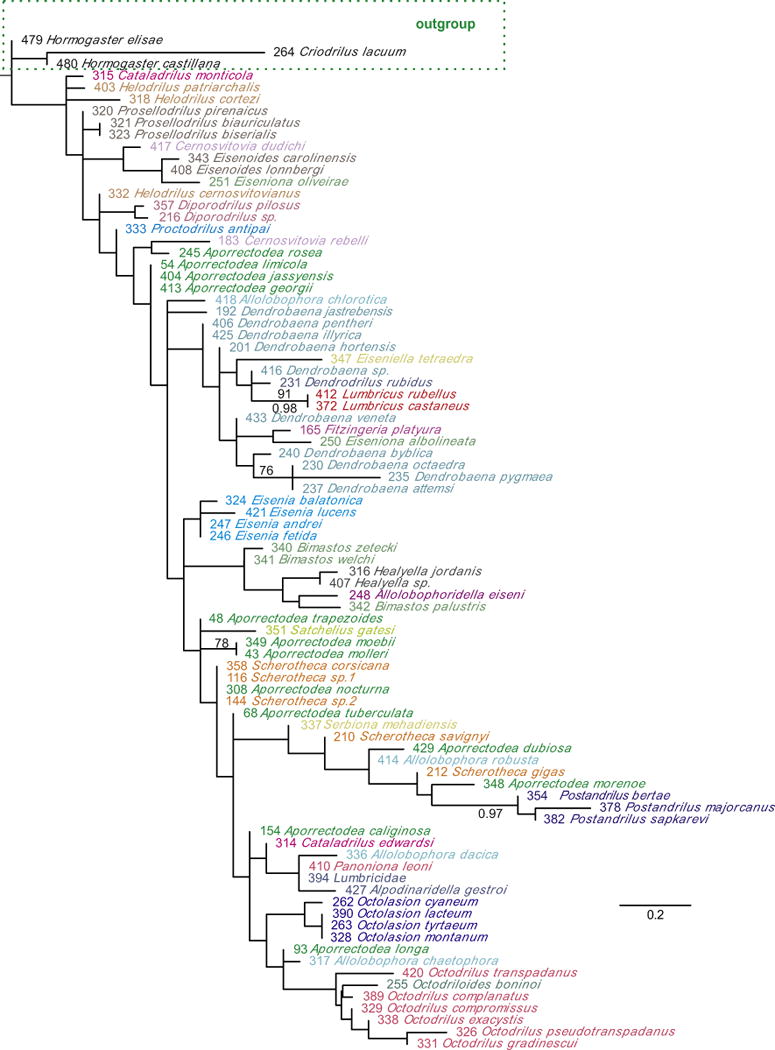
Lumbricidae maximum likelihood morphological tree. Bootstrap proportions (if ≥70%) and Bayesian posterior probabilities (if ≥80%) are shown above and below the branches, respectively. Code numbers are provided for each specimen.
Fig. 2.
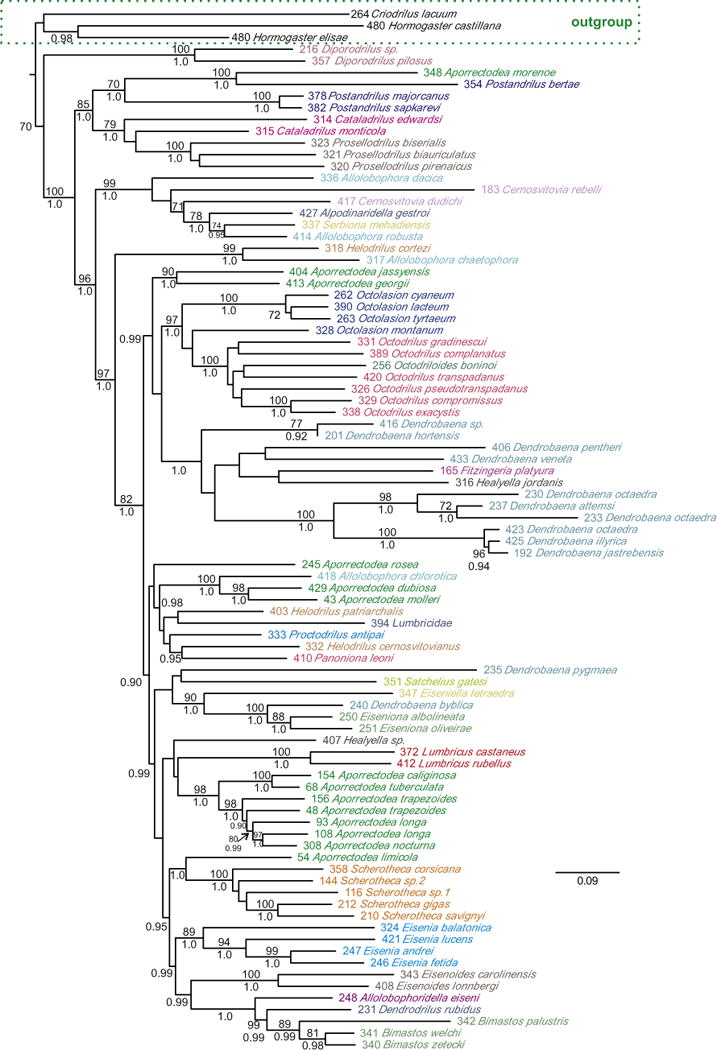
Lumbricidae maximum likelihood molecular + morphological tree. Bootstrap proportions (if ≥70%) and Bayesian posterior probabilities (if ≥95%) are shown above and below the branches, respectively. Selected taxa were pruned to facilitate visualisation of lumbricid relationships. The full tree is shown in Fig. S4 in supplementary material. Code numbers are provided for each specimen.
Monophyly of Lumbricidae genera represented by ≥2 species varied between data sets. In the DNA and DNA + morphology trees (Figs. 1 and 2), ten genera (Allolobophora, Aporrectodea, Cataladrilus, Cernosvitovia, Dendrobaena, Healyella, Helodrilus, Octodrilus, Octolasion, and Postandrilus) were para or polyphyletic as currently defined, while eight genera were monophyletic (Bimastos, Diporodrilus, Eiseniona, Eisenia, Eisenoides, Lumbricus, Prosellodrilus, and Scherotheca). In the morphological trees (Fig. 3), ten genera were para or polyphyletic (Allolobophora, Aporrectodea, Bimastos, Cataladrilus, Cernosvitovia, Dendrobaena, Eiseniona, Helodrilus, Octodrilus and Scherotheca) and seven genera were monophyletic (Eisenia, Eisenoides, Healyella, Lumbricus, Octolasion, Postandrilus, and Prosellodrilus). Over all trees Eisenia, Eisenoides, Lumbricus and Prosellodrilus were always monophyletic while Allolobophora, Aporrectodea Cataladrilus, Cernosvitovia, Dendrobaena, Helodrilus and Octodrilus were always non-monophyletic. Molecular and morphological trees showed disparities in their topologies regarding the positions of Diporodrilus, Octodrilus, Bimastos, Octolasion and Eisenia. Consequently, DNA and morphology trees were significantly different according to the S–H test (P < 0.001) and Bayesian (PP < 0.001) topological tests.
The DNA + morphology tree (Fig. 2) and DNA tree (Fig. 1) showed very similar topologies, except for the relationships of some taxa in clade F (Fig. 1) and the position of Satchellius. Clade support was also similar between these two trees, although some internal nodes (including the most recent common ancestor of clade H) were significantly supported (BP ≥ 70% and/or PP ≥ 0.95) in the DNA tree but not in the DNA + morphology tree and vice versa. The morphology tree showed support for only four shallow clades.
The DNA trees also suggest the existence of at least one potential new species (Dendrobaena octaedra, clade E) based on the length (i.e. genetic divergence) of the branches connecting them and their phylogenetic position. The taxonomic status and evolution of the Aporrectodea species complex has already been discussed elsewhere (Pérez-Losada et al., 2009, 2012; Fernández et al., 2012). Sample 394 Lumbricidae may also be a new species; however, the specimens were all juveniles and although they were morphologically different from any other adults collected in the same area, we were not able to identify them.
3.2. Evolution of morphological and ecological characters
The phylogenetic trees and Bayesian BEAST analysis of the spermathecae (Fig. 5, left) did not show a trend towards reduction of the number of spermathecae from ≥8 to 0. Our analyses strongly support that the ancestor to all the current lumbricids had two pairs of spermathecae (root PP = 0.99) and that both athecate taxa and those with variable numbers of spermathecae pairs evolved multiple times from 2-paired ancestors (PP > 0.95 for most of the ancestral nodes). The phylogenetic and BEAST analyses of the ecotypes (Fig. 5, right) showed that the endogeic ecotype was the first to evolve in the Lumbricidae (root PP = 1), while the epigeic and anecic ecotypes evolved multiples times from endogeic ancestors (PP > 0.95 for most of the ancestral nodes).
Fig. 5.
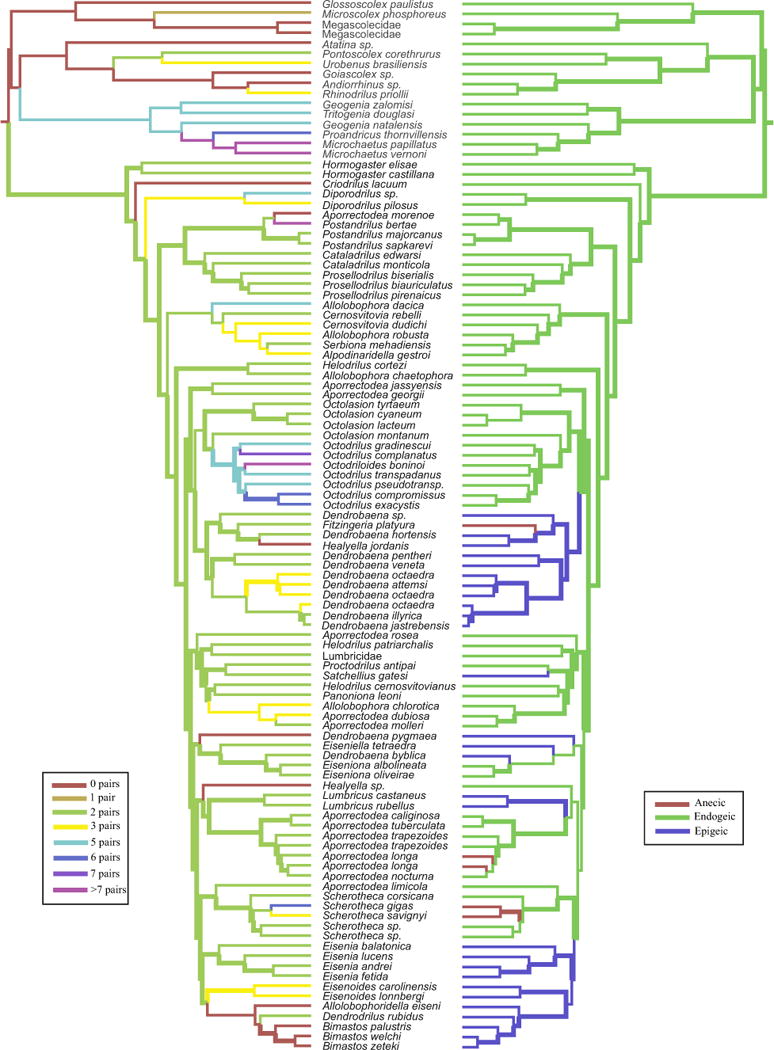
Evolution of Lumbricidae spermathecae (left) and ecotypes (right). Clades with character state posterior probabilities (PP) ≥0.95 are indicated with thick branches, and all other clades are indicated with thin branches. The most likely state for each branch on the tree is coloured. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
4.1. Lumbricidae systematics
The ML and Bayesian phylogenies (Fig. 1) currently represent the most comprehensive hypotheses of the evolutionary relationships of the Lumbricidae in terms of data (5866 aligned DNA sites and 22 morphological characters) and taxon (28 genera) sampling. The phylogenetic trees presented here support some long-held systematic hypotheses regarding lumbricid earthworms and reject others. Some differences in branching order within the outgroup families were found here compared to that in James and Davidson (2012), but those discrepancies could be due to taxon and/or gene sampling differences and should not affect our conclusions. In the full trees (Figs. S3–S6 in supplementary material), Criodrilidae was depicted as the sister taxon to Lumbricidae + Hormogastridae. Stephenson (1930) also suggested a close morphological relationship between Lumbricidae and Criodrilidae, while Qiu and Bouché (1998a) considered Criodrilus lacuum Hoffmeister, 1845 a hormogastrid secondarily adapted to aquatic life. Additionally, molecular phylogenetic analyses by Jamieson (1988) placed Criodrilus in Almidae, while James and Davidson (2012) showed Hormogastridae as the closest relative to Lumbricidae followed by Criodrilidae. All of these authors (except Jamieson, 1988) suggest a close relationship between Lumbricidae, Hormogastridae and Criodrilidae. These three families also share overlapping geographical distributions in Western Europe, which suggests the possibility of a common origin for all and their inclusion in the same taxonomic group.
Previous studies and the present study have confirmed need to reassess the status of many lumbricid genera. Our phylogenetic analyses revealed that at least ten Lumbricidae genera as currently defined do not form monophyletic assemblages, indicating the need for revision of both the taxonomy and the underlying hypothesis of character evolution. Previous attempts (Sims, 1983; Reynolds, 1995; Gates, 1980) to resolve Lumbricidae systematics and more recent phylogenetic studies (Briones et al., 2009; Pérez-Losada et al., 2012; Klarica et al., 2012) have consistently demonstrated that the Lumbricidae classification needs extensive revision. Consensus has not been reached for the placement (or validity) of even some of the most commonly encountered lumbricids (Csuzdi and Zicsi, 2003; Blakemore, 2008).
Within the Lumbricidae, our molecular trees depicted Diporodrilus (Diporodrilinae Bouché, 1970) as the most basal lumbricid (Lumbricinae Rafinesque-Schmaltz, 1815), in agreement with Qiu and Bouché (1998a), who consider this genus a subfamily or even a family. Our Lumbricidae tree largely disagrees with previous phylogenetic hypotheses; such discrepancy is likely caused by the stochastic error (i.e. limited sequence length) and the low phylogenetic signal of the genes analyzed (e.g. Cech et al., 2005; Pop et al., 2003; Briones et al., 2009) and/or their narrower taxonomic scope (Pérez-Losada et al., 2005; Pérez-Losada et al., 2009; Domínguez and Pérez-Losada, 2010; Pérez-Losada et al., 2011; Klarica et al., 2012). Previous phylogenetic analyses of the Lumbricidae generated trees with low support (BP < 70% and PP < 0.95) for most or all clades above the genus level, or were biased because of the few genera (<10) or congeneric species included.
The morphology-based phylogeny confirmed the taxonomic status of seven Lumbricidae genera (monophyla, e.g. Eisenia) and rejected the validity of ten others (paraphyla; e.g. Allolobophora); these assemblages were fairly consistent with those in our molecular phylogenies. Moreover, the morphology trees were also able to separate the lumbricids from the other Lumbricina families, but they produced relationships within the Lumbricidae very different (and unsupported) from those generated in the molecular trees; none of the associations among the backbone clades in the molecular trees were recovered. This highlights the homoplasy or the low phylogenetic signal of the morphological features commonly used in earthworm alpha-taxonomy. Although these characters are useful for identifying Lumbricidae species, they seem unreliable to reconstruct Lumbricidae evolutionary relationships or to delimit supraspecific taxa. Earthworms do not have hard parts, and other features of their internal anatomy may be more informative for those purposes; the study of those features is technically demanding, time consuming, and often prone to producing odd artefacts. Some of these problems could be circumvented by applying non-invasive and non-destructive three-dimensional reconstruction techniques such as micro-computed tomography (e.g. Fernández et al., 2014).
4.2. Phylogeography and time divergence
Our phylogenies clearly separate a Holarctic clade composed of the Lumbricidae, Criodrilidae and Hormogastridae, from the other Crassiclitellata families, which have different geographical distributions. The divergence time estimation analysis (BEAST: Fig. 4) suggests that such clade diversified (crown age) in the lower Cretaceous 125.2 (114.2–137.1) Mya. Similarly the Lumbricidae have a Palearctic origin, given that the basal members of the clade in our tree are in Europe only. The chronogram also indicates that the monophyletic Lumbricidae genera diversified (crown ages) from the lower Miocene 20.5 (14.6–27.1) Mya (Eiseniona) to the Paleocene 61.5 (53.5–68.6) Mya (Eisenia). Our time estimates are not significantly different from those reported by Novo et al. (2011) for the diversification of Hormogastridae-Lumbricidae clade (83–124 Mya), who used multiple time estimators (including BEAST) and one calibration (the separation of the Corso-Sardinian microplate from continental Europe ca. 33 Mya). However, the authors also acknowledged that their time estimates represent a minimum age for the family and that its origin may be older. Both ours and Novós time estimates are older than those proposed by Bouchè (1972) for the origin of the Lumbricoidea families (Late Cretaceous – Early Paleogene; 100–55 Mya) based on current earthworm distributions; but not as old as those proposed by Omodeo (2000), who based on the opening of the South Atlantic, suggested that some sister pairs of confamilial Lumbricoidea genera in Africa and South America have diverged 180 Mya. Ancient patterns of diversification have also been suggested for other widespread terrestrial groups like the Onychophorans (Murienne et al., 2014), which are thought to have diversified prior to the break-up of Pangaea.
The reliability of these time estimates needs to be considered. Fossils are the main source of external information for calibrating molecular phylogenies of species. Unfortunately, as far as we know, no Lumbricidae fossils have been found. Additionally, molecular rates of substitution may vary across taxa (Novo et al., 2012). Nonetheless, the molecular time estimates appear to be supported by additional evidence from at least one taxon. The BEAST time estimate of the split of the Corsican earthworms S. corsicana and Scherotheca sp1 and sp2 from the Spanish S. gigas and French S. savignyi (28.4–44.9 Mya) partly overlaps with the geological time (30–28 Mya) estimated for the separation of Corsica (at the time part of larger microplate) from the proto-Iberian Peninsula (Alvarez et al., 1974; Rosenbaum et al., 2002). Thus, if we assume that this vicariant event caused speciation of these taxa as seen in Postandrilus (Pérez-Losada et al., 2011), the timing would be consistent with our molecular estimate. Other earthworm genera in the analyses also have an insular/continental distribution (e.g. Octolasion and Octodrilus; see Table S1 in supplementary material), but they are considered peregrine earthworms and the effect of human transport may mask the effect of geological processes and/or animal dispersion.
Our phylogenetic analyses suggest correlations between genealogical lineages and geographical distributions for at least 15 of the 34 clades shown in Fig. 4. Even if we exclude the peregrine species, at least 12 of those associations may result from tectonic processes and/or geographic isolation. Clade 1 clusters species from Corsica and Sardinia, while clades 2 and 10 include species from Spain, France, Majorca and Corsica. All five geographical regions were combined before formation of the islands in the Late Oligocene (30–28 Mya) (Alvarez et al., 1974; Rosenbaum et al., 2002). Clade 3 includes species from the nearby countries of Romania and Serbia; clade 4 from southern France; clade 5 from Hungary and Romania; clade 6 from Serbia; clade 7 from Turkey; clade 8 from Hungary and Serbia; clade 9 from Spain and clade 10 from Spain, Corsica and South France and clades 11 and 12 from USA.
From this, we can conclude that the evolution of Lumbricidae in Europe is largely geographically structured, since genera that have been sampled in closer areas are more likely to share a common ancestor. This is not surprising considering the limited dispersal ability of earthworms (Novo et al., 2010; Pérez-Losada et al., 2011) and the small areas of occurrence of many of the analyzed taxa. Previous studies have highlighted that isolation by distance and allopatric speciation are common mechanisms of differentiation in earthworms (Chang et al., 2008; King et al., 2008; Novo et al., 2009, 2010). Speciation related to plate movement and continental masses has been already described in other earthworms from Europe (Novo et al., 2011; Pérez-Losada et al., 2011) and New Zealand (Buckley et al., 2011). Similarly, climate and sea level changes have also been invoked to explain earthworm geographic structuring in European (Pérez-Losada et al., 2011; Fernández et al., 2012) and/or Japanese (Minamiya et al., 2009) earthworms.
Our molecular time estimates also suggest that North American (Bimastos and Eisenoides) and Eurasian lumbricids may have been separated as the result of the final split of Laurasia. The Bimastos-Eisenoides clade also containing Allolobophoridella and Dendrodrilus would have split from their closest relatives (Eisenia) in the Upper Cretaceous 72.6 (69.2–76.1) Mya, about the time of the final split of Laurasia into Eurasia and North America (Cox and Moore, 2010). Since it is unlikely that earthworms migrated (or were transported) across oceans 30 Mya after the split of Laurasia, these results suggest that, either both Allolobophoridella and Dendrodrilus went extinct in North America (with current records based on accidental transport by humans) and/or Bimastos species (e.g., Bimastos syriacus; ordinarily assigned to Healyella) occur(ed) also in Eurasia but have not been found yet or went extinct. Although such a vicariant event requires further confirmation based on additional taxon sampling, the good concordance between molecular and geologic estimates seems to support the vicariance hypothesis.
4.3. Evolution of morphological and ecological characters
The phylogenetic trees and Bayesian analysis of the number of spermathecae suggest that the primitive lumbricid was likely hermaphroditic (all taxa in clades 1 and 2 of Fig. 4 are hermaphroditic), and had two pairs of spermathecae (Fig. 5). From that ancestor, taxa with more (3–8 pairs) or fewer (0 and 1 pair) spermathecae evolved multiple times. This rules out a previous hypothesis suggesting a gradual reduction of the number of spermathecae (Gates, 1972). Although polymorphic degradation of male reproductive structures is often common in parthenogenetic organisms, according to the scientific literature at least seven species in our trees are parthenogenetic and have multiple spermathecae [Dendrobaena octaedra (3 pairs), Aporrectodea trapezoides (2 pairs), Aporrectodea rosea (2 pairs), Octolasion tyrtaeum (2 pairs), Octolasion lacteum (2 pairs), Dendrodrilus rubidus (2 pairs) and Eiseniella tetraedra (2 pairs)]. Our chronogram also shows that athecate male-sterile earthworms arose for the first time in the Lumbricidae ~75 Mya and then four more times between ~70 and ~25 Mya. Previous molecular studies (Fernández et al., 2011, 2012) had already suggested that parthenogenesis emerged twice in the Aporrectodea trapezoides complex during the Late Miocene – Pleistocene epochs (6.4–1.1 Mya in their analyses) from sexual ancestors, which seem to have a strong evolutionary potential to switch from sex to parthenogenesis or vice versa. Similarly, electrophoretical analyses by Jaenicke and Selander (1979) and the extensive morphological work of Gates (1972, 1973, 1974a,b,c, 1977) suggested that parthenogenesis has evolved multiple times and rather recently in the lumbricids Octolasion cyaneum and Octolasion tyrtaeum. Our more comprehensive study indicates that some lumbricid genera also seem to have the same evolutionary potential and/or to exhibit different number of spermathecae (Aporrectodea, Diporodrilus, Allolobophora, Postandrilus, Cernosvitovia, Octodrilus, Scherotheca and Dendrobaena). The present findings also indicate that spermathecae variation is a highly unreliable character for earthworm systematics and taxonomy. Further studies are needed to decipher the evolution of parthenogenesis in earthworms and the relationship between this mode of asexual reproduction and the maintaining of non-functional reproductive structures.
Our phylogenetic trees and Bayesian ancestral state analysis also indicate that ancient lumbricids were endogeic, and that epigeic and anecic earthworms evolved at least 5 and 3 times, respectively. The epigeic ecotype seems to have arisen first (~80 Mya), while the first anecic ecotype evolved more recently (~45 Mya).
Regardless of the timings, the multiple independent origins of epigeic and anecic lifestyles suggests that adaptive diversification did not consist of the unique evolution of epigeic (or endogeic) niche exploitation, followed by speciation within a purely epigeic (or endogeic) clade. Instead, the adaptive radiations occurred independently, and there are even indications of reversals. Some of the Eisenia species of Siberia, for example, show pigmentation and morphology indicative of endogeic living (Perel, 1977) by earthworms of a typically epigeic genus.
If one were to propose that a simpler feeding and digestive system is likely to have arisen before a more complicated one, evidence available would support a later evolution of the endogeic niche. Mutualistic digestive systems involving mucopolysaccharide priming of bacteria have been described in endogeic earthworms (Lavelle and Spain, 2001), while epigeic and anecic earthworm are litter transformers that ingest mixtures of decaying organic matter and microorganisms with direct digestion (Lavelle and Spain, 2001). Consequently one would expect the endogeic to be derived, and not ancestral, as we found in our work. Another possibility is that the hypothetical endogeic ancestor of all Lumbricidae had already established the mutualistic digestive system- after all it was descended from a long-extant lineage of earthworms. An investigation of the digestive processes in epigeic and anecic lumbricid lineages could show if such species can function without mutualistic gut bacteria.
5. Conclusions
Despite more than 130 years of research on earthworms, no robust hypothesis of Lumbricidae evolutionary relationships has yet been established. This study yielded the first well-supported phylogeny of the family and estimated its divergence time. Using this phylogenetic hypothesis, we validated some of the existing taxonomic groups but rejected others. We also studied the patterns of geographical diversification and the evolution of reproductive strategies and ecotypic adaptations. We conclude that lumbricids emerged in the Lower Cretaceous and that their current taxonomic classifications must be revised and new morphological features included. We also showed that both geological processes and geographic isolation may have affected geographical diversification, that parthenogenesis arose multiple times in the group, and that several non-related lumbricid genera developed similar feeding and burrowing habits.
Supplementary Material
Acknowledgments
This research was funded by the Spanish Ministerio de Ciencia e Innovación (projects CGL2006-11928 and CTM2009-08477), Xunta de Galicia (CN2012/305), the United States National Science Foundation Award DEB-0516439 (James) and a “Projeto de investigação Exploratória” from the FCT Investigator Program to MP-L. We gratefully acknowledge Grzegorz Gryziak, Konstantin Gongalsky, Jari Haimi, Martin Holmstrup, Mervi Niemen, Nicolas Bottinelli, Veikko Huhta, Pascal Jouquet, Sonja Migge-Kleian, Yvan Capowiez, Danuta Plisko, Marta Novo, Darío Díaz Cosín and Alexander Feijoo for generously providing earthworm samples. We also thank Maigualida Ricoy, Cristina Lazcano, María Gómez, Marta Lores, Cristóbal Pérez, Pablo González and Fernando Monroy for collaborating in collecting earthworms to complete this study and Alberto Velando for critical discussions and helpful revisions of previous drafts of the manuscript.
Appendix A. Supplementary material
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ympev.2014.10.024.
References
- Alvarez W, Cocozza T, Wezel FC. Fragmentation of Alpine orogenic belt by microplate dispersal. Nature. 1974;248:309–314. [Google Scholar]
- Blakemore RJ. Cosmopolitan earthworms – an eco-taxonomic guide to the peregrine species of the world. second. VermEcology; Japan: 2006. [Google Scholar]
- Blakemore RJ. An updated list of valid, invalid and synonymous names of Criodriloidea and Lumbricoidea (Annelida: Oligochaeta: Criodrilidae, Sparganophilidae, Ailoscolecidae, Hormogastridae, Lumbricidae, Lutodrilidae) Ito MT, Kaneko N, editors. A series of searchable texts on earthworm biodiversity, ecology and systematics from various regions of the world Yokohama University. 2008:1–80. < http://bioeco.eis.ynu.ac.jp/eng/database/earthworm/>.
- Bouché MB. Lombriciens de France, ecologie et systematique. Institut National de la Rcherche Agronomique; Paris: 1972. [Google Scholar]
- Bouché MB. Stratégies lombriciennes. In: Persson T, Lohm U, editors. Soil Organism as Components of Ecosystems Oikos Editorial Office. 1977. pp. 122–132. [Ecological Bulletins, vol. 25] [Google Scholar]
- Briones MJI, Moran P, Posada D. Are the sexual, somatic and genetic characters enough to solve nomenclatural problems in lumbricid taxonomy? Soil Biol Biochem. 2009;41:2257–2271. [Google Scholar]
- Buckley TR, James S, Allwood J, Bartlett S, Howitt R, Prada D. Phylogenetic analysis of New Zealand earthworms (Oligochaeta: Megascolecidae) reveals ancient clades and cryptic taxonomic diversity. Mol Phylogenet Evol. 2011;58:85–96. doi: 10.1016/j.ympev.2010.09.024. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Cech G, Csuzdi C, Marialigeti K. Remarks on the molecular phylogeny of the genus Dendrobaena (sensu Pop, 1941) based on the investigation of 18S rDNA sequences. In: Pop VV, Pop AA, editors. Advances in Earthworm Taxonomy II Cluj. University Press; Cluj-Napoca: 2005. pp. 155–165. [Google Scholar]
- Chang CH, Lin SM, Chen JH. Molecular systematics and phylogeography of the gigantic earthworms of the Metaphire formosae species group (Clitellata, Megascolecidae) Mol Phylogenet Evol. 2008;49:958–968. doi: 10.1016/j.ympev.2008.08.025. [DOI] [PubMed] [Google Scholar]
- Cox CB, Moore PD. Biogeography: an ecological and evolutionary approach. Blackwell; Oxford: 2010. [Google Scholar]
- Csuzdi CS, Zicsi A. Earthworms of Hungary (Annelida, Oligochaeta, Lumbricidae) Hungary Natural History Museum; Budapest: 2003. [Google Scholar]
- Domínguez J, Aira M, Gómez Brandón M. Vermicomposting: earthworms enhance the work of microbes. In: Insam H, Franke-Whittle I, Goberna M, editors. Microbes at Work: From Wastes to Resources. Springer; Berlin Heidelberg: 2010. pp. 93–114. [Google Scholar]
- Domínguez J, Pérez-Losada M. Eisenia fetida (Savigny, 1826) y Eisenia andrei Bouché, 1972 son dos especies diferentes de lombrices de tierra. Acta Zoológica Mexicana. 2010;2:321–331. [Google Scholar]
- Domínguez J, Velando A. Sexual selection in earthworms: mate choice, sperm competition, differential allocation and partner manipulation. Appl Soil Ecol. 2013;69:21–27. [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CA, Bohlen PJ. Biology and Ecology of Earthworms. Chapman and Hall; London: 1996. [Google Scholar]
- Edwards CA. Earthworm Ecology. CRC Press; Boca Ratón: 2004. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fernández RM, Almodóvar A, Novo M, Gutiérrez M, Díaz Cosín DJ. A vagrant clone in a peregrine species: phylogeography, high clonal diversity and geographical distribution in the earthworm Aporrectodea trapezoides (Dugès, 1828) Soil Biol Biochem. 2011;43:2085–2093. [Google Scholar]
- Fernández R, Almodóvar A, Novo M, Simancas B, Díaz Cosín DJ. Adding complexity to the complex: new insights into the phylogeny, diversification and origin of parthenogenesis in the Aporrectodea caliginosa species complex (Oligochaeta, Lumbricidae) Mol Phylogenet Evol. 2012;64:368–379. doi: 10.1016/j.ympev.2012.04.011. [DOI] [PubMed] [Google Scholar]
- Fernández R, Kvist S, Lenihan J, Giribet G, Ziegler A. Sine systemate chaos? A versatile tool for earthworm taxonomy: non-destructive imaging of freshly fixed and museum specimens using micro-computed tomography. PLoS ONE. 2014;9:e96617. doi: 10.1371/journal.pone.0096617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, Suchard MA. Bayesian analysis of elapsed times in continuous-time Markov chains. Can J Stat. 2008;36:355–368. [Google Scholar]
- Gates GE. Burmese earthworms – an introduction to the systematics and biology of megadrile oligochaetes with special reference to Southeast Asia. Trans Am Philos Soc. 1972;62:1–326. [Google Scholar]
- Gates GE. The earthworm genus Octolasion in America. Bull Tall Timbers Res Station. 1973;14:29–50. [Google Scholar]
- Gates GE. Contribution to a revision of the Lumbricidae. X. Dendrobaena octaedra (Savigny) 1826, with special reference to the importance of its parthenogenetic polymorphism for the classification of earthworms. Bull Tall Timbers Res Station. 1974a;15:15–57. [Google Scholar]
- Gates GE. Contributions to a revision of the family Lumbricidae. XI. Eisenia rosea (Savigny, 1826) Bull Tall Timbers Res Station. 1974b;16:9–30. [Google Scholar]
- Gates GE. On a new species of earthworm in a southern portion of the United States. Bull Tall Timbers Res Station. 1974c;15:1–13. [Google Scholar]
- Gates GE. Contributions to a revision of the earthworm family Lumbricidae. XX. The genus Eiseniella in North America. Megadrilogica. 1977;3:71–79. [Google Scholar]
- Gates GE. Contributions to a revision of the earthworm family Lumbricidae. XXV. The genus Allolobophora Eisen 1874 in North America. Megadrilogica. 1980;3:177–184. [Google Scholar]
- Huelsenbeck JP, Larget B, Miller RE, Ronquist F. Potential applications and pitfalls of Bayesian inference of phylogeny. Syst Biol. 2002;51:673–688. doi: 10.1080/10635150290102366. [DOI] [PubMed] [Google Scholar]
- Jaenicke J, Selander K. Evolution and ecology of parthenogenesis in earthworms. Am Zool. 1979;19:729–737. [Google Scholar]
- James SW. Planetary processes and their interactions with earthworm distributions and ecology. In: Edwards CA, editor. Earthworm Ecology. second. St Lucie Press; Boca Raton: 2004. pp. 53–62. [Google Scholar]
- James SW, Davidson SK. Molecular phylogeny of earthworms (Annelida: Crassiclitellata) based on 28S, 18S and 16S gene sequences. Invertebrate Syst. 2012;26:213–229. [Google Scholar]
- Jamieson BGM. On the phylogeny and higher classification of the Oligochaeta. Cladistics. 1988;4:367–410. doi: 10.1111/j.1096-0031.1988.tb00520.x. [DOI] [PubMed] [Google Scholar]
- Jamieson BGM. Non-leech clitellata (with contributions by Marco Ferraguti) In: Pleijel J, Rouse G, editors. Reproductive Biology and Phylogeny of Annelida. Enfield Science Publishers; 2006. pp. 235–392. [Google Scholar]
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HM, editor. Mammalian Protein Metabolism. Academic Press; New York: 1969. pp. 21–32. [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T. Recent developments in the MAFFT multiple sequence alignment program. Briefings Bioinformatics. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- King RA, Tibble AL, Symondson OC. Opening a can of worms: unprecedented sympatric cryptic diversity within British lumbricid earthworms. Mol Ecol. 2008;17:4684–4698. doi: 10.1111/j.1365-294X.2008.03931.x. [DOI] [PubMed] [Google Scholar]
- Klarica J, Kloss-Brandstätter A, Traugott M, Juen A. Comparing four mitochondrial genes in earthworms – implications for identification, phylogenetics, and discovery of cryptic species. Soil Biol Biochem. 2012;45:23–30. [Google Scholar]
- Lavelle P, Spain AV. Soil Ecology. Kluwer Academic Publishers; London: 2001. [Google Scholar]
- Lavelle P, Barros E, Blanchart E, Brown G, Desjardins T, Mariani L, Rossi JP. SOM management in the tropics: why feeding the soil macrofauna? Nutr Cycl Agroecosyst. 2001;61:53–61. [Google Scholar]
- Lee KE. Earthworms: Their Ecology and Relationships with Soils and Land Use. Academic Press; Sydney: 1985. [Google Scholar]
- Lee KE. Earthworm classification and biogeography: Michaelsen’s contribution, with special reference to southern lands. Mitteilungen aus dem Hamburg Zoologischen Museum und Institut. 1994;89:11–21. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. Proceedings of the Gateway Computing Environments Workshop (GCE) New Orleans: 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees; pp. 1–8. [Google Scholar]
- Minamiya Y, Yokoyama J, Fukuda T. A phylogeographic study of the Japanese earthworm, Metaphire sieboldi (Horst, 1883) (Oligochaeta: Megascolecidae): inferences from mitochondrial DNA sequences. Eur J Soil Biol. 2009;45:423–430. [Google Scholar]
- Mrsic N. Monograph on earthworms (Lumbricidae) of the Balkans. Slovenian: Academy of Sciences and Arts, Ljubljana; 1991. [Google Scholar]
- Murienne J, Daniels SR, Buckley TR, Mayer G, Giribet G. A living fossil tale of Pangaean biogeography. Proc Royal Soc B. 2014;281:20132648. doi: 10.1098/rspb.2013.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novo M, Almodóvar A, Díaz Cosín DJ. High genetic divergence of Hormogastridae earthworms (Annelida, Oligochaeta) in central Iberian Peninsula. Evolutionary and demographic implications. Zoologica Scripta. 2009;38:537–552. [Google Scholar]
- Novo M, Almodóvar A, Fernández R, Trigo D, Díaz Cosín DJ. Cryptic speciation of hormogastrid earthworms revealed by mitochondrial and nuclear data. Mol Phylogenet Evol. 2010;56:507–512. doi: 10.1016/j.ympev.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Novo M, Almodóvar A, Fernández R, Giribet G, Díaz Cosín DJ. Understanding the biogeography of a group of earthworms in the Mediterranean basin – The phylogenetic puzzle of Hormogastridae (Clitellata: Oligochaeta) Mol Phylogenet Evol. 2011;61:125–135. doi: 10.1016/j.ympev.2011.05.018. [DOI] [PubMed] [Google Scholar]
- Novo M, Almodóvar A, Fernández R, Trigo D, Díaz Cosín DJ, Giribet G. Appearances can be deceptive: different diversification patterns within a group of Mediterranean earthworms (Oligochaeta, Hormogastridae) Mol Ecol. 2012;21:3776–3793. doi: 10.1111/j.1365-294X.2012.05648.x. [DOI] [PubMed] [Google Scholar]
- Omodeo P. Contributo alla revisione dei Lumbricidae. Archivio Zoologico Italiano. 1956;41:129–212. [Google Scholar]
- Omodeo P. Evolution and biogeography of megadriles (Annelida, Clitellata) Italian J Zool. 2000;67:179–201. [Google Scholar]
- Perel TS. Differences in lumbricid organization connected with ecological properties. Ecol Bull. 1977;25:56–63. [Google Scholar]
- Pérez-Losada M, Eiroa J, Mato S, Domínguez J. Phylogenetic species delimitation of the earthworms Eisenia fetida (Savigny, 1826) and Eisenia andrei Bouché, 1972 (Oligochaeta, Lumbricidae) based on mitochondrial and nuclear DNA genes. Pedobiologia. 2005;49:317–324. [Google Scholar]
- Pérez-Losada M, Høeg JT, Crandall KA. Unraveling the evolutionary radiation of the Thoracican barnacles using molecular and morphological evidence. A comparison of several divergence time estimation approaches. Syst Biol. 2004;53:244–264. doi: 10.1080/10635150490423458. [DOI] [PubMed] [Google Scholar]
- Pérez-Losada M, Ricoy M, Domínguez J, Marshall J. Phylogenetic assessment of the earthworm Aporrectodea caliginosa species complex (Oligochaeta, Lumbricidae) based on mitochondrial and nuclear DNA sequences. Mol Phylogenet Evol. 2009;52:293–302. doi: 10.1016/j.ympev.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Pérez-Losada M, Breinholt JW, Porto PG, Aira M, Domínguez J. An earthworm riddle: systematics and phylogeography of the Spanish lumbricid Postandrilus. PLoS ONE. 2011;6:e28153. doi: 10.1371/journal.pone.0028153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Losada M, Bloch R, Breinholt JW, Pfenninger M, Domínguez J. Taxonomic assessment of Lumbricidae (Oligochaeta) earthworm genera using DNA barcodes. Eur J Soil Biol. 2012;48:41–47. [Google Scholar]
- Pop V. Zur Phylogenie und Systematik der Lumbriciden. Zoologische Jahrbiicher. 1941;74:487–522. [Google Scholar]
- Pop AA, Wink M, Pop VV. Using 18S, 16S rDNA and Cytochrome c oxidase sequences in earthworm taxonomy (Oligochaeta, Lumbricidae) Pedobiologia. 2003;47:428–433. [Google Scholar]
- Pop AA, Cech G, Csuzdi CS, Wink M, Pop VV. An attempt to reconstruct the molecular phylogeny of the genus Allolobophora Eisen, 1874 (sensu lato, Pop, 1941) using 16S rDNA and COI sequences (Oligochaeta, Lumbricidae) In: Pop VV, Pop AA, editors. Advances in Earthworm Taxonomy II Cluj. University Press; Cluj-Napoca: 2005. pp. 155–165. [Google Scholar]
- Posada D. Selection of models of DNA evolution with JModelTest. Methods Mol Biol. 2009;537:93–112. doi: 10.1007/978-1-59745-251-9_5. [DOI] [PubMed] [Google Scholar]
- Posada D, Buckley TR. Model selection and model averaging in phylogenetics: Advantages of Akaike Information Criterion and Bayesian approaches over Likelihood Ratio Tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Qiu JP, Bouché M. Révision des taxons supraspécifiques de Lumbricoidea [Revision of Lumbricoidea supraspecific taxa] Doc pedozoologiq integrologiq. 1998a;3:179–216. [Google Scholar]
- Qiu JP, Bouché M. La decouverte de Postandrilus ge. nov. (Oligochaeta:Lumbricidae) et remarques sur la reproduction des lombriciens. Doc pedozoologiq integrologiq. 1998b;4:65–72. [Google Scholar]
- Qiu JP, Bouché MB. Liste classé e des taxons valides de Lombriciens (Oligochaeta: Lumbricoidea) apre‘ s l’etude des trios cinquie‘ me d’entre-eux. Doc pedozoologiq integrologiq. 1998c;4:181–200. [Google Scholar]
- Rambaut A, Drummond AJ. Tracer: MCMC trace analysis tool Institute of Evolutionary Biology. Edinburgh; 2009. < http://tree.bio.ed.ac.uk/software/tracer/>. [Google Scholar]
- Reynolds JW. Status of exotic earthworm systematics and biogeography in North America. In: Hendrix Paul F, editor. Earthworm ecology and biogeography. Lewis Publishers; Boca Raton, FL: 1995. pp. 1–28. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rosenbaum G, Lister GS, Duboz C. Reconstruction of the tectonic evolution of the western Mediterranean since the Oligocene. J Virtual Explorer. 2002;8:107–130. [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- Sims RW. The scientific names of earthworms. In: Satchell JE, editor. Earthworm Ecology, from Darwin to Vermiculture. Chapman and Hall; London: 1983. pp. 467–474. [Google Scholar]
- Sims RW, Gerard BM. Earthworms. The Linnean Society of London and the Estuarine and Coastal Sciences Association; London: 1999. (Synopses of the British Fauna (New Series) N° 31). [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Stephenson J. The Oligochaeta. Clarendon Press; Oxford: 1930. [Google Scholar]
- Tavaré S. Some probabilistic and statistical problems on the analysis of DNA sequences. Lecture Notes Math Life Sci. 1986;17:57–86. [Google Scholar]
- Velando A, Eiroa J, Domínguez J. Brainless but not clueless: earthworms boost their ejaculates when they detect fecund nonvirgin partners. Proc Royal Soc B. 2008;275:1067–1072. doi: 10.1098/rspb.2007.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens J. Combining data sets with different phylogenetic histories. Syst Biol. 1998;47:568–581. doi: 10.1080/106351598260581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


