Abstract
The tree model and tree-based methods have played a major, fruitful role in evolutionary studies. However, with the increasing realization of the quantitative and qualitative importance of reticulate evolutionary processes, affecting all levels of biological organization, complementary network-based models and methods are now flourishing, inviting evolutionary biology to experience a network-thinking era. We show how relatively recent comers in this field of study, that is, sequence-similarity networks, genome networks, and gene families–genomes bipartite graphs, already allow for a significantly enhanced usage of molecular datasets in comparative studies. Analyses of these networks provide tools for tackling a multitude of complex phenomena, including the evolution of gene transfer, composite genes and genomes, evolutionary transitions, and holobionts.
Keywords: introgression, gene transfer, graph theory, bipartite graph, symbiosis, evolution
Trends
Introgressive processes shape the microbial world at all levels of organisation.
This reticulated evolution is increasingly studied by sequence-similarity networks.
They provide an inclusive accurate multilevel framework to study the web of life.
Networks enhance analyses of microbial genes, genomes, communities, and of symbiosis.
New Methods for Studying the Web of Life
The tree model has been largely and rightly used in evolutionary analyses since Darwin's seminal work [1]. The genealogical relationships between evolving objects are indeed critical to explain life's diversity, not only from a processual perspective (where common ancestry explains some similarities), but also as a powerful pattern to classify all related evolved forms [2]. However, the tree structure, especially when assumed to be universal, strongly constrains our description of the evolution of life 3, 4, 5. By definition, a tree can only describe divergence from a last common ancestor (often with dichotomies, or with polytomies describing fast radiations). In vertical descent, the genetic material of a particular evolutionary unit is propagated by replication inside its own lineage. When such lineages split, and become genetically isolated from one another, this produces a tree. By contrast, in introgressive descent, the genetic material of a particular evolutionary unit propagates into different host structures and is replicated within these host structures [4]. However, a tree with a single ancestor for each object cannot represent such a merging of distinct lineages into a novel common host structure. Typically, organisms produced by sexual reproduction in eukaryotes originate from two parents which merged their genetic material. Genealogical trees with a single ancestor do not describe relationships within eukaryotic sexual populations. Indeed, this genuine genealogical relationship cannot be depicted with a traditional tree representation since this pattern would impose that one considers an offspring either more closely related to only one of its parents, or to be the progenitor of its own ascendants [4].
The distinction between vertical and introgressive descent is not a minor one; introgression (see Glossary) affects all levels of biological organization: from molecules, when sequences legitimately or illegitimately recombine, to genomes, when sequences enter genomes by lateral gene transfer, and to holobionts, when organisms form a collective system (such as the tight association observed between host and endosymbionts) 6, 7, 8 (Box 1). Introgressive descent does not always imply lateral gene transfer: for example, independently replicated gene families, each having their own tree, can merge, and this results in a novel composite gene family. Since even introgressive descent is descent, it encompasses a vertical dimension. The tree representation emphasizes how entities evolve ex unibus plurum, whereas the network representation emphasizes how entities evolve ex pluribus unum. Of course, evolution progresses in both dimensions. Thus, the tree of life and the network of life are not mutually exclusive models. When lineages that evolved in a tree-like fashion merge, this creates reticulation between branches of trees; likewise, after a reticulation event, phylogenetically composite entities can undergo a tree-like evolution: a tree starts growing on the ground of an initial reticulation. Consequently, future synthetic representations could aim at displaying simultaneously both vertical and lateral parts of biological evolution.
Box 1. Mosaicism of Life.
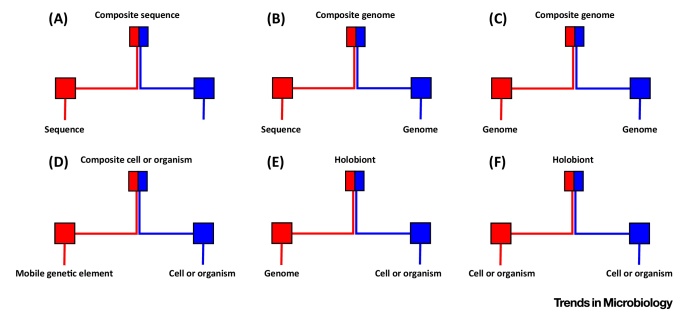
Introgression, the merging of entities from different lineages, affects multiple levels of biological organization. For example, Figure IA describes the introgression of genes from distinct gene families, which results in composite genes, such as multidomain genes [46]. These sequences can come from within a given genome, or when they come from different genomes, such as the genomes of an endosymbiont and of its host, the resulting composite gene, composed partly of material from an endosymbiont, is therefore a symbiogenetic gene. Figure IB describes the introgression of a gene into a host genome, occurring for instance during a lateral or an endosymbiotic gene transfer, which results in a composite genome [63], or when sequences transfer across mobile genetic elements, producing mosaic mobile elements [37]. Of note, more than one gene can be so acquired by a genome 59, 60. Figure IC describes the introgression of a genome into another genome, occurring for instance when the genome of Wolbachia pipiensis becomes inserted into the genome of a Drosophila ananassae, or when the genome of a virophage such as Sputnik becomes inserted into the genome of a giant virus such as Mimivirus, which results in a composite genome 87, 88, 89. Figure ID describes the introgression of a mobile element, such as a plasmid, within a host cell (or organism), occurring for instance when a symbiotic plasmid carrying hypermutagenesis determinants (e.g., the imuABC cassettes) invades soil bacteria, enhancing the ex planta phenotypic diversification of these novel composite cells 90, 91. Figure IE describes the introgression of a genome into a host cell, occurring for instance during events of kleptoplasty 92, 93 or as a result of an extreme reductive evolution after secondary or tertiary plastid acquisition, which results in a (transient or persistent) composite organism [7]. Figure IF describes introgression of cells or organisms, occurring for instance during the evolution and growth of multispecies biofilms [94], endosymbiosis 8, 95, during the development and speciation of animals 82, 83. Typically, sequence-similarity networks can be used to investigate for A; genome networks for B, C, and E; multiplex genome networks for B, C, and E; and bipartite networks for B, D, E, and F.
Importantly, not all evolving objects entertain genealogical relationships: for instance, viruses and cells, both critical players of biological evolution, are not assumed to be related in this way 9, 10, 11, 12, 13, 14, nor are plasmid and plasmid-like transferable objects, as integrative-conjugative elements (ICEs). Cells, viruses, plasmids, and ICEs lack recognized genealogical relationships, either because they genuinely evolved from separate roots, or because their putative common ancestor(s) cannot be inferred from the data, for example, if other descendants of such ancestors became extinct, or have not been sequenced to date. This apparent genealogical disconnection does not exclude vertical evolution within lineages of mobile elements. There is, for example, evidence for both vertical and introgressive descent in plasmids and ICEs of firmicutes [15]. But it means that one genealogical tree cannot represent all the evolutionary history [6]. Therefore, a traditional approach to analyzing evolution incurs the risk of missing explananda (many phenomena that are not described by a genealogical tree) and missing explanans (many evolutionary processes responsible for life's diversity). Trees and networks are representations that allow for scientific analysis. Consistently, tree-thinking has already largely been exploited, and it is now timely and heuristic to turn to network-thinking to illuminate additional and complex aspects of the biology. In this review, we argue that sequence-similarity networks, already used to investigate the evolution of protein coding genes, can also be used to analyze many mosaicisms of life, such as bacterial genome evolution, prokaryotes’ and protists’ organismal evolution, and the evolution of holobionts and communities in which microbes play a role, in particular as symbionts (Box 1). We explain introgression results in at least three major phenomena: (i) microbial social life, understood here as genetic transfers between different genomes, (ii) chimerism (occasionally implying major evolutionary transitions), and (iii) holobionts. All three examples resist classic tree-based analyses and challenge our evolutionary knowledge. A tree model alone does not describe these introgressive processes, that is, the fact that they involve multiple lineages, and their outcomes, that is, the fact that they produce collective, composite, entities. We describe how and why these phenomena can be studied using three classes of networks [sequence-similarity networks, genome networks, and bipartite graphs (Figure 1, Key Figure)], enlarging the analytical toolkit of evolutionary microbiologists. On the one hand, the display of large networks will constitute a challenge for the future development of network-thinking. On the other hand, in terms of interpretation, even very large and dense networks can be effectively simplified, for example, using twin analyses. Thus, we expect a network-thinking era to soon be at the forefront in microbiology.
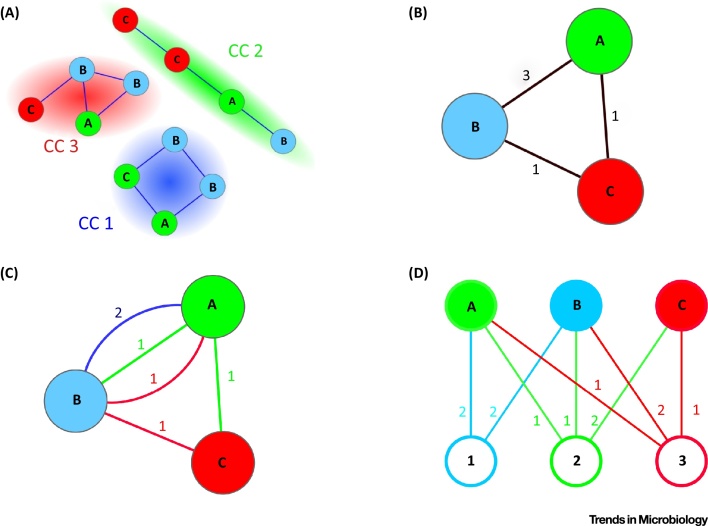
Figure 1.
Key Figure: Different Graph Representations of the Same Gene Sharing among Genomes
(A) Sequence-similarity network (SSN): each node (circle) represents a protein-coding gene sequence; the color and the label of the node represent the genome where the gene is found. Two nodes are connected by an edge (a line linking two nodes) if the pair of sequences fulfills given similarity criteria such as a minimum percentage identity and coverage (i.e., the ratio between the length of the matching parts and the total length of any two sequences). Sequence-similarity networks are analyzed as a partition into connected components (CCs, highlighted as color halos). This partition defines groups of putative gene families, when reciprocal sequence coverage and identity percentage are high [68]: for instance, we can interpret CC1 as a gene family for which two copies are present both in genomes A and B. (B) Genome networks (GNs) can be obtained from SSNs: nodes are genomes (described by color and label); edges connect genomes that share at least one gene family; GNs can be weighted: weights count the number of gene families shared by the two genomes. In the example, A and B share three gene families, but the graph does not specify which ones. (C) Multiplexed networks (MNs) can be, in turn, obtained from GNs by labelling edges in order to identify what gene families are shared: nodes represent genomes; multi-edges represent distinct shared gene families (same color code as the CCs in the SSN); weights count the number of shared genes in each family: the blue edge between A and B corresponds to CC1 in (A) and has therefore weight 2. (D) Bipartite graphs can also be obtained from SSNs; top nodes are genomes; bottom nodes are gene families; edges connect a genome to a gene family if that genome contains at least one representative of the corresponding gene family; weights count the number of genes of that family present in that genome: in the example, node 1 corresponds to CC1 in (A), and has therefore edges incident to genomes A and B, each of weight 2.
Investigating Microbial Social Life with Genome Networks
Gene transfer between prokaryotic organisms and mobile genetic elements (i.e., viruses and plasmids) has largely shaped cellular genome content, as illustrated by the observation of prokaryotic pangenomes 15, 16, for which the collection of gene families used by the members of a given species is larger than the number of gene families present in any individual genome from that species. The flow of genes between genomes, often mediated by mobile genetic elements, explains this observation, but complicates classic inferences about the past (such as genome reconstruction attempts) 4, 5, 17, 18, 19, 20. For a given lineage, the contents of ancestral genomes may be largely different from the union of extant genomes because prokaryotic genomes act as ‘read–write’ storage organelles rather than ‘read–only’ memories [21], and genomes can lose genes. Thus, describing evolution requires not only the tracking of mutations that accumulate within gene families, or loss of gene families [22], but also genes that are gained by introgression [23]. The latter encourages exploring horizontal gene transfer within prokaryotic communities. This brings forward difficult questions 20, 24, 25, 26, 27, 28, 29, 30 since there are many routes through which genes pass from one microbial host to the other, that is, multiple channels [31] for gene transmission. For example, is gene transmission random in terms of cellular, viral, or plasmidic targets (however producing asymmetrical results due to some further host selection acting on the incoming genetic material)? Is it random in terms of what gene families are transmitted? Can we find groups of cotransmitted genes?
Shared gene networks were introduced precisely to tackle these issues (Figure 1) 17, 19, 32. These networks represent which genomes share genetic material, without prejudice regarding the processes involved (vertical descent, but also introgressions 19, 33, 34). In genome networks, all entities are not necessarily genealogically related, allowing for simultaneous analysis of mobile genetic elements and cellular evolution. In that respect, the social microbial network is more inclusive than the tree of life, which is restricted to one type of relationship between one fraction of the biological diversity [6]. Two genomes with a direct connection in such a graph are similar in the sense that they share at least one gene family, whereas two genomes connected only by an indirect path are not similar in terms of gene content. These genome networks display some structure. First of all, plasmids are more central (higher betweenness [35] for a given degree) and viruses more peripheral, testifying that plasmids are general couriers for gene transmission amongst microbes [19]. Second, genome networks have several connected components, that is, several sets of genomes for which there is always an interconnecting path. Each of these connected components groups genomes with exclusive, non-overlapping sets of gene families, and thus corresponds to pools of genes uniquely associated with these genomes [19]. The existence of different connected components suggests the existence of restrictions to introgression.
Within a connected component, a genome network only shows that genomes share genes, but not what the shared genes are. Typically, a triangle of three connected genomes (A, B, C) may result from the sharing of different genes for each pair (AB, BC, AC) within this triangle [4] (see Figure 1B,C). Thus, genomes may form tightly clustered communities [20] in these graphs while sharing different genes. Genome networks provide general information about barriers to transmission and about genetic partnerships, suggesting clubs of genomes enjoying public genetic goods 4, 20. These genome networks require, however, further specifications (for example, on their edges) to address detailed questions about gene transmission and its barriers. A more informative representation displays the identity of shared genes along each edge of a genome network, like in [36], which showed some gene sharing between bacteriophages (as early as 1999), or as in [37] that unraveled genetic transmission between mobile genetic elements of giant viruses (as recently as 2013). Such multiplex graphs are unquestionably attractive and rather natural representations of genetic sharing. However, their display becomes rapidly complex for large datasets, and from an analytical point of view, other graphs can offer practical advantages to analyze gene transmission beyond the genome network framework.
Introducing Bipartite Graphs in Evolutionary Studies
The information on the identity of shared edges (here, gene families) can be conserved in a less cluttered fashion by using bipartite ‘gene families–genomes’ graphs. In these graphs, the precise information regarding gene sharing is directly encoded as edges between these two kinds of nodes. Multiplex genome networks can be seen as unimodal projections [38] of such bipartite ‘gene families–genomes’ graphs (Figure 1D). Bipartite graphs include the same diversity of genomes as the genome networks described above, but they are more accurate. Importantly, simple specific bioinformatic treatments of these multilevel graphs allow one to rapidly identify which groups of genes are shared by which groups of genomes [39], and to display and compare different channels of gene transmission, that is, the routes across generations through which hereditary resources or information pass from parent to offspring [31].
As in genome networks, connected components produce an informative partition of the data. This partition can moreover be examined at different levels of similarity by tuning, for example, the sequence identity percentage. When the data consist of all the protein sequences from all the complete viral (3749), plasmidic (4350), and archaeal (152) genomes, together with a representative subsample of the eubacteria (230) from NCBI, we get the numbers shown in Table 1.
Table 1.
Statistics of the Prokaryote–Virus–Plasmid Gene Families–Genomes Bipartite Graphsa
| Minimal identity percentage to connect sequences | 30% | 60% | 90% |
| Number of connected components (CC) | 156 | 375 | 488 |
| Number of CC having only plasmids | 25 | 73 | 155 |
| Number of CC having only viruses | 130 | 299 | 297 |
| Size of the giant connected component (number of nodes) | 6362 | 5143 | 2769 |
For reciprocal 80% length cover, and different identity thresholds.
The data consist of all protein sequences from all complete plasmidic, viral, and archaeal genomes from NCBI (as of 11/2013), as well as one complete eubacterial complete genome for each family. The identity percentage describes the similarity, in terms of the conservation of primary sequences, between pairs of molecules. The higher this ‘identity threshold’ the more similar pairs of sequences must be to be directly connected in a sequence-similarity network. For high ‘identity threshold’, connected components consist of highly conserved sequences. In a first molecular clock-like approximation, higher ‘identity thresholds’ define groups of sequences that diverged more recently from one another than groups defined with lower ‘identity threshold’.
Assuming a rough molecular clock, these thresholds are useful for investigating events of different ages. Sequences with ≥90% identity have a relatively weak divergence with respect to sequences with 30% identity; indeed, these latter have likely diverged faster or for a longer period of time.
This representation of gene families–genomes bipartite graphs is explicitly multilevel. Interestingly, its analysis does not require any graph clustering algorithm (whose results tend to vary considerably with their implementation). Genetic transmission among microbes can be investigated by simple topological notions of bipartite graphs that result in biologically relevant observations: twins and articulation points [40] that we detail below.
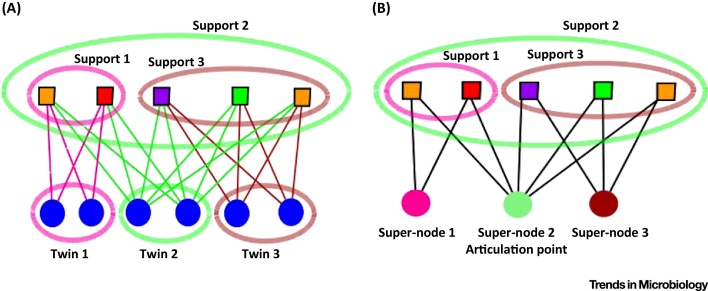
We apply here these notions only to gene family nodes. ‘Twin’ is a notion of graph theory; applied to gene families–genomes graphs, it singles out ‘fellow travellers’: gene families are twins when they are present in exactly the same set of genomes. In the language introduced in [34], the support of such a twin defines a club of genomes. Clubs of genomes, when composed of individuals pertaining to different species, could encourage further studies of ‘kin-coevolution’, for example, the fact that genetic divergence affecting multiple ecologically coexisting lineages, that exchanged genes at some point of their evolution, produces multilineage persistent clubs. The bipartite graph can be simplified by grouping together sets of gene families that are shared by exclusive groups of genomes, and by replacing each such group of gene families by a super-node. Nodes that remain untouched by this reduction process are considered as trivial twin classes (and result in trivial super-nodes). Technically, there is no difference between trivial and non-trivial twins, although, from the biological perspective, the latter correspond to groups of gene families that are more likely to be transmitted together. The resulting quotient graph is reduced, because every club of genomes is now defined by one super-node (individual gene family or group of gene families hosted in this club of genomes) while no information is lost (Figure 2). This property means that even very large graphs can be investigated. In the dataset presented in Table 1, we typically find clubs, such as the one composed of the firmicute Enterococcus faecalis and nine plasmids (present in lactococci or enterococci) that simultaneously and exclusively share the following gene families (at 90% identity): ribose 5-phosphate isomerase RpiB, galactose mutarotase and related enzymes, β-glucosidase/6-phospho-β-glucosidase/β-galactosidase, and phosphotransferase system cellobiose-specific component IIA. These shared mobilized gene families are involved in neighbor pathways of sugar metabolisms (specifically in glycolysis and in the pentose phosphate metabolic pathways), which likely explains their collective mobilization in plasmids.
Figure 2.
Twins and Articulation Points in a Bipartite Graph. (A) Top nodes in this bipartite graph are genomes and bottom nodes gene families. Nodes in each colored ellipse at the bottom form a twin class, since their sets of neighbors (supports encircled by similarly colored ellipses on the top level) are identical (as highlighted by the coloring of their incident edges). (B) Collapsing twin nodes into super-nodes yields a reduced graph, without further bottom twin nodes. The supported groups of host genomes are unchanged, and are now defined as the neighbors of a single super-node. Due to the graph reduction, the green super-node is now an articulation point, since its removal disconnects the nodes in the pink and brown supports.
Articulation points in a gene families–genomes bipartite graph correspond to gene families shared by many genomes with otherwise totally distinct gene contents (for a given similarity threshold). Although strictly topological, the notion of an articulation point is thus expected to help detect public genetic goods [34], that is, genetic material that is being shared by taxonomically distant genomes, which possibly benefit from the properties they confer, for some reason other than genealogy (i.e., genes coding for environmental adaptation or hitch-hiking with those). However, an articulation point can also detect selfish genes, such as the abundant transposases [41], which are spreading across multiple distantly related genomes (Box 2).
Box 2. Articulation Points Reveal Potential Public Genetic Goods.
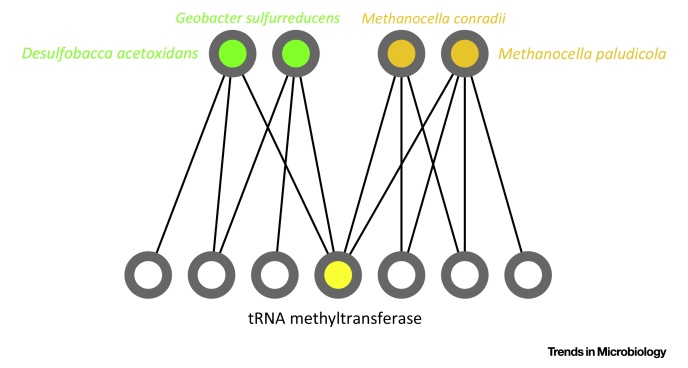
In a prokaryote–virus–plasmid dataset, we typically find clubs of genomes, such as the one (represented in Figure I) composed of two mesophilic sulphur-reducing acetate-metabolizing Proteobacteria (Geobacter sulfurreducens and Desulfobacca acetoxidans) and two thermophilic hydrogenotrophic methanogen Euryarchaeota (Methanocella conradii and Methanocella paludicola). These taxa are linked by an articulation point, which indicates the sharing of a conserved gene family (at >90% identity), functionally annotated as a tRNA (1-methyladenosine) methyltransferase. This kind of association between sulphate-reducer and methanogens is well-documented in the literature 96, 97. The sharing of genes between different prokaryotes suggested by this network analysis makes sense, since these prokaryotes are found in common anoxic environments, such as rice paddy soils [98]. Also, G. sulfurreducens and M. paludicola contain a laterally-transferred two-gene cluster, hgcAB, related to the ability to methylate mercury [99]. Thus, a graph analysis produces a novel testable hypothesis, namely, to see if the shared tRNA methyltransferase is involved in the adaptation to the environment of these taxa, or if it hitch-hiked with other genes transferred between these taxa, such as the hcgAB cluster.
Investigating Composite Genes, Organisms, and Evolutionary Transitions with Sequence-Similarity Networks
Introgression can also be investigated below the gene level and above the organismal level. For instance, composite genes, such as the genes produced by evolutionary tinkering [42], famous for encoding multidomain proteins, are well documented in cellular genomes 43, 44, 45, and have been reported in viruses and plasmids 46, 47. Such genes are composed of genetic fragments (e.g., components, which can be domains or full genes) that are otherwise found in separate gene families [48]. The fusion of a receptor-binding protein with a tail fiber protein in the lactococcal bacteriophage bIBB29, producing a composite gene involved in host specificity, offers a good example of this sort of molecular mosaic [49]. While many substitution models have been developed to account for gradual evolution by point mutation in phylogenetic inferences, models describing the rules and rates of emergence (or fission) of composite genes are still rare 50, 51, 52, especially for unicellular organisms and mobile genetic elements 46, 47. However, many gene families are not just evolving gradually within the boundaries of a single gene family [53]. The accretion of two protein domains into a novel host structure constitutes a case of saltatory molecular evolution by introgression. The rules of evolution and fragment combination largely remain to be discovered 54, 55. Sequence-similarity networks could contribute to this task. Indeed, these graphs can: (i) provide a systematic description of both composite and component genes in genomes (and metagenomes); (ii) be used to polarize fusion and fission events (by comparing the taxonomical distribution of genes hosts in associated component and composite gene families); (iii) be directly used to compare the relative conservation of overlapping component and composite sequences, for example, to determine whether domains found in different combination have different rates of evolution. The detection of composite genes using sequence-similarity networks can further contribute to understanding the rules of evolution of other biological networks, such as protein–protein interaction networks [56]. For instance, when, as a result of exon- or domain-shuffling, composite genes produce novel combinations of domains of interaction, composite genes can introduce novel nodes and edges in protein–protein interactions. Likewise, composite genes can impact the robustness of protein–protein interaction networks, when genes coding for separate proteins involved in a functional interaction become fused, ‘crystallizing’ an edge of the protein–protein interaction network.
This issue takes on particularly fundamental importance in organisms hosting genes from multiple origins. These introgressed genes have distinct evolutionary past histories and, hence, possibly different future evolvabilities. For example, eukaryotic genomes 57, 58 as well as major archaeal lineages are composed of genes from both bacterial and archaeal origins 59, 60. Some studies have focused on the evolution of complete genes of distinct origins in these mosaic taxa [i.e., contrasting the essentiality or centrality of genes from bacterial and archaeal origins in regulatory or metabolic eukaryotic networks [61], or simply performing classic phylogenetic analyses of these genes to identify endosymbiotic gene transfer (or EGT) [8]]. A common fate for proteins derived from such transferred organellar genes is to be targeted back to the compartment of origin to perform their original function, but not only [62]. Regarding these proteins and genes, the study of composite organisms has opened the door to an exciting evolutionary question that, we argue, networks can now better address: what happens after distinct genetic material becomes integrated into a new host? Genes from distinct origins could have different propensities to be lost or to diverge during subsequent evolution of their novel composite host lineage [63]. Likewise, at the infragenic level, the evolutionary impact of introgression deserves consideration. Do composite organisms host novel symbiogenetic composite genes with components from different phylogenetic origins that could only be born in such genetic melting-pots as a result of the original mixing of gene fragments? A positive answer, that is, the detection of such novel composite genes in composite organisms, could revolutionize our understanding of the origins of biological traits. A negative answer, that is, the lack of novel composite genetic material from different organismal sources despite their new physical proximity, would indicate strong selective pressures preventing the birth of novel gene families in spite of changes in their genomic context. Thus, it would be worth testing if introgression at one level of biological organization (i.e., between cells) can favor introgression at another level (i.e., between genes). For example, organisms with composite genomes, or holobionts, might be composed of more composite symbiogenetic genes than organisms devoid of endosymbionts, or less subjected to gene transfer.
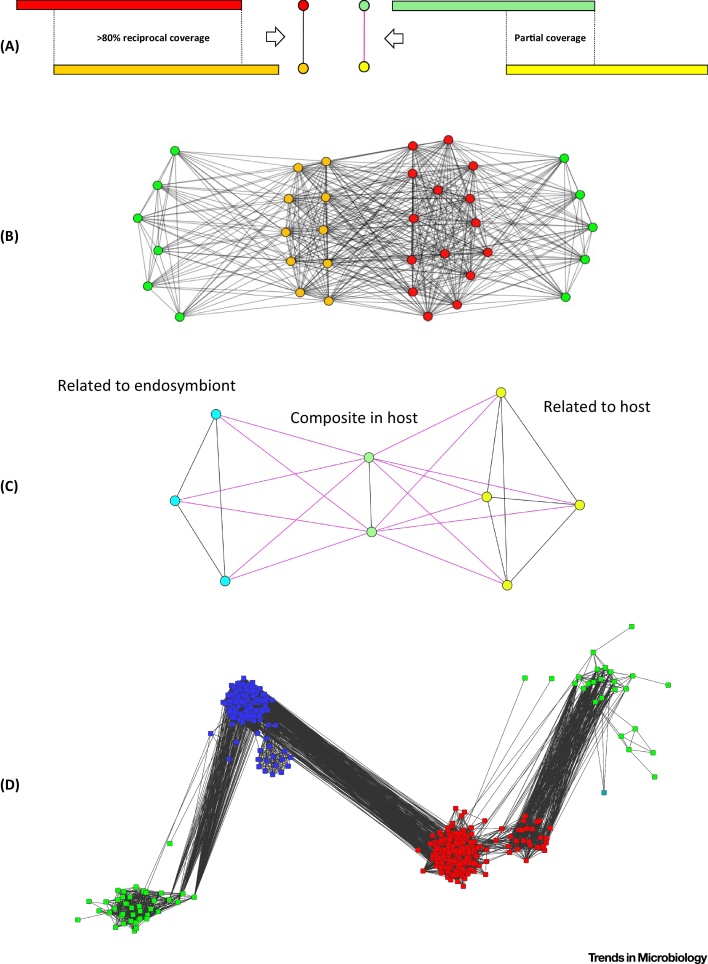
Sequence-similarity networks are ideal tools for investigating these issues (Figure 3). These very inclusive graphs 47, 53 allow for comparative analyses of massive datasets without the need for multiple sequence alignments 4, 64, 65, 66. Similarity is typically detected in a BLAST all-versus-all analysis to produce a table of pairwise hits [67]. Sequence-similarity networks are displayed and analyzed as a set of connected components (Figure 1A) [68]. When the coverage between sequences is high, this partition of the nodes defines groups of putative homologous sequences or gene families. Thus, sequence-similarity networks have been used with relatively stringent criteria (i.e., hits between two sequences must show >30% identity, cover ≥80% of both sequences length, and have a maximal E-value of 10–5 in BLAST comparative analyses) coupled with clustering methods to identify clusters of nodes corresponding to homologous gene families 69, 70, 71. In the past 20 years, sequence-similarity networks have indeed mainly been used to investigate the evolution of protein-coding genes 4, 64, 65, 66, 71, 72, 73, 74, 75, and to perform functional annotation. For instance, the COG categories correspond to groups of similar sequences (with remarkable topological properties in sequence-similarity networks) that have likely evolved from a single ancestral gene. In comparative analyses, COG are often used as proxy for functional annotations because their remarkable conservation suggests that sequences from the same COG may have preserved some common functions [71]. This standard approach, however, would not readily detect composite genes [76]. Using less stringent thresholds for mutual sequence coverage (Figure 3A) or identity percentage, sequence-similarity networks can be used to detect superfamilies 66, 77, 78, 79, divergent homologues, or composite genes [when, for example, the length coverage condition is relaxed to take into account (partial) similarity (Figure 3C), such as domain sharing, between sequences] [46].
Figure 3.
Typical Patterns for Candidate Endosymbiotic Gene Transfer (EGT) and Composite Genes in Sequence-Similarity Networks. (A) Sequence-similarity networks can be used for the detection of distant homologues in eukaryotic genomes. Complete (left) and partial (right) sequence similarity, and how they are translated as different types of edges in the sequence-similarity network (SSN). In black, the percentage of reciprocal cover is high; the sequences are homologous over their entire length. In purple, the cover percentage is low; the sequences are only partly similar, that is, they share a homologous domain. (B) Shortest-path analysis in a sequence-similarity graph can be used for detecting possible endosymbiotic gene transfer (EGT). Indeed, EGT results in a characteristic network pattern: an indirect short path along which all edges indicate homology, connecting two nodes corresponding to diverged sequences present in a given host organism. Green nodes represent eukaryotic sequences; red, bacterial sequences; and yellow, archaeal sequences. Black edges denote complete sequence similarity (>80% length). All shortest paths between eukaryotic sequences that pass through the bacterial and archaeal components are likely candidates for EGT, because this indicates that a first type of eukaryotic sequence has affinities to bacterial sequences while a second type has affinities to archaeal ones. (C) Sequence-similarity networks with edges for complete and partial coverage are also useful for the detection of composite genes. The figure shows a pattern associated with the detection of composite genes. Black edges denote complete (>80% cover) and purple edge denote partial (<80% cover) sequence similarity. The green family is a candidate symbiogenetic composite gene, derived from endosymbiotic lateral gene transfer, since it displays one part with similarity to host-related sequences (yellow) and another part with similarity to endosymbiont-related (blue) genes. (D) A concrete example of a possible EGT: archaeal sequences are represented in blue, eubacterial in red, and eukaryotic genes in green (there is also a single plasmidic sequence in blue-green on the right). Eukaryotic sequences clearly form two groups, one closer to archaea, one more related to eubacteria. All the sequences have a generic annotation as RNA-pseudouridine synthase, but while the eubacterial (and related eukaryotic) sequences are exclusively tRNA synthases (thus putatively of mitochondrial origin), on the archaeal side (thus possibly of host origin) we find tRNA- as well as rRNA-pseudouridine synthases. It indeed turns out that this family contains two pseudouridine synthase genes that are both present in Saccharomyces cerevisiae, having a similar function but acting on a different substrate: one on the archaeal side, coding for Cbf5p that acts on large and small rRNA 100, 101, and the other on the eubacterial side, coding for Pus4, that acts on mitochondrial and cytoplasmic tRNA-uridine [102].
These kinds of analyses with flexible definitions confirm that not all eukaryotic gene families have homologs in prokaryotes. When they do, sequence-similarity network analyses indicate that eukaryotic gene families homologous to those of bacteria (for which sequences of eukaryotes exclusively cluster with sequences from bacteria [63]) and eukaryotic gene families homologous to those of archaea (for which sequences of eukaryotes exclusively cluster with sequences from archaea) have different rates of evolution. For example, eukaryotic gene families with bacterial origins are more easily expanded or lost when eukaryotic genomes expand or shrink, while the number of eukaryotic gene families with archaeal origins is much more stable 22, 63.
Moreover, sequence-similarity networks demonstrated their efficiency to unravel distant homologues in eukaryotic genomes, that is, gene families for which some present-day eukaryotes possess a version that originated from a bacterial progenitor, while other present-day eukaryotes possess an homologous version that originated from an archaeal progenitor, or when the same eukaryotes possess both diverged versions in its nuclear genome, one from a bacterial origin, the other from an archaeal origin [63] (Figure 3B). The latter presence of such distant homologues characterizes the occurrence of EGT 7, 59, an introgressive process where a gene from an organelle (such as mitochondria or plastids) has been imported into the eukaryotic nuclear DNA, where an homologous nuclear copy from archaebacterial origin was already present (Figure 3D). These networks are promising to look for possibly still-hidden EGT, and past endosymbioses when they are applied to new genomic data.
Sequence-similarity networks are also most useful for identifying composite genes (Figure 3C), and their use for detecting genes composed of parts from different origins will likely soon aid reticulate evolution analyses 46, 47, 53. Indeed, the level of molecular intricacy between hosts and symbionts may well exceed whole gene introgression in the genome of composite organisms. Preliminary results show that photosynthetic eukaryotes contain some novel nuclear composite genes, featuring unique couplings of domains from plastid origin, without any counterpart in the prokaryotic world. For example, photosynthetic dinophytes contain a composite gene coding for a protein consisting of two domains: one SufE domain of cyanobacterial origin (i.e., probably originating from the chloroplast genome) and a tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase of proteobacterial origin. Interestingly, SufE displays desulfurylase activity [80], while the tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase posesses a thiol group (R-S-H) containing a sulfur atom. It is possible that the sulfur atom required for the thiol group is provided by the activity of SufE to the new physical coupling of these domains in a symbiogenetic gene. Such findings encourage experimental studies to establish whether and which biological properties emerged from the physical coupling of domains in a novel eukaryotic gene with endosymbiotic origin.
Understanding the entanglement of molecular building blocks, below and above the gene level, is probably the next step required to analyze molecular processes going on during evolutionary transitions mediated by the merging of lineages 4, 57, 58, 59, 60.
Concluding Remarks: Networks Enhance Our Comprehension of Life's Complexity
The complexity and diversity of phenomena acknowledged and investigated by evolutionary biologists is striking, and growing: it now goes well beyond the identification of lineage divergence from a single common ancestor, enhancing what is considered as the Darwinian paradigm. When pushed to its limits, introgression might result in the integration of laterally acquired features into a sustainable structure, controllable by regulatory systems, which may themselves be the result of introgression. A technical and theoretical transition has accompanied this broadening of scope within the evolutionary paradigm. Namely, network models and methods, never truly absent in biological studies [81], have been developed and implemented. Hence, they now offer powerful complementary approaches to evolutionary studies, which will enhance the exploitation of molecular datasets in multiple directions. The routes and genetic goods of microbial social life, the origins and combination rules of composite genes, and the genetic transformation coupled with major evolutionary transitions, can readily be investigated using powerful, inclusive, comparative network-based tools. The diversity of such tools is itself constantly increasing: the multi-thresholded sequence-similarity networks, (multiplex) genome networks, and the bipartite graphs presented here, allow one to perform multi-agent and multilevel comparative analyses, and may become as familiar to evolutionary biologists as phylogenetic trees in the near future. Importantly, these network tools have not yet been used at their full potential (see Outstanding Questions). In particular, they could also be used to analyze the evolution of communities of synthetic microorganisms, biofilms, and holobionts. These latter collective systems encompass a challenging complexity. For example, holobionts rely on a multiplicity of interacting transmission systems and channels for their development and evolution that differ in the microbes and in their hosts. This heterogeneity complicates the understanding of the causes of holobionts’ collective phenotypes by traditional methods, even in the metazoan world [82]. Applying a network analytical framework to holobiont studies may be an innovative way to decipher what traits, long held as characteristic of a single animal (i.e., species incompatibility, self-immunity, or possibly behavior 83, 84), or of an individual organism/biofilm (i.e., health conditions 85, 86 or drug resistance), originate from complex interactions, at multiple biological levels, and how these involve microbes and their genes. More generally, network-thinking has lots to contribute to microbiology.
Outstanding Questions.
What are the rules of domain and gene shuffling in microbes? Sequence-similarity networks provide fast and effective means for systematic analyses of the evolution of composite genes, by simultaneously detecting families of components contributing to composite gene families. The phylogenetic origins and the functional categories of these components will show whether microbes are using transferred genes to create new composite genes in their genomes. For example, do the notoriously mosaic haloarchaeal genomes harbor composite genes of bacterial origin? Does the proportion of composite genes in microbes change with the environment? Can one introduce models of nucleotide substitution into sequence-similarity networks in order to make them more realistic with regard to sequence evolution?
Is every gene everywhere? Gene-similarity networks applied to large-scale metagenomic data and gene-sharing networks featuring environments instead of genomes as their nodes will provide inclusive novel ways to address this important question. These graphs will show whether similar sequences are found in geographically or ecologically similar environments, and serve to detect ubiquitous and endemic genes sets.
What phenotypes in holobionts have multiple origins, that is, did not evolve within a single phylum but emerged from a biological collective? Bipartite graphs with microbial taxa or microbial gene families as bottom nodes and with animal or human hosts as top nodes will immediately allow for the identification of phylogenetically heterogeneous groups of microbes, or groups of gene families in microbes, always associated with a particular host-level phenotype.
How do processes of molecular evolution occurring at the level of the microbiota affect eukaryotic hosts? The microbial gene families–eukaryotic host bipartite graphs described above can be refined to take into account information about the molecular evolution of the gene families (e.g., their rate of evolution, or whether to what extent and by what mobile elements each gene family was eventually transferred). This adds an explicit evolutionary dimension to the bottom-level nodes, allowing one to evaluate, for example, the impact of lateral gene transfer, operating at the microbial level, on the phenotypes of the eukaryotic host. For example, it becomes easy to test whether laterally transferred genes, mobilized by a broader range of mobile elements, are more largely distributed in human hosts than are resident gene families of the microbiome.
Can one extend the methods from bipartite to tripartite graphs, to account for more levels of biological organization? This defines, as a realistic objective, the implementation of genes–genomes–environments tripartite graphs, which can then be clustered to provide a global yet accurate representation of the structure of genetic diversity on Earth in a single comparative analysis.
Figure I.
Several Illustrations of Mosaicism through Merging Events. (A) Composite genes result from the fusion of different gene domains. (B) Composite genomes can result from the introgression of a gene into a genome, or (C) from the introgression of a genome into a genome. (D) Composite organisms can arise from the introgression of a mobile genetic element. Holobionts result from the introgression of a genome (E) or of another cell (F) into a cell.
Figure I.
Excerpt of a Typical Reduced Gene Familes–Genomes Bipartite Graph around an Articulation Point. The top nodes compose the club defined by the sharing of a conserved tRNA methyltransferase (bottom node in yellow). For simplicity, only the direct neighbors of the members of the club have been included in the picture of the graph. The removal of the articulation point (in yellow) isolates the two taxonomically homogeneous groups from each other.
Acknowledgments
E.C. and E.B. are funded by FP7/2007-2013 Grant Agreement #615274.
Glossary
- Articulation point (or cut-vertex)
node in a graph whose removal increases the number of connected components of the resulting graph.
- Betweenness
centrality measure for a node in a graph, namely, the proportion of shortest paths between all pairs of nodes that pass through this specific node. Nodes having a betweenness close to 1 are said to be more central, and those close to 0, more peripheral.
- Bipartite graph
a graph with two types of node (top nodes and bottom nodes) such that an edge only connects nodes of one type with nodes of the other type.
- Club of genomes
a coalition of entities replicating in separate events and exploiting some common genetic material that does not necessarily trace back to a single last common ancestor.
- Community
in graph theory, groups of nodes that are more connected between themselves than with the rest of the graph. This technical meaning should not be confused with its use in expressions such as ‘microbial communities’.
- Connected component
set of nodes in a graph for which there is always an interconnecting path.
- Degree
number of incident edges to a given node.
- Introgression
descent process through which the genetic material of a particular evolutionary unit propagates into different host structures and is replicated within these host structures.
- Multiplex graph
a graph having possibly several edges of different types between two nodes.
- Neighbors
nodes that are directly connected by an edge.
- Public genetic goods
the common genetic material shared by a club of phylogenetically distant genomes.
- Quotient graph
simplified graph whose nodes represent disjoint subsets of nodes of the original graph; an edge in this new graph connects two such new nodes whenever an edge in the original graph connects at least one element of a new node with at least one from the other.
- Support
the common set of neighbors of a twin class.
- Twins
nodes in a graph that have exactly the same set of neighbors.
References
- 1.Darwin C. John Murray; 1859. On the Origin of Species by Means of Natural Selection. [Google Scholar]
- 2.O’Hara R.J. Population thinking and tree thinking in systematics. Zool. Scr. 1997;26:323–329. [Google Scholar]
- 3.Doolittle W.F., Bapteste E. Pattern pluralism and the Tree of Life hypothesis. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2043–2049. doi: 10.1073/pnas.0610699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bapteste E. Evolutionary analyses of non-genealogical bonds produced by introgressive descent. Proc. Natl. Acad. Sci. U.S.A. 2012;109:18266–18272. doi: 10.1073/pnas.1206541109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doolittle W.F. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 6.Bapteste E. The origins of microbial adaptations: How introgressive descent, egalitarian evolutionary transitions and expanded kin selection shape the network of life. Front. Microbiol. 2014;5:1–4. doi: 10.3389/fmicb.2014.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Archibald J.M. Genomic perspectives on the birth and spread of plastids: Fig 1. Proc. Natl. Acad. Sci. U.S.A. 2015;112:10147–10153. doi: 10.1073/pnas.1421374112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lane C.E., Archibald J.M. The eukaryotic tree of life: endosymbiosis takes its TOL. Trends Ecol. Evol. 2008;23:268–275. doi: 10.1016/j.tree.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Claverie J-M., Ogata H. Ten good reasons not to exclude giruses from the evolutionary picture. Nat. Rev. Microbiol. 2009;7:615. doi: 10.1038/nrmicro2108-c3. [DOI] [PubMed] [Google Scholar]
- 10.Koonin E.V. Compelling reasons why viruses are relevant for the origin of cells. Nat. Rev. Microbiol. 2009;7:615. doi: 10.1038/nrmicro2108-c5. [DOI] [PubMed] [Google Scholar]
- 11.Moreira D., López-García P. Ten reasons to exclude viruses from the tree of life. Nat. Rev. Microbiol. 2009;7:306–311. doi: 10.1038/nrmicro2108. [DOI] [PubMed] [Google Scholar]
- 12.Navas-Castillo J. Six comments on the ten reasons for the demotion of viruses. Nat. Rev. Microbiol. 2009;7:615. doi: 10.1038/nrmicro2108-c2. [DOI] [PubMed] [Google Scholar]
- 13.Raoult D. There is no such thing as a tree of life (and of course viruses are out!) Nat. Rev. Microbiol. 2009;7:615. doi: 10.1038/nrmicro2108-c6. [DOI] [PubMed] [Google Scholar]
- 14.Villarreal L.P., Witzany G. Viruses are essential agents within the roots and stem of the tree of life. J. Theor. Biol. 2010;262:698–710. doi: 10.1016/j.jtbi.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 15.Lukjancenko O. Comparison of 61 sequenced Escherichia coli genomes. Microb. Ecol. 2010;60:708–720. doi: 10.1007/s00248-010-9717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tettelin H. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dagan T. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10039–10044. doi: 10.1073/pnas.0800679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dagan T., Martin W. Ancestral genome sizes specify the minimum rate of lateral gene transfer during prokaryote evolution. Proc. Natl. Acad. Sci. U.S.A. 2007;104:870–875. doi: 10.1073/pnas.0606318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halary S. Network analyses structure genetic diversity in independent genetic worlds. Proc. Natl. Acad. Sci. U.S.A. 2010;107:127–132. doi: 10.1073/pnas.0908978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skippington E., Ragan M.A. Lateral genetic transfer and the construction of genetic exchange communities. FEMS Microbiol. Rev. 2011;35:707–735. doi: 10.1111/j.1574-6976.2010.00261.x. [DOI] [PubMed] [Google Scholar]
- 21.Shapiro J.A. Bacteria are small but not stupid: cognition, natural genetic engineering and socio-bacteriology. Stud. Hist. Philos. Biol. Biomed. Sci. 2007;38:807–819. doi: 10.1016/j.shpsc.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Ku C. Endosymbiotic origin and differential loss of eukaryotic genes. Nature. 2015;524:427–432. doi: 10.1038/nature14963. [DOI] [PubMed] [Google Scholar]
- 23.Lobkovsky A.E. Estimation of prokaryotic supergenome size and composition from gene frequency distributions. BMC Genomics. 2014;15:S14. doi: 10.1186/1471-2164-15-S6-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kloesges T. Networks of gene sharing among 329 proteobacterial genomes reveal differences in lateral gene transfer frequency at different phylogenetic depths. Mol. Biol. Evol. 2011;28:1057–1074. doi: 10.1093/molbev/msq297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popa O., Dagan T. Trends and barriers to lateral gene transfer in prokaryotes. Curr. Opin. Microbiol. 2011;14:615–623. doi: 10.1016/j.mib.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 26.Popa O. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res. 2011;21:599–609. doi: 10.1101/gr.115592.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain R. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl. Acad. Sci. U.S.A. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park C., Zhang J. High expression hampers horizontal gene transfer. Genome Biol. Evol. 2012;4:523–532. doi: 10.1093/gbe/evs030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorek R. Genome-wide experimental determination of barriers to horizontal gene transfer. Science. 2007;318:1449–1452. doi: 10.1126/science.1147112. [DOI] [PubMed] [Google Scholar]
- 30.Cohen O. The complexity hypothesis revisited: connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol. Biol. Evol. 2011;28:1481–1489. doi: 10.1093/molbev/msq333. [DOI] [PubMed] [Google Scholar]
- 31.Lamn E. Inheritance systems. In: Zalta E.N., editor. The Stanford Encyclopedia of Philosophy. Winter 2014. 2014. http://plato.stanford.edu/archives/win2014/entries/inheritance-systems. [Google Scholar]
- 32.Lima-Mendez G. Reticulate representation of evolutionary and functional relationships between phage genomes. Mol. Biol. Evol. 2008;25:762–777. doi: 10.1093/molbev/msn023. [DOI] [PubMed] [Google Scholar]
- 33.Halary S. EGN: a wizard for construction of gene and genome similarity networks. BMC Evol. Biol. 2013;13:146. doi: 10.1186/1471-2148-13-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McInerney J.O. The public goods hypothesis for the evolution of life on Earth. Biol. Direct. 2011;6:41. doi: 10.1186/1745-6150-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brandes U. On variants of shortest-path betweenness centrality and their generic computation. Soc. Netw. 2008;30:136–145. [Google Scholar]
- 36.Hendrix R.W. Evolutionary relationships among diverse bacteriophages and prophages: All the world's a phage. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yutin N. Virophages, polintons, and transpovirons: a complex evolutionary network of diverse selfish genetic elements with different reproduction strategies. Virol. J. 2013;10:158. doi: 10.1186/1743-422X-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahn Y-Y. Flavor network and the principles of food pairing. Sci. Rep. 2011;1:196. doi: 10.1038/srep00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rivera C.G. NeMo: network module identification in cytoscape. BMC Bioinformatics. 2010;11(Suppl. 1):S61. doi: 10.1186/1471-2105-11-S1-S61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diestel R. Springer Science & Business Media; 2006. Graph Theory. [Google Scholar]
- 41.Aziz R.K. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 2010;38:4207–4217. doi: 10.1093/nar/gkq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Derouiche A. Evolution and tinkering: what do a protein kinase, a transcriptional regulator and chromosome segregation/cell division proteins have in common? Curr. Genet. 2015 doi: 10.1007/s00294-015-0513-y. Published online August 19, 2015. [DOI] [PubMed] [Google Scholar]
- 43.Kawashima T. Domain shuffling and the evolution of vertebrates. Genome Res. 2009;19:1393–1403. doi: 10.1101/gr.087072.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chothia C. Evolution of the protein repertoire. Science. 2003;300:1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 45.de Souza S.J. Domain shuffling and the increasing complexity of biological networks. Bioessays. 2012;34:655–657. doi: 10.1002/bies.201200006. [DOI] [PubMed] [Google Scholar]
- 46.Jachiet P.A. MosaicFinder: Identification of fused gene families in sequence similarity networks. Bioinformatics. 2013;29:837–844. doi: 10.1093/bioinformatics/btt049. [DOI] [PubMed] [Google Scholar]
- 47.Jachiet P. Extensive gene remodeling in the viral world: new evidence for non-gradual evolution in the mobilome network. Genome Biol. Evol. 2014;6:2195–2205. doi: 10.1093/gbe/evu168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng S. Sequence similarity network reveals the imprints of major diversification events in the evolution of microbial life. Front. Ecol. Evol. 2014;2:1–13. [Google Scholar]
- 49.Hejnowicz M.S. Analysis of the complete genome sequence of the lactococcal bacteriophage bIBB29. Int. J. Food Microbiol. 2009;131:52–61. doi: 10.1016/j.ijfoodmicro.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 50.Pasek S. Gene fusion/fission is a major contributor to evolution of multi-domain bacterial proteins. Bioinformatics. 2006;22:1418–1423. doi: 10.1093/bioinformatics/btl135. [DOI] [PubMed] [Google Scholar]
- 51.Kummerfeld S.K., Teichmann S.A. Relative rates of gene fusion and fission in multi-domain proteins. Trends Genet. 2005;21:25–30. doi: 10.1016/j.tig.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 52.Snel B. Genome evolution. Trends Genet. 2000;16:9–11. doi: 10.1016/s0168-9525(99)01924-1. [DOI] [PubMed] [Google Scholar]
- 53.Haggerty L.S. A pluralistic account of homology: adapting the models to the data. Mol. Biol. Evol. 2014;31:501–516. doi: 10.1093/molbev/mst228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patthy L. Modular assembly of genes and the evolution of new functions. Genetica. 2003;118:217–231. [PubMed] [Google Scholar]
- 55.Nakamura Y. Rate and polarity of gene fusion and fission in Oryza sativa and Arabidopsis thaliana. Mol. Biol. Evol. 2007;24:110–121. doi: 10.1093/molbev/msl138. [DOI] [PubMed] [Google Scholar]
- 56.Dohrmann J. Global multiple protein–protein interaction network alignment by combining pairwise network alignments. BMC Bioinformatics. 2015;16:S11. doi: 10.1186/1471-2105-16-S13-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McInerney J.O. The hybrid nature of the Eukaryota and a consilient view of life on Earth. Nat. Rev. Microbiol. 2014;12:449–455. doi: 10.1038/nrmicro3271. [DOI] [PubMed] [Google Scholar]
- 58.Williams T.A. An archaeal origin of eukaryotes supports only two primary domains of life. Nature. 2013;504:231–236. doi: 10.1038/nature12779. [DOI] [PubMed] [Google Scholar]
- 59.Nelson-Sathi S. Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc. Natl. Acad. Sci. U.S.A. 2012;109:20537–20542. doi: 10.1073/pnas.1209119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nelson-Sathi S. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature. 2015;517:77–80. doi: 10.1038/nature13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alvarez-Ponce D., McInerney J.O. The human genome retains relics of its prokaryotic ancestry: human genes of archaebacterial and eubacterial origin exhibit remarkable differences. Genome Biol. Evol. 2011;3:782–790. doi: 10.1093/gbe/evr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deane J.A. Evidence for nucleomorph to host nucleus gene transfer: light-harvesting complex proteins from cryptomonads and chlorarachniophytes. Protist. 2000;151:239–252. doi: 10.1078/1434-4610-00022. [DOI] [PubMed] [Google Scholar]
- 63.Alvarez-Ponce D. Gene similarity networks provide tools for understanding eukaryote origins and evolution. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E1594–E1603. doi: 10.1073/pnas.1211371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yona G. ProtoMap: automatic classification of protein sequences and hierarchy of protein families. Nucleic Acids Res. 2000;28:49–55. doi: 10.1093/nar/28.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frickey T., Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 66.Atkinson H.J. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS ONE. 2009;4:e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Forster D. Testing ecological theories with sequence similarity networks: marine ciliates exhibit similar geographic dispersal patterns as multicellular organisms. ISME J. 2014;13:1–16. doi: 10.1186/s12915-015-0125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bittner L. Some considerations for analyzing biodiversity using integrative metagenomics and gene networks. Biol. Direct. 2010;5:47. doi: 10.1186/1745-6150-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Altenhoff A.M. The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements. Nucleic Acids Res. 2015;43:D240–D249. doi: 10.1093/nar/gku1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sayers E.W. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tatusov R.L. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 72.Bapteste E. Networks: expanding evolutionary thinking. Trends Genet. 2013;29:439–441. doi: 10.1016/j.tig.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 73.Enright A.J., Ouzounis C.A. GeneRAGE: a robust algorithm for sequence clustering and domain detection. Bioinformatics. 2000;16:451–457. doi: 10.1093/bioinformatics/16.5.451. [DOI] [PubMed] [Google Scholar]
- 74.Li L. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Enright A.J. Protein families and TRIBES in genome sequence space. Nucleic Acids Res. 2003;31:4632–4638. doi: 10.1093/nar/gkg495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Song N. Sequence similarity network reveals common ancestry of multidomain proteins. PLoS Comput. Biol. 2008;4:e1000063. doi: 10.1371/journal.pcbi.1000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sasson O. ProtoNet: hierarchical classification of the protein space. Nucleic Acids Res. 2003;31:348–352. doi: 10.1093/nar/gkg096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matsui M. Comprehensive computational analysis of bacterial crp/fnr superfamily and its target motifs reveals stepwise evolution of transcriptional networks. Genome Biol. Evol. 2013;5:267–282. doi: 10.1093/gbe/evt004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rappoport N. ProtoNet: charting the expanding universe of protein sequences. Nat. Biotechnol. 2013;31:290–292. doi: 10.1038/nbt.2553. [DOI] [PubMed] [Google Scholar]
- 80.Ollagnier-de-Choudens S. Mechanistic studies of the SufS-SufE cysteine desulfurase: evidence for sulfur transfer from SufS to SufE. FEBS Lett. 2003;555:263–267. doi: 10.1016/s0014-5793(03)01244-4. [DOI] [PubMed] [Google Scholar]
- 81.Ragan M.A. Trees and networks before and after Darwin. Biol. Direct. 2009;4:43. doi: 10.1186/1745-6150-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Selosse M-A. Microbial priming of plant and animal immunity: symbionts as developmental signals. Trends Microbiol. 2014;22:607–613. doi: 10.1016/j.tim.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 83.Brucker R.M., Bordenstein S.R. Speciation by symbiosis. Trends Ecol. Evol. 2012;27:443–451. doi: 10.1016/j.tree.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 84.Brucker R.M., Bordenstein S.R. The hologenomic basis of speciation: gut bacteria cause hybrid lethality in the genus Nasonia. Science. 2013;341:667–669. doi: 10.1126/science.1240659. [DOI] [PubMed] [Google Scholar]
- 85.Hur K.Y., Lee M-S. Gut microbiota and metabolic disorders. Diabetes Metab. J. 2015;39:198–203. doi: 10.4093/dmj.2015.39.3.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gilbert S.F. A symbiotic view of life: we have never been individuals. Q. Rev. Biol. 2012;87:325–341. doi: 10.1086/668166. [DOI] [PubMed] [Google Scholar]
- 87.Dunning Hotopp J.C. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317:1753–1756. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- 88.La Scola B. The virophage as a unique parasite of the giant mimivirus. Nature. 2008;455:100–104. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]
- 89.Boyer M. Mimivirus shows dramatic genome reduction after intraamoebal culture. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10296–10301. doi: 10.1073/pnas.1101118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lanza V.F. The plasmidome of Firmicutes: impact on the emergence and the spread of resistance to antimicrobials. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.PLAS-0039-2014. PLAS-0039-2014. [DOI] [PubMed] [Google Scholar]
- 91.Remigi P. Transient hypermutagenesis accelerates the evolution of legume endosymbionts following horizontal gene transfer. PLoS Biol. 2014;12:e1001942. doi: 10.1371/journal.pbio.1001942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Serôdio J. Photophysiology of kleptoplasts: photosynthetic use of light by chloroplasts living in animal cells. Philos. Trans. R. Soc. Lond. B: Biol. Sci. 2014;369:20130242. doi: 10.1098/rstb.2013.0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rauch C. Why it is time to look beyond algal genes in photosynthetic slugs. Genome Biol. Evol. 2015;7:2602–2607. doi: 10.1093/gbe/evv173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ereshefsky M., Pedroso M. Rethinking evolutionary individuality. Proc. Natl. Acad. Sci. U.S.A. 2015;112:10126–10132. doi: 10.1073/pnas.1421377112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Martin W.F. Endosymbiotic theories for eukaryote origin. Philos. Trans. R. Soc. Lond. B: Biol. Sci. 2015;370:20140330. doi: 10.1098/rstb.2014.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Orphan V.J. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 2001;67:1922–1934. doi: 10.1128/AEM.67.4.1922-1934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ozuolmez D. Methanogenic archaea and sulfate reducing bacteria co-cultured on acetate: teamwork or coexistence? Front. Microbiol. 2015;6:492. doi: 10.3389/fmicb.2015.00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sun M. Microbial community analysis in rice paddy soils irrigated by acid mine drainage contaminated water. Appl. Microbiol. Biotechnol. 2015;99:2911–2922. doi: 10.1007/s00253-014-6194-5. [DOI] [PubMed] [Google Scholar]
- 99.Liu Y.R. Patterns of bacterial diversity along a long-term mercury-contaminated gradient in the paddy soils. Microb. Ecol. 2014;68:575–583. doi: 10.1007/s00248-014-0430-5. [DOI] [PubMed] [Google Scholar]
- 100.Lafontaine D.L. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zebarjadian Y. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell. Biol. 1999;19:7461–7472. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Becker H.F. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of psi55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res. 1997;25:4493–4499. doi: 10.1093/nar/25.22.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]