Abstract
Understanding how to achieve efficient transduction of hematopoietic stem and progenitor cells (HSPCs), while preserving their long-term ability to self-reproduce, is key for applying lentiviral-based gene engineering methods. SAMHD1 is an HIV-1 restriction factor in myeloid and resting CD4+ T cells that interferes with reverse transcription by decreasing the nucleotide pools or by its RNase activity. Here we show that SAMHD1 is expressed at high levels in HSPCs cultured in a medium enriched with cytokines. Thus, we hypothesized that degrading SAMHD1 in HSPCs would result in more efficient lentiviral transduction rates. We used viral like particles (VLPs) containing Vpx, shRNA against SAMHD1, or provided an excess of dNTPs or dNs to study this question. Regardless of the method applied, we saw no increase in the lentiviral transduction rate. The result was different when we used viruses (HR-GFP-Vpx+) which carry Vpx and encode GFP. These viruses allow assessment of the effects of Vpx specifically in the transduced cells. Using HR-GFP-Vpx+ viruses, we observed a modest but significant increase in the transduction efficiency. These data suggest that SAMHD1 has some limited efficacy in blocking reverse transcription but the major barrier for efficient lentiviral transduction occurs before reverse transcription.
Keywords: SAMHD1, dNTP pools, Transduction, Hematopoietic stem and progenitor cells, (HSPCs), Gene engineering, Lentiviral vectors
1. Introduction
Gene engineered hematopoietic stem and progenitor cells (HSPCs) are the basis of a promising therapeutic approach for a variety of diseases, including HIV/AIDS, Wiskott–Aldrich syndrome and metachromatic leukodystrophy (Cartier & Aubourg, 2010; Sheridan, 2011; Aiuti et al., 2013; meBiffi et al., 2013). Gene engineering has become more feasible since retroviruses were found to integrate into the host chromosome and retroviral vectors were developed (Sheridan, 2011).
Gene engineering of HSPCs seems straightforward, but challenging technical issues remain, including efficient transduction of HSPCs without disrupting their multi-potency, ex vivo or in vivo selection of gene engineered cells, generation of sufficient numbers of gene engineered cells and ultimately homing and efficient engraftment (Millington et al., 2009; Glimm et al., 2000; Barquinero et al., 2000). Transduction is most efficient when cells are activated and dividing. In that case, HSPCs will differentiate and lose their multi-potency. Unlike retroviral vectors, lentiviral vectors benefit from HIV’s capacity to enter the nucleus of non-dividing cells, while using the envelope of unrelated viruses to widen their tropism (Frimpong & Spector, 2000; Cronin et al., 2005). Although lentiviral vectors eliminate the strict requirement for cell division, they still need HSPCs to be stimulated to progress into the late G1 phase for efficient transduction (Miller et al., 1990). Thus, despite many efforts to achieve higher transduction rates for HSPCs, the lentiviral transduction efficiency is still low (20–30%) when avoiding harsh activation and proliferation, and this is a major limitation for efficient gene engineering. In addition, multiple integration events may result in disruption of cell homeostasis, and so, achieving single integration events is a significant goal.
The sterile alpha motif (SAM) domain and HD domain-containing protein 1 (SAMHD1) were identified as a potent restriction factor for HIV (Hrecka et al., 2011; Laguette et al., 2011). Its deoxyguanosine-triphosphate (dGTP)-dependent triphosphate triphosphohydrolase activity hydrolyses the deoxynucleoside triphosphates (dNTPs) to deoxynucleoside (dNs) and inorganic triphosphate (Lahouassa et al., 2012). SAMHD1 was originally proposed to limit HIV-1 infection by depleting the intracellular levels of dNTPs to below those required for reverse transcription. Very recent data challenge this proposed mechanism and present SAMHD1’s RNase activity as crucial for HIV restriction (Ryoo et al., 2014). This mechanism appears to be plausible since the phosphorylated form of SAMHD1 is unable to block HIV-1 infection, but still actively depletes the intracellular dNTP pools (White et al., 2013).
SAMHD1 is expressed in cells of the myeloid lineage, such as dendritic cells (DCs), monocytes and macrophages, as well as CD4+ T cells, although SAMHD1 expression was reported as missing in T cell lines (Hrecka et al., 2011; Laguette et al., 2011; Descours et al., 2012; Baldauf et al., 2012). Moreover, the accessory viral protein Vpx, which is encoded by HIV-2 and SIV variants, but not HIV-1, targets SAMHD1 for proteasomal degradation in myeloid and resting CD4+ T cells. Therefore, Vpx facilitates HIV-1 reverse transcription (Hrecka et al., 2011; Laguette et al., 2011). These findings strongly implicate SAMHD1 as a key restriction factor that inhibits HIV-1 replication in myeloid cells and resting CD4+ T cells at the level of reverse transcription.
We hypothesized that SAMHD1 acts to restrict HIV-1-based lentiviral HSPC transduction. To address this hypothesis, we assessed the efficiency of lentiviral transduction of HSPCs under the following conditions: 1) Vpx-mediated degradation of SAMHD1, 2) shRNA against SAMHD1, 3) an excess of dNTPs or dNs, and 4) HR-GFP-Vpx+/− lentivirus, which carries Vpx and encodes GFP.
2. Material and methods
2.1. Ethics statement
The use of human cord blood was approved by the ethical committee of the University of Zurich. Written informed consent was obtained from the parents before collection.
2.2. Cells and reagents
Human peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats, obtained from the local blood donation center in Zurich, by Ficoll (Axis-Shield PoC AS, Oslo, Norway) gradient centrifugation. Monocytes were purified using CD14 MicroBeads (Miltenyi Biotec, Bergish Gladbach, Germany); the purity was >90%. Monocyte-derived macrophages (MDMs) were generated as described (Schlaepfer et al., 2014). HSPCs were isolated from human cord blood with immunomagnetic beads (Miltenyi Biotec, Bergish Gladbach, Germany) with an yield of 0.5–4 × 106 HSPCs from one donation (purity > 90%). HSPCs and “non-target” fractions were stored frozen in liquid nitrogen until use. THP-1, SupT-1, MT-2, U937 cells were cultured in RPMI 1640 (BioWhittaker) supplemented with 10% FBS, 1% glutamine (Invitrogen Life Technologies), and antibiotics (Pen Strep). SAMHD1 shRNA, encoded within a SIN vector cassette (St. Louis, MO, TRCN0000145408), and control mammalian non-target shRNA (SHC002) were purchased from Sigma-Aldrich. THP-1 cells transduced with lentiviral vectors encoding SAMHD1 shRNA were selected by culturing in a medium containing 0.8 μg/ml puromycin. dNs consisted of a mixture of dA (D8668), dC (D0776), dG (D0901) and dT (T1895; all from Sigma-Aldrich). dNTPs (R0192) were purchased from Fermentas (Glen Burnie, MD). Cytokine cocktail for HSPC transduction consisted of a mixture of stem cell factor (SCF) at 100 ng/ml, Fms-related tyrosine kinase 3 ligand (Flt3L) at 100 ng/ml, interleukin-3 (IL-3) at 60 ng/ml and thrombopoietin (TPO) at 10 ng/ml (Immunotools, Friesoythe, Germany).
2.3. Viruses, and transduction procedure
Viruses were produced by transfection of 293T cells with polyethylenimine (PEI) (Sigma-Aldrich) with the various constructs. Media were replaced 16 h post-transfection, and supernatants were collected and filtered 48 h post-transfection; viruses were concentrated 100-fold with PEG-it™ (System Biosciences, Mountain View, CA). Len-EF1α-GFP: To generate lentiviral particles encoding GFP, 293T cells were co-transfected with the self-inactivating (SIN) vector pLen-EF1α-GFP (kindly provided by D. Boden, Tibotec, Belgium), pCMVΔR8.91 (containing the coding sequences for gag/pol, rev and tat under the control of the CMV promoter) and pMD2.G (coding for the vesicular-stomatitis virus (VSV-G) envelope) (both provided by D. Trono, Switzerland). HR-GFP virus with Vpx incorporated: The HR-GFP virus carrying Vpx was produced by co-transfection of the proviral HIV-1 GFP plasmid HR-GFP, Vpx239+ (Vpx expression construct), pR8.9NdeltaSB and pMD2.G; the Gag P6 in pR8.9NdeltaSB is modified for better incorporation of Vpx (Baldauf et al., 2012). Vpx+ and Vpx− viral like particles (VLPs) were produced by co-transfection with pSIV3+ (Vpx+) or pSIV3+ Δvpx (Vpx−) (provided by A. Cimarelli, Lyon) and pMD2.G in the absence of a viral genome. Lentiviruses containing SAMHD1 shRNA and control mammalian non-target shRNA were produced by co-transfection of lentiviral vectors which encode SAMHD shRNA or control shRNA with pCMVΔR8.91 and pMD2.G. All viruses encoding GFP were titrated on SupT-1 cells by measuring the frequency of GFP+ cells by flow cytometry (van Lent et al., 2010).
Transduction of HSPCs was done essentially as described (Amsellem et al., 2002). Briefly, HSPCs were resuspended in a HP01 medium (Macopharma, Mouvaux, France) containing a mixture of cytokines (see above). Three days later, transduction efficacy was determined by measuring the frequency of GFP+ cells by flow cytometry.
2.4. Flow cytometry
HSPCs and monocytes were incubated with PBS containing 2% FBS, 2 mM EDTA (Invitrogen Life Technologies) and 0.1% sodium azide (Sigma-Aldrich) for staining cell surface markers. Intracellular SAMHD1 (Abcam, Cambridge, UK) staining was performed as described (Baldauf et al., 2012). Depending on the purpose of the experiment, the following antibodies were used for staining HSPCs in various combinations: Lin-, CD34, CD133, and CD38 (BD Biosciences, San Jose). Samples were acquired on a CyAn™ ADP Analyzer (Beckman Coulter, Pasadena, CA) and analyzed by FlowJo software (TreeStar, Ashland, OR). Gating strategy is shown in Fig. 2A.
Fig. 2.
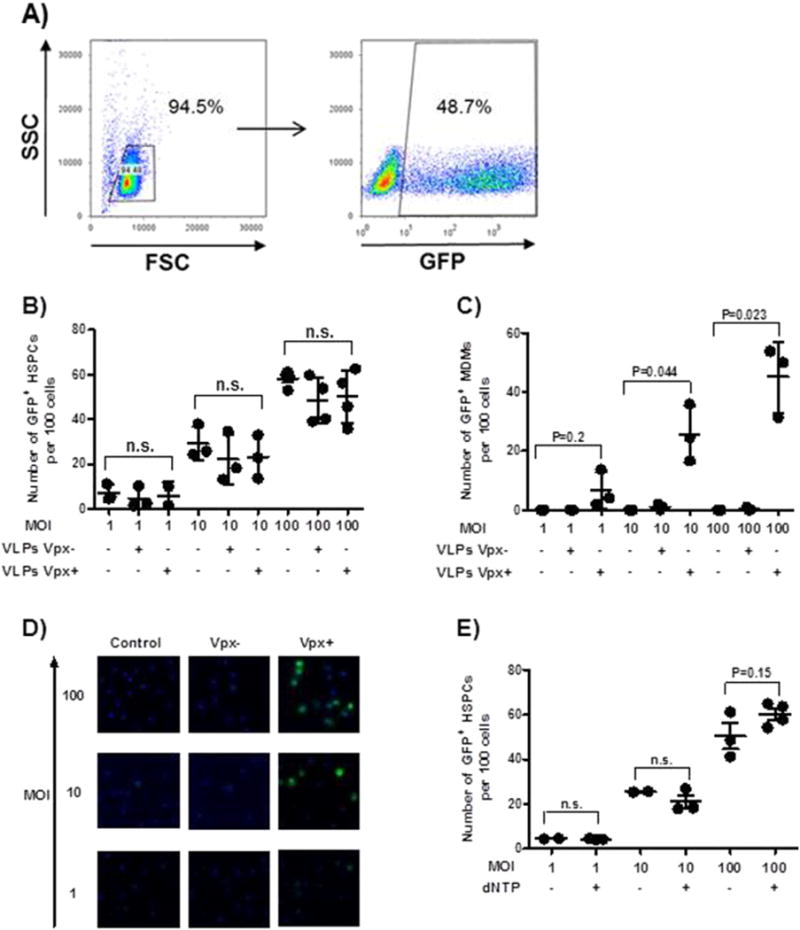
Vpx+ virus pretreatment has no effect on transduction efficiency. (A) Freshly isolated cord blood-derived HSPCs were cultured in the presence of cytokines as mentioned in Materials and methods section, HSPCs were challenged with Len-EF1α-GFP at an MOI of 100. Transduction efficiency was determined by quantifying the percentage of GFP+ HSPCs by flow cytometry. A representative dataset of three independent experiments is shown. HSPCs (B) or MDMs (C and D) were pretreated for 2 h with Vpx+/− viruses. In (E), HSPCs were exposed to 0.01 mM dNTPs. Transduction was performed with Len-EF1α-GFP at MOIs of 1, 10 and 100. Transduction efficiency was determined by quantifying the percentage of GFP+ HSPCs by flow cytometry (B and E) and by immunohistochemistry (C and D). Data are from three independent donors of MDMs and HSPCs. Dots represent three individual measurements with mean ± SEM indicated. Statistical analysis used the two-tailed paired t-test.
2.5. Quantitative PCR for measuring SAMHD1 mRNA
RNA from 2 × 106 of primary cells or cell lines was isolated using an RNeasy kit (Qiagen, Hilden, Germany). Reverse transcription was performed essentially as described (Audige et al., 2004). SAMHD1 mRNA was quantified using commercially available primers and probes (Assays-on-demand; Applied Biosystems, Foster City, CA) by real-time quantitative PCR (RT-qPCR) analysis. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Applied Biosystems) was used as a housekeeping gene. Data generated by RT-qPCR were analyzed as described (Schlaepfer et al., 2014). For each sample, the mean normalized gene expression (MNE) was determined with the software application Q-Gene.
2.6. Western blot analysis
MDMs and HSPCs were pretreated with Vpx+/− viruses for 2 h and cultured for 3 days before the preparation of whole-cell extracts. Western blotting was performed essentially as described (Miller et al., 2011) by using mouse anti-SAMHD1 (Abcam, Cambridge, UK), rabbit anti-pSAMHD1 directed against the phosphorylated threonine (T592) (White et al., 2013), and rabbit anti-β actin (Cell Signalling, Danvers, MA) antibodies in blocking buffer overnight at 4 °C. After three washes, the membranes were incubated in HRP-conjugated anti-mouse or anti-rabbit IgG secondary antibody (Cell Signalling) in a washing buffer for 1 h at room temperature. After three washes in a washing buffer, membranes were incubated for 1 min with ECL detection reagents (Amersham Biosciences, Little Chalfont, UK). Signals were quantified with Adobe Photoshop CS3 software.
2.7. Quantification of proviral DNA with Alu-PCR
Alu-PCR was performed with specific primers to human Alu sequences and to HIV-1-based lentiviral vector sequences as described (Schlaepfer et al., 2014; Althaus et al., 2010). Results were considered as valid only if the same results were obtained in at least three separate experiments.
2.8. Quantification of viral DNA intermediates
DNA was extracted from MDMs or HSPCs transduced with HR-GFP-Vpx+ or HR-GFP-Vpx− lentiviruses with a QIAamp DNA Mini kit (Qiagen) according to the manufacturer’s instructions. Samples treated with 1 μM efavirenz (EVF) or 30 μM raltegravir (RTV) (EVF and RTV were provided by the NIH AIDS Reagent Program, Germantown, MD), which block the reverse transcription and integration step, respectively, were used as negative controls. Real-time PCR was performed by using primers and probes as described (Butler et al., 2001). Reactions were carried out in 50 μl volumes containing 400 ng of DNA in the presence of 600 nM of each primer and 80 nM of probes. Results were obtained from three independent experiments using three donors.
2.9. Quantification of whole cell dNTP pools
MDMs and HSPCs were exposed for 2 h to Vpx+/− viruses/dNs/dNTPs and cultured for 3 days before the preparation of whole cell extracts. dNTPs were quantified as described (Diamond et al., 2004).
2.10. Immunofluorescence
Adherent differentiated MDMs were exposed for 2 h to Vpx+/− viruses, and infected with Len-EF1α-GFP lentiviral viruses for 6 h. At day 5, MDMs were rinsed three times with phosphate-buffered saline (PBS, Sigma-Aldrich). The adherent cells were fixed in culture plates with methanol at −20 °C (VWR International, Radnor, PA) for 5 min and blocked with PBS plus 10% normal human serum (Pel-Freez, Rogers, AR) for 20 min. Three rinses with PBS followed all incubations, and all steps were performed at room temperature. Fluorescence was detected as described (Kuster et al., 2000). In every experiment, background FITC fluorescence was determined from identically processed uninfected cell lines and expressed as total gray values (TGV) per cell. The FITC background was calculated as [mean (log10 TGV) plus 3 SD] and then subtracted from measurements on the MDMs. To define the total area of the cells, the nuclear signal was measured with the DAPI filter.
2.11. Statistics
For statistical analysis, the statistical software GraphPad Prism (Version 5) was used. The statistical test used for a given experiment is indicated in the figure legends.
3. Results
3.1. SAMHD1 is highly expressed in HSPCs cultured in a medium enriched with cytokines
We first characterized HSPCs freshly isolated from cord blood by staining for lineage markers and the cell surface markers CD34, CD133 and CD38 overtime. In one representative of three analyses (performed with cells from three different donors), 87.2% of the cells were CD34+Lin− and 84.3% of these cells were CD133+CD38+ (Fig. 1A). Lin−CD34+CD133+CD38+ cells shifted partially after 12 h of cultivation to Lin−CD34+CD133−CD38+ cells, a process which continued over the entire observation time. We observed also a loss of the Lin− CD34+CD133+CD38− cells over time; The Lin−CD34+CD133+ cell population correlates best with HSPC SCID repopulation capacity (Drake et al., 2011).
Fig. 1.
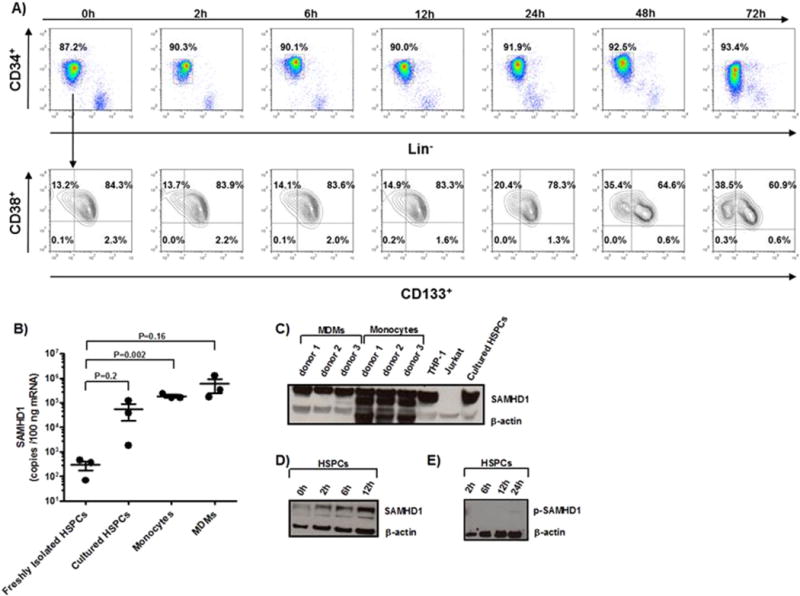
Characterization of HSPCs and GFP expression in lentiviral-transduced HSPCs by flow cytometry and analysis of SAMHD1 expression in cultured HSPCs. (A) Freshly isolated cord blood-derived HSPCs were cultured in the presence of cytokines as mentioned in “Materials and methods” section, and characterized by the cell surface expression of lineage markers, CD34, CD133, CD38 over time. A representative dataset of three independent experiments is shown. Expression levels of mRNA (B) and protein of SAMHD1 (C and D) and p-SAMHD1 (E) were measured in freshly isolated HSPCs, HSPCs cultured for 3 days (B and C), 0–12 h (D) or 2–24 h (E) in a cytokine-enriched milieu, as well as in freshly isolated monocytes and MDMs (C). THP-1 cells and Jurkat cells served as positive and negative controls, respectively, for Western blotting (C). RNA was extracted from 1 × 106 cells, cDNA was synthesized, and the levels of SAMHD1 expression were analyzed by RT-qPCR. Data are presented as absolute copy numbers/100 ng of mRNA (n = 3). Dots represent individual measurements with mean ± SEM indicated. Statistical analysis was done using two-tailed unpaired t-test. Protein levels of SAMHD1 and p-SAMHD1 were determined by Western blotting; β-actin served as loading control. A representative dataset of three independent experiments is shown.
SAMHD1 mRNA was expressed at much lower levels in freshly isolated HSPCs than in HSPCs cultured for 3 days (Fig. 1B). SAMHD1 protein expression was already markedly increased at 2 h in culture and progressively increased over the next 3 days (Fig. 1C and D). Notably, retroviral restriction by SAMHD1 is regulated by its phosphorylation. p-SAMHD1 is unable to block HIV-1 infection, but it still depletes intracellular dNTP pools (White et al., 2013; Cribier et al., 2013). In the present study, increased expression of SAMHD1 after 12 h of cell culture did not lead to any concomitant increase of p-SAMHD1 expression (Fig. 1E). Thus, we hypothesized that the increased levels of SAMHD1 when culturing HSPCs limit their susceptibility to transduction with lentiviral vectors.
3.2. Transduction efficacy of HSPCs with lentiviral vectors is not affected by Vpx+ viruses, dNs or SAMHD1 shRNA pretreatment, and dNTPs only marginally increase the transduction efficacy of HSPCs
We optimized the transduction procedure for our purposes based mainly on protocols previously reported in the literature (for details see “Material and methods” section). HSPCs were challenged with SIN vector pLen-EF1α-GFP-derived lentivirus at MOI of 100. The transduction efficiency was 30–50% as shown in Fig. 2A. Here, we explored whether SAMHD1 affects the lentiviral-based transduction rate in HSPCs.
We first validated all the methods we used by documenting the Vpx-mediated degradation of SAMHD1 in MDMs and the subsequent increase of transduction rates with lentiviral vectors. Experiments examining the role of SAMHD1 in HSPCs were always done in parallel with MDMs as positive controls. HSPCs were pretreated with either Vpx+/− viruses from SIV for 2 h or were mock-treated and then transduced with Len-EF1α-GFP-based viruses. Pretreatment with Vpx+ viruses did not result in a higher transduction rate than pretreatment with Vpx− viruses or mock-treatment as determined by the number of GFP+ cells 3 days post-transduction (Fig. 2B–D).
In previous work, the actions of SAMHD1 were counteracted in MDMs and resting CD4+ T cells by adding dNs as dNTP precursors to the culture medium 2 h before infection (Lahouassa et al., 2012; Baldauf et al., 2012). Overall, this procedure did not affect the transduction rate of HSPCs (Figs. 2E and S1). Moreover, combining Vpx+ viruses and dNTPs had no effect on the transduction efficiency of HSPCs (data now shown).
3.3. Vpx+ virus pretreatments do not result in an accumulation of integrated provirus and viral DNA intermediates in HSPCs
A GFP-based readout does not define at which level a potential block exists in the HIV replication cycle, or may be even misleading when studying restriction factors acting at various levels of the HIV replication cycle. To address this issue, we first quantified the integrated provirus 3 days post-infection by Alu-PCR. We found that Vpx significantly enhanced HIV-1-based lentiviral vector integration in MDMs by more than 15-fold over the Vpx− virus-treated controls; however, there was no positive effect of Vpx+ viruses in HSPCs (Fig. 3A). We wondered whether Vpx might have an effect on earlier steps of the replication cycle. However, this was not the case in HSPCs when examining early, intermediate, or late transcripts (Fig. 3B).
Fig. 3.
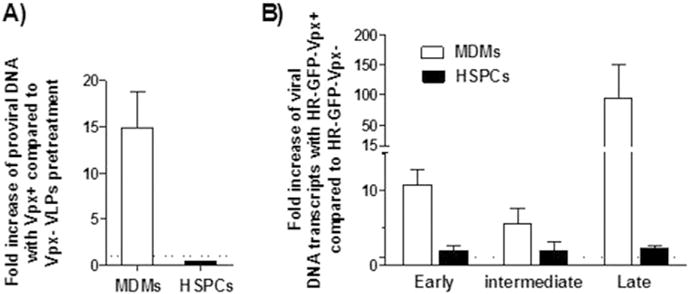
Vpx+ virus treatment does not increase the proviral DNA or the viral DNA intermediates in HSPCs. MDMs and HSPCs were pretreated for 2 h with Vpx+/− viruses, followed by Len-EF1α-GFP transduction at an MOI of 100, and subsequently cultured for 3 days before harvesting. Data are presented as fold increase of samples pretreated with Vpx+ and Vpx− viruses (A). MDMs or HSPCs transduced with HR-GFP-Vpx+ (HR-GFP-Vpx+ carries Vpx within the virions and its lentiviral gene encodes GFP) or HR-GFP-Vpx− at an MOI of 100, and subsequently cultured for 3 days before harvesting. Fold inductions of early, intermediate and late transcripts from HR-GFP-Vpx+ were determined by the ratio of MNEs of samples exposed to HR-GFP-Vpx+ and HR-GFP-Vpx−. (B). The dashed line indicates fold increase of 1. Quantification of proviral DNA and viral DNA intermediates were done as described in “Materials and methods” section. Dots represent three individual measurements with mean ± SEM indicated.
3.4. HSPCs pretreated with Vpx+ viruses have lower amounts of SAMHD1 protein than mock-treated control
We wondered whether the absence of effect of Vpx on transduction efficacy in HSPCs is due to a lack of its effect on SAMHD1 degradation. We, thus, pretreated freshly isolated HSPCs from three donors with Vpx+ viruses for 2 h, kept them for 3 days in a transduction medium, and eventually harvested them for immunoblotting of SAMHD1. The protein levels of SAMHD1 were comparable in cultured HSPCs and MDMs; Vpx degraded SAMHD1 in HSPCs, but the absolute Vpx-mediated decreases were significantly lower in HSPCs than in MDMs (Fig. 4A and B). Thus, the remaining SAMHD1 levels in HSPCs might be sufficient to block transduction. It also appears that HSPCs have higher levels of p-SAMHD1 protein than MDMs subsequent to treatment with Vpx+ viruses (Fig. 4C and D), pointing to a more efficient Vpx-dependent degradation of p-SAMHD1 in MDMs. It has to be noted that the efficiency of Vpx+ viruses to degrade SAMHD1 in MDMs is also donor dependent (Fig. 4A and C). Irrespective whether the phosphorylated or unphosphorylated form of SAMHD1 is anti-HIV active, we speculated that the degradation pathway is saturated with the residual SAMHD1 in HSPCs.
Fig. 4.
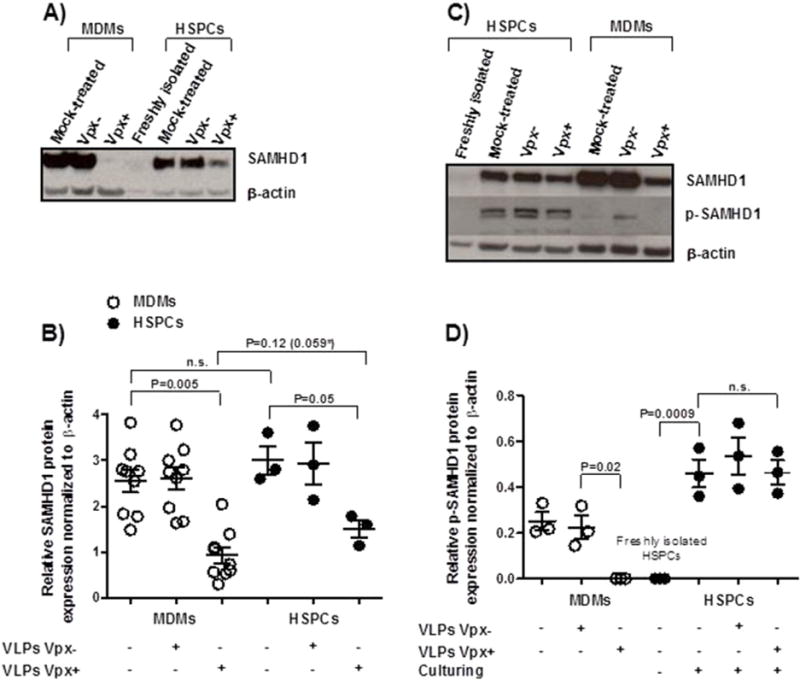
Vpx+ virus treatment more effectively reduces SAMHD1 and p-SAMHD1 in MDMs than HSPCs. MDMs from nine donors and HSPCs from three donors were pretreated for 2 h with Vpx+/− viruses before cell lysis. Protein levels of SAMHD1 and p-SAMHD1 were determined by Western blotting; β-actin served as loading control. A representative Western blot is shown in (A and C) and semi-quantitative analysis of experiments is shown in (B and D). Dots represent individual measurements with mean ± SEM indicated in (B and D). Statistical analysis was done using two-tailed paired t-test when comparing the same cell types from the same donor and unpaired one for comparison of distinct cell types (*P value if we use a one sided test).
3.5. Vpx viruses, dN and dNTP-pretreatment influence the intracellular dNTP pools in HSPCs
The intracellular dNTP concentrations of HSPCs and whether they will be affected by SAMHD1 are unknown. Thus, we determined the intracellular dNTP levels in HSPCs at baseline and subsequent to treatment with Vpx+/− viruses from three independent donors in a single nucleotide incorporation assay 3 days later. dNTP concentrations in both Vpx+ virus-pretreated HSPCs and MDMs were greater than in Vpx− virus-treated cells (Fig. 5A and B). Cellular dNTP concentrations were significantly different in HSPCs and MDMs. HSPCs had 10–30 times lower dNTP concentrations than MDMs before and after Vpx+ virus treatment. Similarly, we evaluated whether adding dNs or dNTPs changed the intracellular dNTP pool in HSPCs and MDMs. Indeed, it did so in both cell types and there was a more prominent increase in intracellular dNTP concentrations in dN-treated MDMs than in HSPCs (Fig. 5C–F). The increase in dNTPs might still be insufficient to support efficient reverse transcription, suggesting that low dNTPs could be a restricting factor in cultured HSPCs.
Fig. 5.
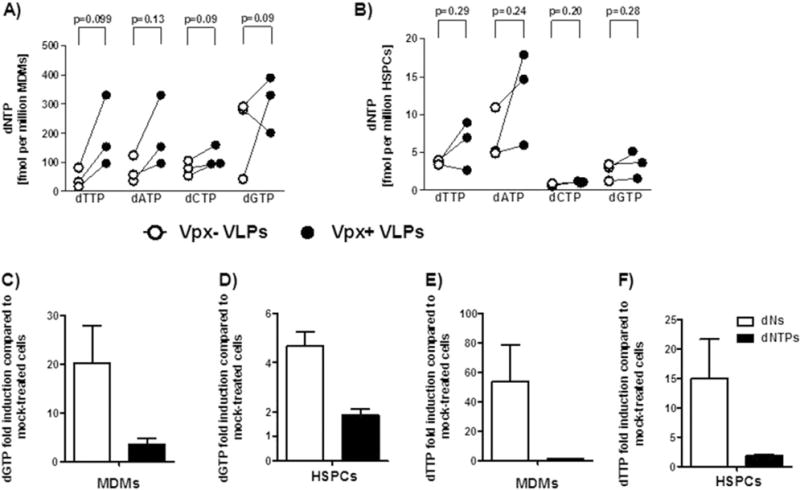
Vpx+ virus, dN and dNTP treatments result in minor increases in the dNTP pool of HSPCs. MDMs (A, C and E) and freshly isolated HSPCs (B, D and F) were pretreated for 2 h with Vpx+/− viruses, dNs or dNTPs, and cultured for 3 days before cell lysis. Quantification of dNTPs was done by a single nucleotide incorporation assay (see “Material and methods” section). Fold induction was calculated based on the absolute values of vero- vs mock-treated samples (C–F). White dots represent Vpx− virus-pretreated samples, black dots represent Vpx+ virus-pretreated samples (A–B). White bars represent dN-pretreated samples, black bars represent dNTP-pretreated samples (C–F). Data are from three donors and bars represent mean ± SEM. Statistical analysis was done using two-tailed paired t-test.
3.6. Assessment of Vpx-mediated degradation of SAMHD1 when using a lentivirus carrying Vpx+ and encoding GFP
To corroborate our data, we used shRNA to silence the expression of SAMHD1. shRNA against SAMHD1 knocked down SAMHD1 expression in THP-1 cells (Fig. S2A). It was less efficient in HSPCs and led only to a partial knockdown of SAMHD1, which did not translate into any increase in the transduction rate (Fig. S2B). We saw no difference, either when SAMHD1 shRNA and Vpx + viruses were combined. We cannot exclude that the lack of an increase in transduction is due to the rather modest silencing or that viruses containing Vpx may enter only a fraction of cells, which are not necessarily targeted by subsequent viral challenge. In the latter case, we may miss a Vpx-mediated effect on lentiviral transduction. Thus, we eventually used a replication-incompetent HIV-1 GFP reporter virus HR-GFP-Vpx +/−, which carries Vpx and encodes GFP, and its control lacking Vpx (HR-GFP-Vpx −). Cells transduced by these virions can be identified by GFP expression, and should be exposed to Vpx’s effects. In one representative example, SAMHD1 was depleted from 4.96% in the mock-transduced to 1.9% in the HR-GFP-Vpx+-transduced group 3 days after challenge, and depletion lasted till day 7. Of note, GFP+ cells were detected mostly in the cell population expressing low levels of SAMHD1 (Fig. 6A and B). GFP+ cells were increased from 1.3% to 4.2% at day 7 compared with HR-GFP-Vpx-transduced control (Fig. 6A). The number of GFP+ cells and mean fluorescence intensity (MFI) of GFP+ cells in HR-GFP-Vpx+ transduced group increased significantly more than in the HR-GFP-Vpx− control (Fig. 6C and D), with the increase being most prominent 7 days post-transduction. The residual cell population expressing high amounts of SAMHD1 remained refractory to lentiviral transduction.
Fig. 6.
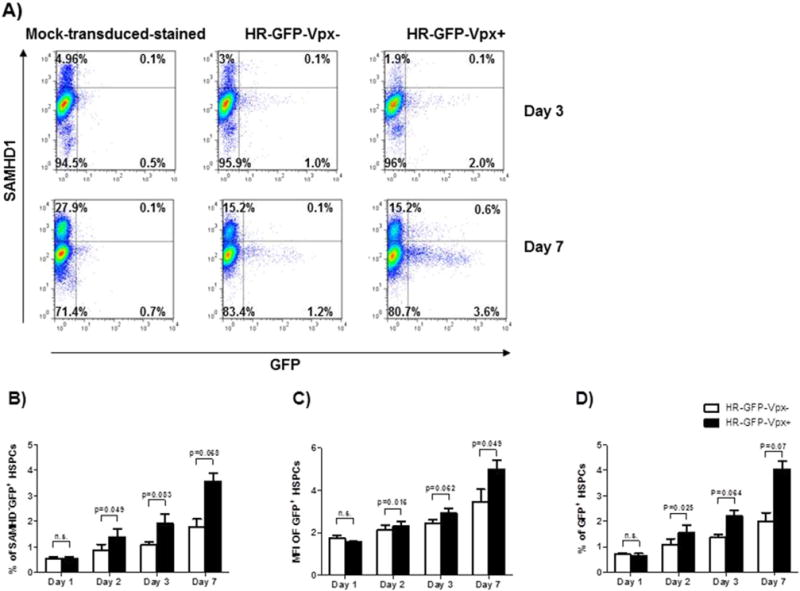
Vpx partially overcomes restriction to HIV-1-based lentiviral transduction in HSPCs. HSPCs were transduced with HR-GFP-Vpx+ or HR-GFP-Vpx− at an MOI of 100, and were subsequently cultured for 7 days. SAMHD1 and GFP expressions were analyzed by flow cytometry at days 3 and 7 (A). The percentage of SAMHD1−GFP+ cells in total GFP+ cells is shown in (B). MFI and transduction efficiency of HR-GFP-Vpx+ or HR-GFP-Vpx− transduced HSPCs are shown in (C and D). A representative dataset of three independent experiments is shown. Statistical analysis was done using two-tailed paired t-test.
We observed a difference in transduction efficacy when Vpx was delivered in trans or in cis. The decrease of SAMHD1 was similar with either virus particle generated (Fig. 7). In fact, the similar decrease in SAMHD1 excludes those of the Vpx-mediated effects observed with the vector system in cis is only due to an overall more potent reduction in SAMHD1; we hold on to our hypothesis that the consecutive challenge with virus particles which first deliver Vpx in trans and second the reporter gene, may target different cells and thus is poorly sensitive. In contrast, the vector system in cis permits to focus on the very small subsets of cells transduced.
Fig. 7.
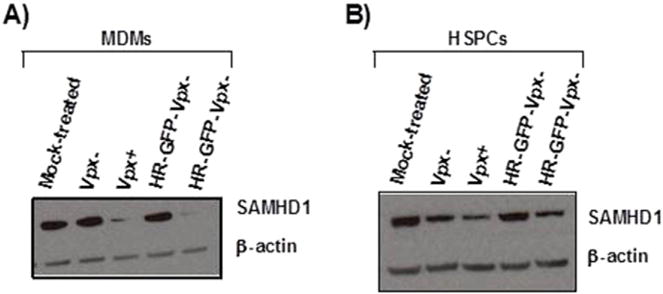
HR-GFP-Vpx+/− and Vpx+/− viruses have similar efficacy in degrading SAMHD1 in both MDMs and HSPCs. MDMs and HSPCs from three donors were transduced with Vpx+/− viruses or with HR-GFP-Vpx+/− viruses before cell lysis. Protein levels of SAMHD1 were determined by Western blotting; β-actin served as loading control. A representative Western blot is shown (A and B).
4. Discussion
In the current study, we examined the role of SAMHD1 in restricting HIV-1-based lentiviral transduction of HSPCs. We made several main findings: i) SAMHD1 was highly expressed in HSPCs cultured with cytokines. In contrast, freshly isolated HSPCs had low SAMHD1 expression. Expression levels of SAMHD1 in cultured HSPCs were even close to those found in myeloid cells, including monocytes and MDMs. ii) While pretreatment of HSPCs with Vpx+ viruses or SAMHD1 shRNA resulted in a significant decrease of SAMHD1, we did not observe any increase in subsequent lentiviral transduction. However, by using viruses carrying Vpx and encoding GFP, which is more accurate for examining the role of SAMHD1 in lentiviral transduction of HSPCs than the consecutive exposure to viruses carrying Vpx and then viruses encoding GFP, we found a significant but modest increase in the number of GFP+ cells in the HSPCs exposed to viruses with Vpx. The GFP+ cells exposed to viruses carrying Vpx showed a decrease in SAMHD1 expression, pointing to Vpx-mediated degradation of SAMHD1 resulting in the increased expression of GFP. Thus, we concluded that Vpx promotes lentiviral-based transduction also in a subset of HSPCs but that other blocks mainly at cell entry are the major limiting step for lentiviral-based transduction of HSPCs.
SAMHD1 emerges as a ubiquitous and potent barrier to productive HIV-1 infection in DCs, myeloid cells, and resting CD4+ T cells. Thus, it appeared likely that SAMHD1 might also be effective when transducing HSPCs with lentiviral vectors. Efficient transduction of HSPCs requires prestimulation with cytokines. We were surprised to find that SAMHD1 levels were very low or even absent in freshly isolated HSPCs, while SAMHD1 was already highly expressed in HSPCs 2 h after the start of culturing. Therefore, we hypothesized that Vpx-mediated SAMHD1 degradation would result in a significant increase in the transduction rate of HSPCs with lentiviral vectors. Surprisingly, this was not the case. We even tested MOIs of 1 and 10 to exclude that the conventionally used high MOI of 100 masked the effects of Vpx+ VLP pretreatment (data not shown). To validate the results we obtained by monitoring GFP expression, we also quantified the number of chromosomally integrated proviral DNA: results were identical with a lack of increase of proviral DNA in HSPCs, while there was a substantial increase in MDMs after pretreatment with Vpx+ viruses. We also quantified the viral DNA intermediates during reverse transcription in HSPCs and MDMs pretreated with Vpx. We observed no increase in HSPCs and a vigorous one in MDMs. Thus, overall, Vpx+ viruses seemed to have no role in relieving the block in lentiviral transduction in HSPCs.
We wondered if Vpx+ viruses participate in the degradation of SAMHD1 in HSPCs. Indeed, that was the case: Vpx degraded SAMHD1 in HSPCs, but residual levels tended to be higher in HSPCs than in MDMs. Considering the data so far, we expected that Vpx had a more modest effect in HSPCs than MDMs and thus a one-sided statistical test can be justified; using a two-sided statistical test gave no significant result and thus we prefer to define it more as a trend than as a significant difference. We further verified to what extent Vpx affected the activity of SAMHD1 or p-SAMHD1. p-SAMHD1 has no anti-HIV activity while still being able to deplete the dNTP pool (White et al., 2013; Cribier et al., 2013). Indeed, phosphorylation of SAMHD1 negatively regulates SAMHD1’s RNase activity and impedes HIV-1 restriction (Ryoo et al., 2014). Taking into account our data so far, we were not surprised that HSPCs had a much higher p-SAMHD1 protein expression than MDMs, and that Vpx had only a minor effect on decreasing p-SAMHD1 in HSPCs. These results might explain that despite the overall substantial Vpx-mediated decrease of SAMHD1 in HSPCs, we did not observe any clear change in the transduction rate.
While the mechanism of SAMHD1’s anti-HIV activity (i.e., dNTPase activity vs RNase activity) is controversial, we investigated whether Vpx-mediated degradation of SAMHD1 affects the dNTP pool in HSPCs. Vpx-mediated degradation of SAMHD1 only slightly increased the dNTP pool in HSPCs, but greatly in MDMs, as reported (Lahouassa et al., 2012). This negative result might be explained by the lack of degradation of p-SAMHD1. We observed substantial increases of the dNTP pools when adding dNs and less prominent ones when adding dNTPs to the two cell types; again, it was much more pronounced in MDMs. Lentiviral reverse transcriptases uniquely remain functional even at the low dNTP concentrations found in non-dividing cells (Amie et al., 2013). Thus, the regulation of the dNTP pools by SAMHD1 may be an epiphenomenon of studying SAMHD1’s anti-HIV activity rather than being causally related. However, we cannot exclude that changes in the dNTP pool may contribute to a minor extent to the reverse transcriptase activity.
The assays used above might confound a minor role of Vpx-mediated degradation of SAMHD1 since the consecutive treatment first with viruses carrying Vpx or silencing SAMHD1 with shRNA and the adding reporter viruses may not necessarily target the same cells. In contrast, this is the case when using viruses carrying Vpx and encoding GFP, and thus, these viruses may even reveal minor effects of Vpx on lentiviral transduction rate (Lahouassa et al., 2012; Baldauf et al., 2012). Indeed, by this approach, we found a minor but significant increase of GFP+ cells going along with a decrease in SAMHD1. Moreover, the residual cell population that expresses high amounts of SAMHD1 remained refractory to lentiviral transduction.
From an evolutionary perspective, the high level of resistance of HSPCs to lentiviruses makes sense, since any major chromosomal vulnerability would have more serious implications than in progeny cells. The high-level expression of SAMHD1 in cultured HSPCs is reminiscent of the one in activated CD4+ T cells (Baldauf et al., 2012). In activated CD4+ T cells, the high expression of SAMHD1 goes along with a high concentration of dNTP and high degree of permissiveness to HIV and HIV-1-based transduction. In HSPCs, we encounter an entirely different situation with a low concentration of dNTPs and a low permissiveness to HIV-1-based lentiviral vectors. Thus, each cellular setting must be examined separately.
5. Conclusions
In summary, HSPCs, in culture, express high levels of SAMHD1. Vpx-mediated decrease of SAMHD1 relieves only marginally the restriction of lentiviral-based transduction. The data imply that other blocks mainly at cell entry are the major limiting step for efficient transduction. Indeed, a recent study showed that the LDL receptor acts as the receptor for VSV-G-pseudotyped particles (Amirache et al., 2014), and lack of it appears to be at the origin of the poor permissiveness of HSPCs to lentiviral transduction.
Supplementary Material
Acknowledgments
We thank D. Boden (Tibotec, Belgium) for providing pLen-EF1α-GFP and the NIH AIDS Reagent Program for providing Raltegravir (Cat # 11680; from Merck & Company). We are grateful to N.R. Landau, O.T. Fackler H.M. Baldauf, V. Vongrad, Y.L. Kok and A. Scherrer for advice. The study was supported by the OPO-Foundation and the clinical research focus program “Human Hemato-Lymphatic Diseases” of the University of Zürich (10/2012–10/2015). RFS is supported by the SNF (SSAJRP; IZLSZ3_149100/1). This work was partially supported by US National Institute of Health grant GM104198 (B.K.).
Abbreviations
- HSPCs
hematopoietic stem and progenitor cells
- VLPs
viral like particles
- IL-2R
interleukin-2 receptor
- SIN
self-inactivating
- SAMHD1
sterile alpha motif domain and HD domain-containing protein 1
- dGTP
deoxyguanosine-triphosphate
- dNTPs
deoxynucleoside triphosphates
- dNs
deoxynucleosides
- DCs
dendritic cells
- PBMCs
peripheral blood mononuclear cells
- MDMs
monocyte-derived macrophages
- SCF
stem cell factor
- Flt3L
Fms-related tyrosine kinase 3 ligand
- IL-3
interleukin-3
- TPO
thrombopoietin
- PEI
polyethylenimine
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- MNE
mean normalized gene expression
- EVF
efavirenz
- RTV
raltegravir
- TGV
total gray values
- MFI
mean fluorescence intensity
Footnotes
Author contribution: DL designed, conducted and analyzed all experiments. ES, AA, M-A.R, SI, C-N.K, BK, O-T.K assisted in some experiment. DL, AA and RS wrote the paper.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.scr.2015.06.012.
References
- Aiuti A, Biasco L, Scaramuzza S, Ferrua F, Cicalese MP, Baricordi C, Dionisio F, Calabria A, Giannelli S, Castiello MC, Bosticardo M, Evangelio C, Assanelli A, Casiraghi M, Di Nunzio S, Callegaro L, Benati C, Rizzardi P, Pellin D, Di Serio C, Schmidt M, Von Kalle C, Gardner J, Mehta N, Neduva V, Dow DJ, Galy A, Miniero R, Finocchi A, Metin A, Banerjee PP, Orange JS, Galimberti S, Valsecchi MG, Biffi A, Montini E, Villa A, Ciceri F, Roncarolo MG, Naldini L. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott–Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althaus CF, Gianella S, Rieder P, von Wyl V, Kouyos RD, Niederost B, Schmid A, Metzner KJ, Joos B, Gunthard HF, Fischer M. Rational design of HIV-1 fluorescent hydrolysis probes considering phylogenetic variation and probe performance. J Virol Methods. 2010;165:151–160. doi: 10.1016/j.jviromet.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Amie SM, Noble E, Kim B. Intracellular nucleotide levels and the control of retroviral infections. Virology. 2013;436:247–254. doi: 10.1016/j.virol.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirache F, Levy C, Costa C, Mangeot PE, Torbett BE, Wang CX, Negre D, Cosset FL, Verhoeyen E. Mystery solved: VSV-G-LVs do not allow efficient gene transfer into unstimulated T cells, B cells, and HSCs because they lack the LDL receptor. Blood. 2014;123:1422–1424. doi: 10.1182/blood-2013-11-540641. [DOI] [PubMed] [Google Scholar]
- Amsellem S, Ravet E, Fichelson S, Pflumio F, Dubart-Kupperschmitt A. Maximal lentivirus-mediated gene transfer and sustained transgene expression in human hematopoietic primitive cells and their progeny. Mol Ther. 2002;6:673–677. [PubMed] [Google Scholar]
- Audige A, Schlaepfer E, Bonanomi A, Joller H, Knuchel MC, Weber M, Nadal D, Speck RF. HIV-1 does not provoke alteration of cytokine gene expression in lymphoid tissue after acute infection ex vivo. J Immunol. 2004;172:2687–2696. doi: 10.4049/jimmunol.172.4.2687. [DOI] [PubMed] [Google Scholar]
- Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med. 2012;18:1682–1687. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquinero J, Segovia JC, Ramirez M, Limon A, Guenechea G, Puig T, Briones J, Garcia J, Bueren JA. Efficient transduction of human hematopoietic repopulating cells generating stable engraftment of transgene-expressing cells in NOD/SCID mice. Blood. 2000;95:3085–3093. [PubMed] [Google Scholar]
- Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- Cartier N, Aubourg P. Hematopoietic stem cell transplantation and hematopoietic stem cell gene therapy in X-linked adrenoleukodystrophy. Brain Pathol. 2010;20:857–862. doi: 10.1111/j.1750-3639.2010.00394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribier A, Descours B, Valadao AL, Laguette N, Benkirane M. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 2013;3:1036–1043. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Cronin J, Zhang XY, Reiser J. Altering the tropism of lentiviral vectors through pseudotyping. Curr Gene Ther. 2005;5:387–398. doi: 10.2174/1566523054546224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descours B, Cribier A, Chable-Bessia C, Ayinde D, Rice G, Crow Y, Yatim A, Schwartz O, Laguette N, Benkirane M. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology. 2012;9:87. doi: 10.1186/1742-4690-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond TL, Roshal M, Jamburuthugoda VK, Reynolds HM, Merriam AR, Lee KY, Balakrishnan M, Bambara RA, Planelles V, Dewhurst S, Kim B. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J Biol Chem. 2004;279:51545–51553. doi: 10.1074/jbc.M408573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake AC, Khoury M, Leskov I, Iliopoulou BP, Fragoso M, Lodish H, Chen J. Human CD34+ CD133+ hematopoietic stem cells cultured with growth factors including Angptl5 efficiently engraft adult NOD-SCID Il2rgamma−/− (NSG) mice. PLoS One. 2011;6:e18382. doi: 10.1371/journal.pone.0018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frimpong K, Spector SA. Cotransduction of nondividing cells using lentiviral vectors. Gene Ther. 2000;7:1562–1569. doi: 10.1038/sj.gt.3301283. [DOI] [PubMed] [Google Scholar]
- Glimm H, Oh IH, Eaves CJ. Human hematopoietic stem cells stimulated to proliferate in vitro lose engraftment potential during their S/G(2)/M transit and do not reenter G(0) Blood. 2000;96:4185–4193. [PubMed] [Google Scholar]
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuster H, Opravil M, Ott P, Schlaepfer E, Fischer M, Gunthard HF, Luthy R, Weber R, Cone RW. Treatment-induced decline of human immunodeficiency virus-1 p24 and HIV-1 RNA in lymphoid tissue of patients with early human immunodeficiency virus-1 infection. Am J Pathol. 2000;156:1973–1986. doi: 10.1016/S0002-9440(10)65070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol. 2012;13:223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- meBiffi A, Montini E, Lorioli L, Cesani M, Fumagalli F, Plati T, Baldoli C, Martino S, Calabria A, Canale S, Benedicenti F, Vallanti G, Biasco L, Leo S, Kabbara N, Zanetti G, Rizzo WB, Mehta NA, Cicalese MP, Casiraghi M, Boelens JJ, Del Carro U, Dow DJ, Schmidt M, Assanelli A, Neduva V, Di Serio C, Stupka E, Gardner J, von Kalle C, Bordignon C, Ciceri F, Rovelli A, Roncarolo MG, Aiuti A, Sessa M, Naldini L. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- Miller DG, Adam MA, Miller AD. Gene transfer by retrovirus vectors occurs only in cells that are actively replicating at the time of infection. Mol Cell Biol. 1990;10:4239–4242. doi: 10.1128/mcb.10.8.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RC, Schlaepfer E, Baenziger S, Crameri R, Zeller S, Byland R, Audige A, Nadal D, Speck RF. HIV interferes with SOCS-1 and -3 expression levels driving immune activation. Eur J Immunol. 2011;41:1058–1069. doi: 10.1002/eji.201041198. [DOI] [PubMed] [Google Scholar]
- Millington M, Arndt A, Boyd M, Applegate T, Shen S. Towards a clinically relevant lentiviral transduction protocol for primary human CD34 hematopoietic stem/progenitor cells. PLoS One. 2009;4:e6461. doi: 10.1371/journal.pone.0006461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo J, Choi J, Oh C, Kim S, Seo M, Kim SY, Seo D, Kim J, White TE, Brandariz-Nunez A, Diaz-Griffero F, Yun CH, Hollenbaugh JA, Kim B, Baek D, Ahn K. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat Med. 2014;20:936–941. doi: 10.1038/nm.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer E, Rochat MA, Duo L, Speck RF. Triggering TLR2, 3, 4, 5 and 8 reinforces the restrictive nature of M1- and M2-polarized macrophages to HIV. J Virol. 2014;88(17):9769–9781. doi: 10.1128/JVI.01053-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C. Gene therapy finds its niche. Nat Biotechnol. 2011;29:121–128. doi: 10.1038/nbt.1769. [DOI] [PubMed] [Google Scholar]
- van Lent AU, Centlivre M, Nagasawa M, Karrich JJ, Pouw SM, Weijer K, Spits H, Blom B, Legrand N. In vivo modulation of gene expression by lentiviral transduction in “human immune system” Rag2−/− gamma c−/− mice. Methods Mol Biol. 2010;595:87–115. doi: 10.1007/978-1-60761-421-0_6. [DOI] [PubMed] [Google Scholar]
- White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, Tuzova M, Diaz-Griffero F. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe. 2013;13:441–451. doi: 10.1016/j.chom.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


