These bacteria may cause paucisymptomatic bacteremia and endocarditis in humans.
Keywords : Bartonella, zoonoses, tickborne infections, ticks, bacteria, vector, vector-borne infections, bacteremia, France
Abstract
Certain Bartonella species are known to cause afebrile bacteremia in humans and other mammals, including B. quintana, the agent of trench fever, and B. henselae, the agent of cat scratch disease. Reports have indicated that animal-associated Bartonella species may cause paucisymptomatic bacteremia and endocarditis in humans. We identified potentially zoonotic strains from 6 Bartonella species in samples from patients who had chronic, subjective symptoms and who reported tick bites. Three strains were B. henselae and 3 were from other animal-associated Bartonella spp. (B. doshiae, B. schoenbuchensis, and B. tribocorum). Genomic analysis of the isolated strains revealed differences from previously sequenced Bartonella strains. Our investigation identifed 3 novel Bartonella spp. strains with human pathogenic potential and showed that Bartonella spp. may be the cause of undifferentiated chronic illness in humans who have been bitten by ticks.
Bartonella spp. cause varied and multifaceted human diseases, including cat scratch disease (B. henselae), Carrion’s disease (B. bacilliformis), trench fever (B. quintana), endocarditis (B. quintana and B. henselae) (1,2), bacillary angiomatosis (B. quintana and B. henselae), and hepatic peliosis (B. henselae). Bartonella spp. can also cause prolonged intra-erythrocytic bacteremia in both humans and animals (3): in humans, B. quintana, B. bacilliformis, and B. rochalimae are known pathogens, and in animals, B. henselae, B. clarridgeiae, and B. koehlerae have been identified in felids; B. grahamii, B. taylorii, B. doshiae, B. birtlesii, and others in rodents; and B. bovis, B. chomelii, B. schoenbuchensis, in ruminants. In humans, chronic bacteremia caused by B. quintana causes few obvious symptoms apart from generalized fatigue and nonspecific leg pain (1,4).
It has been assumed that each Bartonella species infected 1 or a few closely related mammalian reservoir hosts, in which infection caused long-lasting bacteremia. Nonreservoir hosts were considered incidentally infected without bacteria being detected in blood. Recently, these assumptions has been contradicted by studies describing animal-associated Bartonella spp. indirectly associated with bacteremia and a spectrum of diverse symptoms in immune-competent persons who had contact with animals, arthropods, or both, which are natural routes of Bartonella transmission (5–7). In some cases, the source of infection remains unknown; ticks have been suggested as a possible source of animal-associated Bartonella infection in humans (6,8–10).
Related to a patient’s history of tick bites, it is common for physicians to suspect Lyme disease, some rickettsial diseases, or tickborne encephalitis. However, in many cases, the diagnosis is not confirmed by serologic or DNA-based tests. In recent years, alternate interpretations of Lyme disease serology have flourished, leading to considerable discord between formal institutions for infectious disease and patient advocacy associations. Thus, unexplained symptoms after tick bites have become an issue of increasing importance for patients and their physicians (11,12).
In this context, we screened for the presence of Bartonella in the blood of patients reporting tick bites and with unexplained and aspecific symptoms. Here we report the isolation and genomic sequencing of 6 Bartonella strains obtained by blood culture from 66 patients. Three strains were identified as B. henselae, and 3 other strains were identified as different animal–associated species (B. doshiae, B. tribocorum, and B. schoenbuchensis).
Methods
Patients
During January–June 2013, we conducted a study of a cohort of 66 French patients who had consulted their doctors for chronic symptoms appearing after a tick bite. The entire study protocol was approved by the ethics committee of the Institut Federatif de Recherche 48 under reference 13–022–1.
All patients associated symptom onset with tick bites that occurred during 2008–2012 (Table 1). At symptom onset, local doctors were consulted, and serologic tests for Lyme borreliosis were performed. All patient samples tested were seronegative for Lyme borreliosis bacteria; however, since that time, their symptoms had become chronic. The patients completed information forms giving informed consent for the use of their samples in the study. All of the patients lived in the countryside, where ticks were abundant and contact with wild animals was possible. The patients reported that they had not undergone antibacterial drug treatment for >3 months before the study.
Table 1. Patients whose blood cultures were positive for Bartonella spp. that had no previously known zoonotic activity, France.
| Case-patient no./age, y/sex | Bartonella spp. | Tick bite date | Pets | Wild animal contact | Main complaints | Bacteremia, CFU/mL |
|---|---|---|---|---|---|---|
| 1/49/F | B. henselae | Multiple since 2008 | Cats, dogs, horses | Rats, fish | Fatigue, muscle pain, headache | 50 |
| 2/58/M | B. henselae | 2011 | Birds, rabbits | No | Fatigue, muscle pain | 70 |
| 3/47/F | B. henselae | 2012 | Dog, hamster | No | Fatigue, generalized pain, insomnia | 80 |
| 4/45/F | B. doshiae | 2009 | No | No | Fatigue, blurred vision, arthralgia | 50 |
| 5/64/M | B. tribocorum | 2012 | Dog | Game animals (hunter) | Fatigue, muscle pain, headache | 60 |
| 6/40/F | B. schoenbuchensis | 2011 | No | No | Fatigue, muscle pain, fever | 850 |
We collected blood samples from each patient in EDTA-containing sample tubes. For a control population, we used anticoagulated blood samples from 70 anonymous healthy blood donors from Paris (France). All samples (control and patients) were tested simultaneously.
Bartonella Isolation from Blood
To specifically isolate Bartonella spp., 100 μL of blood samples from patients or healthy donors were directly plated onto sheep blood agar plates and incubated at 35°C in a humidified atmosphere with 5% CO2 for 45 days. The plates were assessed daily from days 7–45 before the culture was deemed negative (i.e., absence of colony in the absence of contamination) (1). Colony-forming units (CFU) were counted and bacteremia (UFD/mL of blood) evaluated.
Genome Sequencing, Assembly, and Analysis
We extracted genomic DNA from each isolated strain by using the EZ1 automated extraction system (QIAGEN, Hilden, Germany), following the manufacturer’s recommendations. Bacterial genomic DNA was sequenced by using the Nextera XT DNA sample prep kit (Illumina Inc., San Diego, CA, USA) and a 2×250 paired-end protocol with the MiSeq pyrosequencer (Illumina), according to the manufacturer’s instructions. We aligned each genome by using Mira version 3.2 software in the mapping mode (13). The resulting contigs were combined by using Opera version 1.2 (14) and GapFiller (15) software. Finally, the genomic assemblies were improved with manual refinement by using the CLC Genomics version 4.7.2 software package (CLC Bio, Aarhus, Denmark). Noncoding genes and miscellaneous features were predicted by using RNAmmer (16) and ARAGORN (17). Coding DNA sequences were predicted by using Prodigal (18), and functional annotation was achieved by using BLAST+ (19) and HMMER3 (20) against the UniProtKB database (21). Coding DNA sequences were also annotated by using the Clusters of Orthologous Groups databases (22) with blastp (http://blast.ncbi.nlm.nih.gov/Blast.cgi) default parameters.
Single-nucleotide polymorphisms (SNPs) among genomes were identified by using SNIT software (23). SNPs were searched in regions exhibiting >95% nt sequence identity and the SNIT software was used with default parameters except for the Tandem Repeat Finder filter for avoiding ambiguous SNPs in repeat regions. We also performed in silico DNA–DNA hybridization (DDH) between Bartonella strains by using GGDC software (24).
Taxonomic Classification
To determine the taxonomic classification of the 6 isolates, we used previously proposed criteria (25) in which the gltA and rpoB gene sequences from each strain were compared to those of validated published Bartonella species. These criteria classify Bartonella isolates within a particular species if they share >96% and 95.4% nucleotide sequence similarity for the gltA and rpoB genes, respectively (25). In our study, gltA and rpoB sequences were retrieved from the genomes.
Results
Bartonella spp. Isolation
Bartonella spp. were isolated by prolonged culture from blood samples of 6 of the 66 patients who reported chronic symptoms following a tick bite. In contrast, samples from the 70 healthy blood donors remained negative after 45 days of incubation.
Bacteremia in the Bartonella infected patients increased from 50 to 850 CFU/mL. For 1 patient (case-patient 2), we had access to 2 blood samples that were taken at a 1-month interval. B. henselae was grown from the 2 samples, with similar bacteremia (50 and 60 CFU/mL, respectively), suggesting chronic bacteremia.
The case-patients who tested positive for Bartonella (Table 1) reported tick bites occurred 1–5 years before blood samples were collected. All of them live in the countryside, in contrast to the healthy blood donors, who were all from Paris, France. The main complaint of the case-patients was chronic fatigue, but they also reported other subjective or nonspecific symptoms (or both), such as headaches and myalgia. A qualifying characteristic of the 70 healthy blood donors was absence of chronic fatigue. Even though potential exposure to ticks is difficult to evaluate, because the anonymous blood donors all lived in Paris, we assumed they were not likely to have frequent tick exposure or wild animal contact.
Taxonomic Classification
Of the 6 Bartonella isolates from this study, 3 (MVT01, MVT02, and MVT03) were classified within the B. henselae species on the basis of both their phylogenetic position and gltA and rpoB sequence similarities (Figure; Tables 1, 2). The isolates from samples from case-patient 2 at a 1-month interval shared 100% identity, based on gltA and rpoB gene comparison. Isolates MVT04, MVT05, and MVT07 were classified within the B. tribocorum, B. doshiae, and B. schoenbuchensis species, respectively.
Figure.
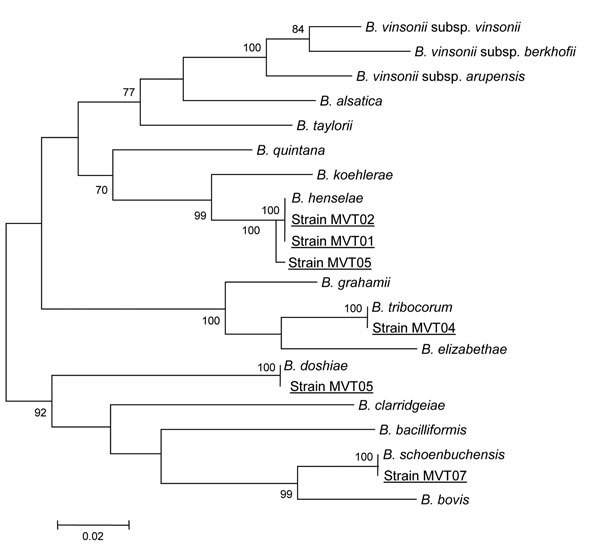
rpoB gene-based phylogenetic tree showing the relationships of 6 Bartonella isolates (underlined). Briefly, rpoB nucleotide sequences were aligned by using ClustalW software (http://www.clustal.org/clustal2/), and phylogenetic relationships were inferred by using the maximum-likelihood strategy and MEGA software (http://www.megasoftware.net). Bootstrap values above 70%, obtained from 500 analyses, are indicated at the nodes. Scale bar represents a 2% nucleotide sequence divergence.
Table 2. Nucleotide similarity of 6 Bartonella isolates from patients in France expressing novel zoonotic activity and their most phylogenetically similar published validated species*.
| Isolate | gltA, % | rpoB, % | Species |
|---|---|---|---|
| MVT01 | 100 | 100 | B. henselae |
| MVT02 | 100 | 100 | B. henselae |
| MVT03 | 99.7 | 99.6 | B. henselae |
| MVT04 | 100 | 100 | B. tribocorum |
| MVT05 | 98.7 | 100 | B. doshiae |
| MVT07 | 100 | 99.9 | B. schoenbuchensis |
*gltA and rpoB sequences were obtained from genomic sequences. Genome sequences of B. henselae strains MVT01, MVT02, and MVT03 were deposited in GenBank under accession numbers HG965802, NZ_LN879429, and HG969191, respectively; the genome sequence of B. tribocorum strain MVT04 was deposited in GenBank under accession numbers HG969192 and HG969193; the genome sequence of B. doshiae strain MVT05 was deposited in GenBank under accession numbers CCBL010000001–CCBL010000013; the genome sequence of B. schoenbuchensis strain MVT07 was deposited in GenBank under accession numbers HG977193–HG977197; the genome sequences of the reference strains B. henselae strain Houston-1, B. tribocorum strain CIP 105476, B. doshiae strain NCTC 12862, and B. schoenbuchensis strain m07a are available in genBank under accession numbers NC_005956, NC_010161, and NC_010160, NZ_JH725094 to NZ_JH725100 and NZ_KB915627-NZ_KB915629, NZ_CM001846-NZ_CM001845, respectively.
The assembly data and main genomic characteristics of each isolated strain are summarized in Table 3; in silico DDH values and SNP numbers are described in Table 1. All studied strains displayed a similar genomic content when compared with reference genomes. The GGDC software we used proposes that DDH values >70% could classify isolates in the same species. Here, intraspecies values ranged from 80.3% to 100% (Table 4). Nevertheless, from 10 to 1,938 SNPs were identified among B. henselae isolates MVT01, MVT02, and MVT03 and from 693 to 2,093 SNPs when comparing these strains to B. henselae Houston-1 (Table 4), confirming that each strain was unique and did not result from cross-contamination or contamination from laboratory strains.
Table 3. Assembly information and main characteristics of 6 sequenced Bartonella genomes from patients in France expressing novel zoonotic activity .
| Genome characteristics | Species and isolate identification |
|||||
|---|---|---|---|---|---|---|
| B. henselae MVT01 | B. henselae MVT02 | B. henselae MVT03 | B. tribocorum MVT04 | B. doshiae MVT05 | B. schoenbuchensis MVT07 | |
| GenBank accession nos. | HG965802 | NZ_LN879429 | HG969191 | HG969192–HG969193 | CCBL010000001–CCBL010000013 | HG977193–HG977197 |
| Size, bp | 1,902,535 | 1,905,383 | 1,975,503 | 2,609,404 | 1,919,109 | 1,734,324 |
| No. contigs | 1 | 1 | 1 | 2 | 13 | 5 |
| Average read coverage | 87 | 94 | 110 | 46 | 15 | 41 |
| Average read length, trimmed | 183 | 190 | 192 | 194 | 168 | 193 |
| Total no. reads, trimmed | 946,882 | 1,034,894 | 1,263,492 | 738,522 | 261,0852 | 666,371 |
| Total no. predicted genes | 1,659 | 1,658 | 1,726 | 2,335 | 1,720 | 1,574 |
| Protein-coding genes | 1,603 | 1,602 | 1,668 | 2,279 | 1,654 | 1,519 |
| rRNA operons | 2 | 2 | 2 | 2 | 2 | 2 |
| tRNAs | 43 | 43 | 45 | 43 | 53 | 41 |
| Other RNAs | 7 | 7 | 7 | 7 | 9 | 8 |
| GC% content | 38.18 | 38.18 | 38.09 | 38.84 | 37.82 | 35.58 |
| Plasmid | 0 | 0 | 0 | 1 | 0 | 1 |
| Genome used as a reference for assembly (accession nos.) | B. henselae Houston-1 (NC_005956) | B. henselae Houston-1 (NC_005956) | B. henselae Houston-1 (NC_005956) | B. tribocorum CIP 105476 (NC_010161, NC_010160) | B. doshiae NCTC 12862 (NZ_JH725094– NZ_JH725100) | B. schoenbuchensis m07a (NZ_KB915627–NZ_KB915629, NZ_CM001846, NZ_CM001845) |
Table 4. Comparisons of 6 sequenced genomes of Bartonella spp. isolated from humans in France expressing novel zoonotic activity showing relationships to each other and to those of closely related strains.
| Compared genomes (GenBank accession nos.) | B. henselae MVT01 | B. henselae MVT02 | B. henselae MVT03 | B. tribocorum MVT04 | B. doshiae MVT05 | B. schoenbuchensis MVT07 | B. henselae Houston-1 | B. tribocorum CIP 105476 | B. doshiae NCTC 12862 | B. schoenbuchensis m07a |
|---|---|---|---|---|---|---|---|---|---|---|
| B. henselae MVT01 (HG965802) | *100% | 100% ± 0.05 | 92.90% ± 1.74 | 28.20% ± 2.43 | 28.10% ± 2.43 | 23.80% ± 2.38 | 98.30% ± 0.64 | 28.20% ± 2.43 | 28.00% ± 2.43 | 23.60% ± 2.38 |
| B. henselae MVT02 (NZ_LN879429) | 10 | 100% | 93.00% ± 1.72 | 28.20% ± 2.43 | 28.10% ± 2.43 | 23.80% ± 2.39 | 98.30% ± 0.64 | 28.20% ± 2.43 | 28.00% ± 2.43 | 23.60% ± 2.38 |
| B. henselae MVT03 (HG969191) | 1,938 | 1,937 | 100% | 28.00% ± 2.43 | 28.00% ± 2.43 | 23.80% ± 2.39 | 92.30% ± 1.82 | 28.00% ± 2.43 | 27.90% ± 2.43 | 23.80% ± 2.39 |
| B. tribocorum MVT04 (HG969192–HG969193) | NA | NA | NA | 100% | 26.50% ± 2.42 | 22.70% ± 2.37 | 28.20% ± 2.43 | 99.30% ± 0.33 | 26.60% ± 2.42 | 22.50% ± 2.36 |
| B. doshiae MVT05 (CCBL010000001–CCBL010000013) | NA | NA | NA | NA | 100% | 23.60% ± 2.38 | 28.10% ± 2.43 | 26.50% ± 2.42 | 81.40% ± 2.72 | 23.60% ± 2.38 |
| B. schoenbuchensis MVT07 (HG977193–HG977197) | NA | NA | NA | NA | NA | 100% | 23.70% ± 2.38 | 22.70% ± 2.37 | 23.60% ± 2.38 | 80.30% ± 2.77 |
| B. henselae Houston-1 (NC_005956) | 693 | 698 | 2,093 | NA | NA | NA | 100% | 28.20% ± 2.43 | 28.00% ± 2.43 | 23.60% ± 2.38 |
| B. tribocorum CIP 105476 (NC_010161, NC_010160) | NA | NA | NA | 51 | NA | NA | NA | 100% | 26.60% ± 2.42 | 22.60% ± 2.37 |
| B. doshiae NCTC 12862 (NZ_JH725094–NZ_JH725100) | NA | NA | NA | NA | >10,000 | NA | NA | NA | 100% | 23.60% ± 2.38 |
| B. schoenbuchensis m07a (NZ_KB915627–NZ_KB915629, NZ_CM001846, NZ_CM001845) | NA | NA | NA | NA | NA | >10,000 | NA | NA | NA | 100% |
*All values are percentage identity + SD. Isolates designated MVT01–07 and MVT07 are isolates from humans in France. Gray shading indicates in silico DNA–DNA hybridization values obtained by using Genome-to-Genome Distance Calculator software (http://ggdc.dsmz.de/); darker shading indicates values >70%. Unshaded values are SNP numbers calculated by using SNP identification for strain typing software (23). NA, not applicable because SNP numbers cannot be estimated for distant genomes.
Of note, B. tribocorum isolate MVT04 and B. schoenbuchensis isolate MVT07 were the only 2 that exhibited plasmids. However, when compared with reference strain m07a, MVT04 and MVT07 carried a large plasmid and not the small plasmid homologous to the cryptic pBGR plasmid harbored by B. grahamii (26).
Discussion
In this study, animal-associated Bartonella isolates were individually cultured from the blood of patients who had been bitten by ticks and reported subjective symptoms, whereas no strains were isolated from healthy blood donors. This report describes the isolation of 3 different animal-associated Bartonella species from human samples, highlighting their potential novel zoonotic properties. Moreover, we found that zoonotic Bartonella spp. can be detected in the blood of afebrile patients, as has been shown for human-specific B. quintana and B. bacilliformis and as was recently reported for Candidatus Bartonella ancashi (27). Chronic bacteremia caused by infection by Bartonella spp. is well-described in many mammals, including humans (4,28). The Bartonella–mammalian host association is considered to be species-specific and attributable to co-evolution between host and pathogen (28). However, we show that animal-associated species can also chronically infect human blood, highlighting the possibility of host shift despite apparent host specificity (28,29).
This work is similar to that of E.B. Breitschwerdt et al. (5–7), who also recovered zoonotic Bartonella spp. from human samples using an in-house technique based on results of blood pre-enrichment followed by PCR detection of Bartonella spp.; members of the same team have investigated many cases of persons who had nonspecific symptoms, including arthralgia, muscle pain, fatigue, headaches, visual blurring, neurocognitive symptoms, and, in 2 case-patients, hemangioendothelioma (30). In total, B. henselae DNA was detected in 47 cases (5,30–33); B. koehlerae (another common agent of feline bacteremia) DNA in 96 cases, including 2 co-infected with B. henselae (31,32,34,35); and B. vinsonii berkhoffii (an agent of canine bacteremia and endocarditis) DNA in 24 cases (31–34), including 16 case-patients with B. henselae and 2 cases of B. melophagi (36). These results have been questioned because minute levels of contamination can result in false positives by PCR. Therefore, we deliberately avoided PCR to overcome this problem, and the resulting strain isolation was consequently straightforward and indisputable. These isolates (Table 2) have been archived in our collection (Collection de Souches de l’Unité des Rickettsies, World Data Center for Microorganisms no. 875, http://www.mediterranee-infection.com/article.php?laref=14&titre=collection-de-souches) and are available upon request under references B546, B547, B548, B549, B550, and B551 for isolates MVT01, MVT02, MVT03, MVT04, MVT05, and MVT07, respectively.
Our findings also confirm studies identifying zoonotic Bartonella in the blood of patients with nonspecific complaints. Among them, B. henselae is well known worldwide as a zoonotic agent infecting both cats and their fleas and has also been found in ticks (10). B. henselae has been detected in the blood of a patient without apparent symptoms 4 months after recovering from cat scratch disease. For this particular case, the sequence of manifestation of cat scratch disease, then bacteremia, followed by endocarditis was proposed because it has been known to occur for B. quintana bacteremia. One of the case-patients in this study owns a cat and may have been infected by this pet.
The 3 other animal-associated species we detected should now be considered zoonotic Bartonella spp. B. doshiae and B. tribocorum are both rodent-associated species; in France and worldwide, these species have mainly been recovered from rats (Microtus agentiis for B. doshiae and Rattus rattus for B. tribocorum). B. schoenbuchensis is normally found in deer, elk, and cattle (37,38).
The zoonotic agents we isolated from patients from France have also been detected in animals in France. Similarly, in the United States and Thailand, Bartonella species known to be prevalent in animals have also been identified in humans: (B. henselae, B. vinsonii berkhoffii, and B. koehlerae in the United States (33,35) and B. tribocorum and B. rattimassiliensis in Thailand (39). Therefore, the zoonotic Bartonella species discovered in humans in this study generally appear to be related to the prevalence among animals.
The significance of these Bartonella spp. in the genesis of the clinical picture is difficult to determine. Bartonella spp. are present in ticks, and we have previously reported Bartonella infections following tick bites, such as SENLAT (scalp eschar and neck lymphadenopathy after tick bite [40]). However, the causal link between the conditions observed here, Bartonella and tick bite, cannot yet be concretely established, especially for persons with tick bites occurring up to 5 years previously, which introduces innumerable potential confounding exposures within the same period, including bites by other arthropods. For instance, 1 of the 3 patients with B. henselae bacteremia reported contact with cats; this contact was a more plausible source of infection than tick bites. Furthermore, it is crucial to determine whether Bartonella played a notable role in the observed pathologies, because treatment for chronic Bartonella bacteremia (as for B. quintana) is particularly arduous and may require 6 weeks of doxycycline treatment together with 3 weeks of gentamicin, as these are the only antimicrobial drugs known to be effective in eradication of Bartonella (1). Many Bartonella spp. can also cause endocarditis, including B. quintana and B. henselae; therefore, reports of rare cases of endocarditis attributed to zoonotic Bartonella such as B. kohlerae, B. alsatica, Candidatus B. mayotimonensis, B. vinsonii, or B. elizabethae may actually be the final manifestation of asymptomatic bacteremia, similar to that reported by our infected patients (28).
In summary, our major finding is the isolation of zoonotic Bartonella other than B. quintana in the blood of patients with poorly qualified syndromes. These results indicate that zoonotic Bartonella spp. infection may cause undifferentiated chronic illness in humans.
Acknowledgments
We thank Evelyne Le Naour and Martine Cotte for their technical support, Lionel Arnaud and the Institut National de Transfusion Sanguine for providing blood samples from anonymous blood donors, and the physicians from the Chronimed network who participated in patient recruitment.
This work was partially funded by the French National Institute of Agronomical Research and by the European Union grant FP7-261504 EDENext and is catalogued by the EDENext Steering Committee as EDENext422 (http://www.edenext.eu). The contents of this publication are the sole responsibility of the authors and do not necessarily reflect the views of the European Commission. This work was also supported by the COST action TD1303 (EurNegVec).
Biography
Dr. Vayssier-Taussat is a senior scientist at the French National Institute of Agronomical Research, where she leads a research team involved in the study of Bartonella and other vectorborne pathogens.
Footnotes
Suggested citation for this article: Vayssier-Taussat M, Moutailler S, Féménia F, Raymond P, Croce O, La Scola B, et al. Identification of novel zoonotic activity of Bartonella spp., France. Emerg Infect Dis. 2015 Mar [date cited]. http://dx.doi.org/10.3201/eid2203.150269
References
- 1.Brouqui P, La Scola B, Roux V, Raoult D. Chronic Bartonella quintana bacteremia in homeless patients. N Engl J Med. 1999;340:184–9. 10.1056/NEJM199901213400303 [DOI] [PubMed] [Google Scholar]
- 2.Raoult D, Fournier PE, Drancourt M, Marrie TJ, Etienne J, Cosserat J, et al. Diagnosis of 22 new cases of Bartonella endocarditis. Ann Intern Med. 1996;125:646–52. 10.7326/0003-4819-125-8-199610150-00004 [DOI] [PubMed] [Google Scholar]
- 3.Eremeeva ME, Gerns HL, Lydy SL, Goo JS, Ryan ET, Mathew SS, et al. Bacteremia, fever, and splenomegaly caused by a newly recognized Bartonella species. N Engl J Med. 2007;356:2381–7. 10.1056/NEJMoa065987 [DOI] [PubMed] [Google Scholar]
- 4.Maurin M, Raoult D. Bartonella (Rochalimaea) quintana infections. Clin Microbiol Rev. 1996;9:273–92 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breitschwerdt EB, Maggi RG, Duncan AW, Nicholson WL, Hegarty BC, Woods CW. Bartonella species in blood of immunocompetent persons with animal and arthropod contact. Emerg Infect Dis. 2007;13:938–41. 10.3201/eid1306.061337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maggi RG, Ericson M, Mascarelli PE, Bradley JM, Breitschwerdt EB. Bartonella henselae bacteremia in a mother and son potentially associated with tick exposure. Parasit Vectors. 2013;6:101. [DOI] [PMC free article] [PubMed]
- 7.Lantos PM, Maggi RG, Ferguson B, Varkey J, Park LP, Breitschwerdt EB, et al. Detection of Bartonella species in the blood of veterinarians and veterinary technicians: a newly recognized occupational hazard? Vector Borne Zoonotic Dis. 2014;14:563–70. 10.1089/vbz.2013.1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotté V, Bonnet S, Le RD, Le NE, Chauvin A, Boulouis HJ, et al. Transmission of Bartonella henselae by Ixodes ricinus. Emerg Infect Dis. 2008;14:1074–80. 10.3201/eid1407.071110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angelakis E, Pulcini C, Waton J, Imbert P, Socolovschi C, Edouard S, et al. Scalp eschar and neck lymphadenopathy caused by Bartonella henselae after tick bite. Clin Infect Dis. 2010;50:549–51 . 10.1086/650172 [DOI] [PubMed] [Google Scholar]
- 10.Angelakis E, Billeter SA, Breitschwerdt EB, Chomel BB, Raoult D. Potential for tick-borne bartonelloses. Emerg Infect Dis. 2010;16:385–91. 10.3201/eid1603.081685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breitschwerdt EB, Linder KL, Day MJ, Maggi RG, Chomel BB, Kempf VA. Koch’s postulates and the pathogenesis of comparative infectious disease causation associated with Bartonella species. J Comp Pathol. 2013;148:115–25. 10.1016/j.jcpa.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raoult D. Lack of knowledge can anger patients with chronic diseases. Lancet Infect Dis. 2012;12:654–5. 10.1016/S1473-3099(12)70185-8 [DOI] [PubMed] [Google Scholar]
- 13.Chevreux B, Wetter T, Suhai S. Genome sequence assembly using trace signals and additional sequence information. In: Proceedings of the German Conference on Bioinformatics; October 4–6, 1999; Hanover, Germany. Hanover: Computer Science and Biology; 1999. p. 45–56. [Google Scholar]
- 14.Gao S, Sung WK, Nagarajan N. Opera: reconstructing optimal genomic scaffolds with high-throughput paired-end sequences. J Comput Biol. 2011;18:1681–91. 10.1089/cmb.2011.0170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boetzer M, Pirovano W. Toward almost closed genomes with GapFiller. Genome Biol. 2012;13:R56. 10.1186/gb-2012-13-6-r56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–8. 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–6. 10.1093/nar/gkh152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eddy SR. Accelerated Profile HMM Searches. PLOS Comput Biol. 2011;7:e1002195. 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The Uniprot Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–9. 10.1093/nar/gkq1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–7 . 10.1126/science.278.5338.631 [DOI] [PubMed] [Google Scholar]
- 23.Vijaya Satya R, Zavaljevski N, Reifman DJ. SNIT: SNP identification for strain typing. Source Code Biol Med. 2011;6:14. 10.1186/1751-0473-6-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. 10.1186/1471-2105-14-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.La Scola B, Zeaiter Z, Khamis A, Raoult D. Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol. 2003;11:318–21. 10.1016/S0966-842X(03)00143-4 [DOI] [PubMed] [Google Scholar]
- 26.Seubert A, Falch C, Birtles RJ, Schulein R, Dehio C. Characterization of the cryptic plasmid pBGR1 from Bartonella grahamii and construction of a versatile Escherichia coli–Bartonella spp. shuttle cloning vector. Plasmid. 2003;49:44–52. 10.1016/S0147-619X(02)00103-8 [DOI] [PubMed] [Google Scholar]
- 27.Mullins KE, Hang J, Jiang J, Leguia M, Kasper MR, Maguina C, et al. Molecular typing of “Candidatus Bartonella ancashi,” a new human pathogen causing verruga peruana. J Clin Microbiol. 2013;51:3865–8. 10.1128/JCM.01226-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacomo V, Kelly PJ, Raoult D. Natural history of Bartonella infections (an exception to Koch's postulate). Clin Diagn Lab Immunol. 2002;9:8–18 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vayssier-Taussat M, Le RD, Deng HK, Biville F, Cescau S, Danchin A, et al. The Trw type IV secretion system of Bartonella mediates host-specific adhesion to erythrocytes. PLoS Pathog. 2010;6:e1000946. 10.1371/journal.ppat.1000946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mascarelli PE, Iredell JR, Maggi RG, Weinberg G, Breitschwerdt EB. Bartonella species bacteremia in two patients with epithelioid hemangioendothelioma. J Clin Microbiol. 2011;49:4006–12. 10.1128/JCM.05527-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maggi RG, Mascarelli PE, Pultorak EL, Hegarty BC, Bradley JM, Mozayeni BR, et al. Bartonella spp. bacteremia in high-risk immunocompetent patients. Diagn Microbiol Infect Dis. 2011;71:430–7. 10.1016/j.diagmicrobio.2011.09.001 [DOI] [PubMed] [Google Scholar]
- 32.Pultorak EL, Maggi RG, Mascarelli PE, Breitschwerdt EB. Serial testing from a 3-day collection period by use of the Bartonella Alphaproteobacteria growth medium platform may enhance the sensitivity of Bartonella species detection in bacteremic human patients. J Clin Microbiol. 2013;51:1673–7. 10.1128/JCM.00123-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breitschwerdt EB, Maggi RG, Lantos PM, Woods CW, Hegarty BC, Bradley JM. Bartonella vinsonii subsp. berkhoffii and Bartonella henselae bacteremia in a father and daughter with neurological disease. Parasit Vectors. 2010;3:29. [DOI] [PMC free article] [PubMed]
- 34.Maggi RG, Mozayeni BR, Pultorak EL, Hegarty BC, Bradley JM, Correa M, et al. Bartonella spp. bacteremia and rheumatic symptoms in patients from Lyme disease-endemic region. Emerg Infect Dis. 2012;18:783–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breitschwerdt EB, Maggi RG, Robert MB, Hegarty BC, Bradley JM, Mascarelli PE. PCR amplification of Bartonella koehlerae from human blood and enrichment blood cultures. Parasit Vectors. 2010;3:76. [DOI] [PMC free article] [PubMed]
- 36.Maggi RG, Kosoy M, Mintzer M, Breitschwerdt EB. Isolation of “Candidatus Bartonella melophagi” from human blood. Emerg Infect Dis. 2009;15:66–8 . 10.3201/eid1501.081080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rolain JM, Rousset E, La Scola B, Duquesnel R, Raoult D. Bartonella schoenbuchensis isolated from the blood of a French cow. Ann N Y Acad Sci. 2003;990:236–8. 10.1111/j.1749-6632.2003.tb07370.x [DOI] [PubMed] [Google Scholar]
- 38.Ko S, Kim SJ, Kang JG, Won S, Lee H, Shin NS, et al. Molecular detection of Bartonella grahamii and B. schoenbuchensis-related species in Korean water deer (Hydropotes inermis argyropus). Vector Borne Zoonotic Dis. 2013;13:415–8. 10.1089/vbz.2012.1105 [DOI] [PubMed] [Google Scholar]
- 39.Kosoy M, Bai Y, Sheff K, Morway C, Baggett H, Maloney SA, et al. Identification of Bartonella infections in febrile human patients from Thailand and their potential animal reservoirs. Am J Trop Med Hyg. 2010;82:1140–5. 10.4269/ajtmh.2010.09-0778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Angelakis E, Billeter SA, Breitschwerdt EB, Chomel BB, Raoult D. Potential for tick-borne bartonelloses. Emerg Infect Dis. 2010;16:385–91. 10.3201/eid1603.081685 [DOI] [PMC free article] [PubMed] [Google Scholar]


