Abstract
Studies conducted in C. elegans over the last decade highlight the ancient and complex origins of body fat regulation. In this critical review, I introduce the major functional approaches used to study energy balance and body fat, the lipid composition of C. elegans, the regulation of cellular fat metabolism and its transcriptional control. Next I describe the influence of the sensory nervous system on body fat and the major regulatory mechanisms that couple food perception in the nervous system with the production of energy via fat metabolism. The final section describes the opportunities for the discovery of neuroendocrine factors that control communication between the nervous system and the metabolic tissues. The coming years are expected to reveal a wealth of information on the neuroendocrine control of body fat in C. elegans.
Keywords: Energy balance, Lipid regulatory network, Neuroendocrine
The C. elegans Model System
C. elegans is a small (~1.5 mm as adult) free-living roundworm that lives worldwide and feeds on a variety of microbes, primarily bacteria. The body plan is anatomically simple with about 1000 somatic cells, and is highly stereotyped across animals (Figure 1A). In a laboratory setting, the life cycle is 3 days at 20 degrees Celsius, and the lifespan is 3-4 weeks. Powerful genetic and genomic tools, rapidly-quantifiable phenotypes for food intake, body fat and energy expenditure, and the capacity to conduct large-scale screens offer an unprecedented opportunity to identify novel genetic pathways and molecular mechanisms underlying the conserved regulatory mechanisms of body fat control.
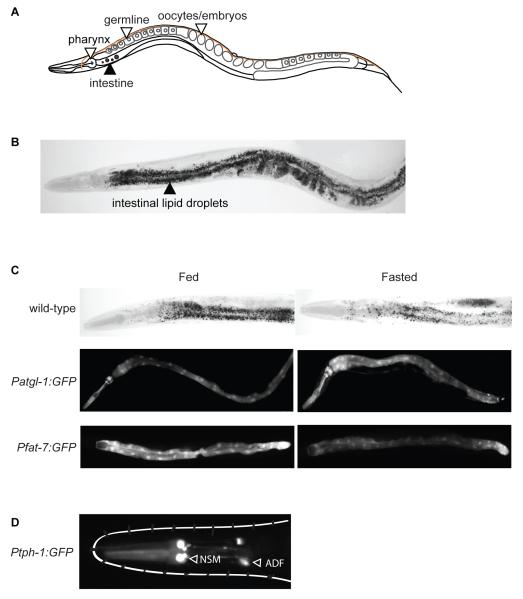
Figure 1. The C. elegans model system for studies of body fat regulation.
A. A schematic depiction of the anatomy of adult C. elegans. The pharynx is the feeding organ, and the intestine is the major organ for fat storage and mobilization. The germline and fertilized oocytes also store fat. B. Image of a fixed adult worm stained with Oil Red O, a lipophilic dye used to visualize and quantify body fat. Intestinal lipid droplets of different sizes can be clearly seen. C. Animals fasted for 4-6 hours deplete their fat reserves (upper panels). Fasted atgl-1::GFP transgenic animals show induction of atgl-1 expression upon fasting, and can be used as a screening tool for regulators of fat breakdown (middle panels). The fatty acid desaturase enzyme FAT-7 is inhibited upon fasting, as seen in fat-7::GFP reporter animals. D. The transparent body of the worm allows direct visualization of reporters in living animals. The rate-limiting enzyme for serotonin synthesis TPH-1, is expressed in two pairs of bilaterally symmetric neurons, NSM and ADF, in the head of the worm, and an additional pair of hermaphrodite-specific neurons called HSN near the vulva (not depicted).
Food Intake and Satiety
C. elegans ingests food via the rhythmic contractions of the pharynx, the feeding organ (Figure 1A). The terminal bulb of the pharynx grinds the ingested bacteria, and the ground material is transferred to the lumen of the intestine from where it is absorbed into the intestinal cells (1). Food intake can be readily measured by counting the rhythmic contractions of the pharynx under a stereomicroscope. The pharynx has a basal intrinsic rhythmic activity off food (100-120 pumps/min). Animals transferred to a food source such as a bacterial lawn will nearly double their pumping rate to approximately 220 pumps/min in young adult animals. The contraction-relaxation cycles of the pharynx that lead to food ingestion are mediated by the pharyngeal nervous system via the actions of the neurotransmitters serotonin, acetylcholine, glutamate and GABA (2). The sensory nervous system modulates the activity of the pharynx based on food availability. Serotonin and Transforming Growth Factor β (TGF β) are two major neuroendocrine effectors that are synthesized in sensory neurons in proportion to food availability, and modulate the activity of the pharynx accordingly (3-5). Defects in insulin signaling via mutations in the insulin receptor DAF-2 also lead to reduced food intake (S. Srinivasan unpublished observations), however the role of insulin signaling in regulating feeding is yet to be fully studied in C. elegans. There are likely to be additional positive and negative regulators of pharyngeal activity and feeding behavior that originate in the sensory nervous system to couple environmental information with feeding behavior.
C. elegans has a strong locomotor response to fluctuations in food availability. At a population level, food-deprived animals move faster than well-fed animals, because locomotion is the major foraging strategy for worms. Individual fasted animals slow down when re-introduced to food, and the extent of the slowing is proportionate to the length of food deprivation. In short-term fasting studies, wild-type animals that have been food-deprived for 30 minutes have a greater reduction in locomotion than well-fed animals. This behavior is called the enhanced slowing response, and is controlled predominantly by serotonin signaling from the sensory nervous system (6). In longer-term fasting studies, wild-type animals that have been food-deprived for 12 hours display rapid re-feeding in the first 30 minutes when re-introduced to food, followed by locomotor quiescence for 3-6 hours. This behavior is regulated predominantly by insulin and TGF-beta signaling from sensory neurons (7). Thus, locomotor behavior on and off food is an effective method to measure satiety.
Fat Storage and Utilization
The intestine is the major metabolic organ for C. elegans, and performs numerous functions including food absorption, incorporation of nutrients into metabolic pathways, fat storage and utilization, detoxification, pathogen defense and more broadly, the management of energy flow over the lifetime of the animal (www.wormatlas.org). The intestine is comprised of 10 pairs of cells that are connected to form a syncytium and span the length of the animal (Figure 1A). A variety of methods have been utilized to visualize and measure body fat and are reviewed elsewhere (8, 9). The predominant method to visualize fat droplets and to quantify body fat is by fixing animals and staining with a lipophilic dye such as Oil Red O or Nile Red, followed by light microscopy and quantification of the intensity of the stain (Figure 1B). Protocols to conduct Oil Red O-based body fat determination on fixed worms have recently been adapted for 96-well format (10), allowing for more convenient screening for regulators of body fat. Biochemical extraction of triglycerides and quantitation by mass spectrometry or thin-layer chromatography shows excellent concordance with dye-based methods of fixed worms. Short-term fasting depletes body fat stores in proportion to the length of food deprivation (Figure 1C). Fasted animals have increased expression of the canonical Adipocyte Triglyceride Lipase (ATGL/atgl-1) to stimulate fat loss, whereas fasted animals re-introduced to food induce the C18 fatty acid desaturase fat-7, presumably to restore fat levels upon re-feeding (Figure 1). Energy expenditure is typically inferred from measuring oxygen consumption, and can be performed in 96-well format (11, 12).
More recently, marker proteins of different lipid subcellular compartments have become known. Transgenic animals stably expressing fluorescently-tagged resident proteins for the various compartments of lipid regulation, including lipid droplets, lysosome-related organelles (LROs), peroxisomes and mitochondria are a valuable resource to visualize dynamic changes in lipid droplet morphology and mobilization (13-15). In principle, forward genetic and genome-wide RNAi-based screens for regulators of body fat can be performed with relative ease using these animals. A small amount of fat in the form of droplets is also visible in the hypodermis, the skin of C. elegans. Whether fat is synthesized locally in the skin or in the intestine and then transported, remains to be determined. However, mitochondrial and peroxisomal β-oxidation genes essential for fat breakdown and energy production are expressed in the skin (12).
Neurons and Neural Circuits
The nervous system of the C. elegans adult hermaphrodite is composed of 302 neurons. The completed wiring diagram of the C. elegans nervous system reveals that there are approximately 6000 chemical synapses, 600 gap junctions and 2000 neuromuscular junctions (www.wormatlas.org). There are sixty ciliated sensory neurons in C. elegans with dendritic endings that project outside the animal, thus information from the environment can be accessed directly (16). In addition, recent evidence suggests that C. elegans neurons are capable of using neuropeptides to communicate in a manner not specified by the wiring diagram, adding greatly to the complexity of possible neural circuit conformations (17, 18). Functional studies of the neurons and neural circuits involved in food sensing and fat regulation are greatly facilitated by the transparent body of the worm, allowing direct visualization of proteins in individual neurons in living animals (Figure 1D).
Composition of Body Fat
As seen in all eukaryotes, fatty acids in C. elegans are composed exclusively of even chains of saturated and un-saturated carbon-carbon bonds. The predominant fatty acids are derivatives of 16-, 18- and 20-carbon chains (19, 20). The fatty acid profile is a diverse array of saturated, mono- and poly-unsaturated fats modified by the actions of fatty acid desaturases whose precise catalytic functions have been delineated (20). Lipid profiling studies show that C. elegans fats are stored predominantly as triglycerides, which are composed of three fatty acid chains anchored by a glycerol backbone (Figure 2A). Diglycerides and monoglycerides are present but not prevalent in wild-type animals. RNA-mediated interference of the C. elegans ortholog of ATGL, a conserved enzyme that catalyzes the conversion of triglycerides to diglycerides and a free fatty acid chain, increases the overall abundance of triglycerides (Figure 2A). Fatty acids obtained from the bacterial diet are converted to triglycerides, which become the major form of stored body fat in C. elegans. Isotope labeling experiments show that in wild-type animals de novo lipogenesis accounts for less than 10% of stored body fat, whereas defects in insulin signaling increase that number to approximately 25% (21). Fatty acid desaturation in C. elegans appears to be a strategy to diversify the lipidome and provide a plethora of substrates for signal transduction pathways that control organismal physiology (22-24). Nuclear magnetic resonance studies reveal that a novel form of pheromones called ascarosides are composed of long-chain fatty acids (26 carbon and up), anchored by a sugar backbone. Current estimates suggest that ascarosides may make up as much as 15-30% of lipids in C. elegans (Frank Schroeder, personal communication). Ascarosides have important modulatory effects on development, animal communication and lifespan (25). Based on their effects on the insulin signaling pathway, ascarosides are likely to play interesting roles in lipid metabolism that are yet to be fully explored.
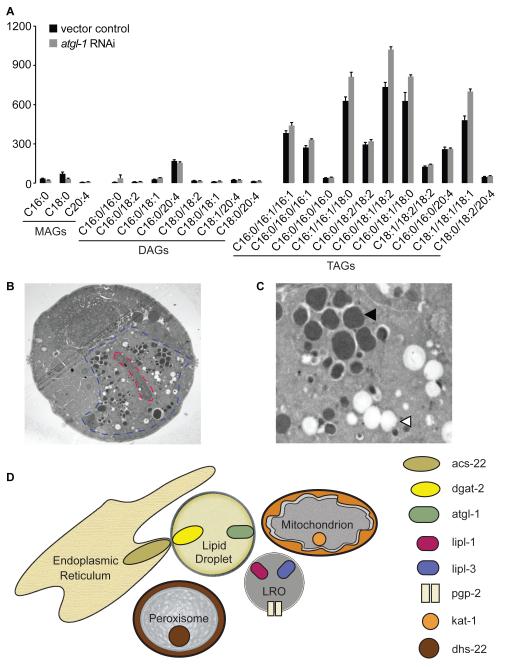
Figure 2. Composition of body fat and lipid droplets in C. elegans.
A. Liquid Chromatography-Mass Spectrometric analysis of monoglycerides (MAGs), diglycerides (DAGs) and triglycerides (TAGs) in C. elegans shows that lipid is stored predominantly as TAGs, as in most metazoans including humans. Relative to wild-type animals (black bars) RNAi-mediated interference of atgl-1 (gray bars), the rate-limiting enzyme for lipid mobilization, shows a preferential accumulation of TAGs. B. Transmission electron microscopic images of a cross-section of the intestine in wild-type animals, 1450X. The intestinal cell is demarcated in blue and the lumen of the intestine, in red. C. At 4000X magnification, at least two lipid containing organelles can be visualized (Personal communication, Malcolm Wood, Microscopy Core, TSRI). One class has a phase-dark core (closed arrowhead) and the other is electron-lucent (open arrowhead). D. Model depicting the lipid regulatory network in C. elegans. Markers and regulatory proteins associated with each organelle are depicted. The lipid droplets and LROs contain fat, whereas the mitochondria and peroxisomes are metabolic organelles and utilize fat to produce energy. The ER and lipid droplets form contacts via ACS-22 and DGAT-2. Whether each organelle is in physical contact with other organelles remains to be determined. However, inhibition of LRO lipases lipl-1 and lipl-3 lead to lipid accumulation in the lipid droplets, and inhibition of mitochondrial and peroxisomal components lead to lipid accumulation in both the lipid droplets and the LROs.
Cellular Fat Metabolism
In C. elegans the intestine serves as the major organ for lipid uptake, synthesis, storage and mobilization. In order to avoid futile cycles of triglyceride synthesis and breakdown within the same tissue, one can envision two complementary cellular strategies that might have evolved: elaborate subcellular compartmentalization and strict cellular regulation of triglyceride accumulation and breakdown. Experimental evidence to support both strategies has been uncovered using a combination of genetic and biochemical approaches.
Lipid Droplet Biology
There are two predominant lipid-containing organelles in the C. elegans intestine that can be visualized by electron microscopy (14, 15, 26, 27) (Figure 2B and C). The majority of intestinal fat is stored in neutral lipid droplets bound by a phospholipid monolayer (28), and some fats are also stored in a specialized organelle called the lysosome-related organelle (LRO) (29). LROs perform diverse cellular functions in different organisms and tissues, and may be co-opted for specific roles in a given organismal context (30). An ABC transporter protein called PGP-2 is selectively localized to the LROs in the intestine, and pgp-2 mutants lack LROs yet retain normal levels of fat, suggesting that although LROs may contain fats, they are not obligate organelles for fat storage. Conversely, RNAi-based gene inactivations that decrease fat staining in LROs do not show defects in LRO biogenesis, suggesting that fat accumulation in LROs is likely to be a regulated process (14).
Genetic screens and cell biological approaches have been valuable sources of information on the genesis of neutral lipid droplets. Two well-conserved genes called acyl CoA synthase-22 (acs-22/fatp-1) and diacylglycerol acyl transferase-2 (dgat-2) emerged as suppressors of a mutant with enlarged lipid droplets, visualized by confocal and electron microscopy (15, 26). Based on their homology, both proteins regulate discrete steps in triglyceride synthesis. Mak and colleagues showed that ACS-22 and DGAT-2 form a protein complex at the interface between the endoplasmic reticulum and lipid droplets, and the extent of co-localization of the two proteins increases with increasing size and number of droplets. These data lend support to the hypothesis that lipid droplets bud off from the ER in proportion to the amount of lipid needed to be stored.
Subcellular fractionation studies have provided information on the biochemical nature of C. elegans lipid droplets. As seen in most other species, lipid droplets are composed predominantly of triglycerides, and range in size from 50 – 3000 nm. The density, size and lipid composition of the C. elegans lipid droplets are in excellent agreement with those of other species (31). Proteomic analysis of the fractionated and purified lipid droplets showed that greater than 60% of the identified proteins have been previously found in lipid droplet proteomes of other species. In addition, the in vivo subcellular localization of the isolated proteins confirmed their association with lipid droplets (13). Some of the most abundantly-expressed lipid droplet proteins are graphically depicted in the model in Figure 2D. Because these lipid droplets have been isolated from intact organisms rather than cells in culture, it will be interesting to determine whether the newly-identified proteins associated with C. elegans lipid droplets have similar roles in mammalian lipid droplet biology. A recent study of fasting-induced lipases reveals an interesting association between lipid droplets and LROs (32). Two fasting-induced lipases LIPL-1 and LIPL-3 belong to the family of ATGL-like patatin-domain containing lipases and are localized to the LROs in the intestine. Although LIPL-1 and LIPL-3 do not associate with neutral lipid droplets, their loss is accompanied by a dramatic increase in neutral lipid droplets and overall body fat. Short-term fasting simultaneously leads to a decrease in neutral lipid droplets (Figure 1C) but an increase in the LRO compartment (29). These data suggest a subcellular conformation in which the LROs may function as a transit compartment for the flux of fatty acids from neutral lipid droplets to the mitochondria and/or peroxisomes for the production of energy. Physical contacts between lipid droplets, LROs, mitochondria and peroxisomes would allow for efficient sequential transfer of fats for energy production. Cellular energy sensors and long-range endocrine effectors would be predicted to govern the proposed lipid regulatory network (Figure 2D) to control the flow of energy in different environmental and physiological contexts.
Regulation of Fat Synthesis and Breakdown
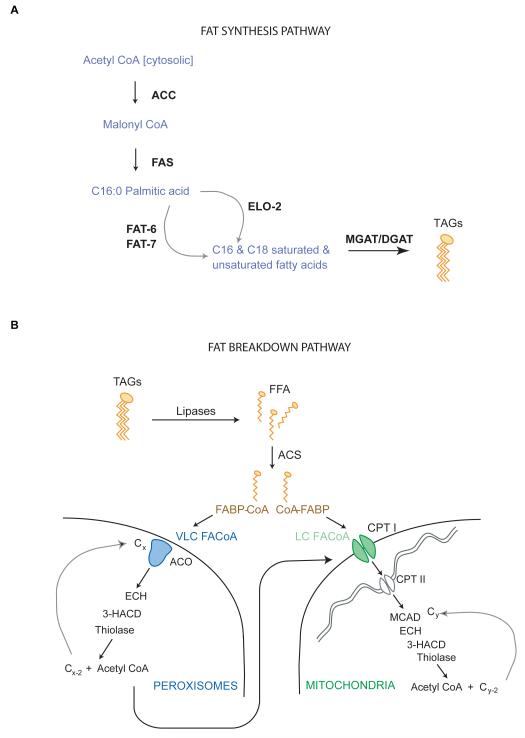
As seen in most organisms, fat synthesis is controlled by a few key enzymes that use acetyl CoA as a substrate to generate malonyl CoA and C16 palmitate (33) (Figure 3A). A model depicting the genes in the C. elegans genome that regulate the various steps of fat synthesis from acetyl CoA is shown in Figure 3A. Hypomorphic mutants of the two rate-limiting enzymes for fat synthesis, acetyl CoA carboxylase (ACC/pod-2) and fatty acid synthase (FAS/fasn-1) lead to early larval arrest. Both genes are expressed throughout the body of the animal and at all developmental stages (34, 35), thus fat synthesis appears to be a key requirement for growth. A second homolog of ACC is also found in the genome but remains uncharacterized. In adults, RNA-mediated interference of ACC/pod-2, the critical regulatory step in fat synthesis, leads to a severe decrease in body fat (E. Witham and S. Srinivasan, unpublished observations). Sterol Regulatory Element Binding Protein (SREBP) is a major conserved transcriptional regulator of body fat. In response to lowered sterol and lipid levels in the cell, SREBP modulates the transcription of hundreds of genes to coordinately restore cellular lipid homeostasis. In mammals, there are two SREBP homologs: SREBP-2 preferentially controls cholesterol and bile acid synthesis, whereas SREBP-1 controls fatty acid, phospholipid and triglyceride synthesis (36, 37). In C. elegans a single SREBP ortholog called SBP-1 is expressed in all metabolic tissues (38). Many functions of SREBP-1 are conserved between C. elegans and mammals, including regulation of fatty acid synthesis and desaturation and the regulation of phosphatidylcholine, a key component of cellular membranes (39, 40).
Figure 3.
Metabolic pathways for fat synthesis (A) and breakdown (B).
The regulatory pathway for fat utilization is β-oxidation, the process by which a cascade of enzymes coordinately cleaves the carbon-carbon bonds of fatty acids to produce energy (Figure 3B). β-oxidation occurs in the mitochondria and peroxisomes, the two major metabolic organelles, and is a conserved mechanism of energy generation across all metazoans (33). In C. elegans RNAi-based and forward genetic screens have shown that inhibition of the rate-limiting steps of β-oxidation leads to the accumulation of fats in both classes of lipid-containing organelles: the LROs and the neutral lipid droplets (12, 26). Numerous acyl CoA synthases (ACS) and dehydrogenases (ACD), which comprise the first two steps in the β-oxidation pathway are also among the most abundant proteins identified in purified lipid droplets (13), suggesting a possible physical association between the organelles of fat storage and mobilization.
With respect to lipid metabolism, an interesting feature of the C. elegans genome is the discrepancy between the numbers of homologs for genes that control lipid synthesis, versus those for lipid breakdown. Fat synthesis from acetyl CoA is controlled by relatively few enzymatic steps, for which the C. elegans genome contains a single, or at most two homologs. In contrast, fat breakdown is intrinsically more complex. Many more enzymatic steps are required, and the number of homologs for each step is greatly increased: the C. elegans genome contains more than 100 transcribed genes for lipases approximately 20 for acyl CoA synthases. Yet, inactivation of any single homolog results in a clear, discernable phenotype in a context-dependent manner. For example, β-oxidation genes essential for serotonin-mediated fat loss do not overlap with those for fasting-induced fat loss (12, 41). Although much work remains to be done to understand the functional consequences of the increased number of homologs for each step controlling fat breakdown, it is tempting to speculate that such a genomic expansion provides a broad genetic template that can be exploited for the sophisticated control of fatty acid breakdown in an environment- and context-specific manner. Relative to fat synthesis, the complexity of fat breakdown in C. elegans makes it a particularly well-suited model for the study of molecular mechanisms that induce sustained fat loss.
Transcriptional Control of Body Fat
Three predominant lines of evidence support a model in which changing energy demands leads to transcriptional induction of genes that re-model the lipid landscape in C. elegans. Lipases, transcription factors themselves, and nuclear receptors are amongst the most highly regulated genes, and have been best-studied in the context of fasting and re-feeding.
Lipases
Lipases are enzymes that catalyze the conversion of glycerol-bound fatty acids (monoglycerides, diglycerides and triglycerides) to free fatty acids and glycerol. Adult C. elegans fasted for six hours increase transcription of numerous lipases (32, 42). An RNAi-mediated screen for transcription factors that mediate the induction of a lipase called LIPL-1 revealed that transcription of LIPL-1 is repressed by the basic helix-loop-helix (bHLH) domain-containing transcription factor MXL-3, when food is available. Chromatin immunoprecipitation experiments showed that MXL-3 binds a predicted cis-regulatory element (43) in the LIPL-1 promoter to repress transcription of fasting-induced LIPL-1 expression. As expected, mxl-3 mutants have increased lipl-1 expression even in the fed state, accompanied by decreased body fat, similar to that of fasted animals. In contrast, when food is withdrawn, a related bHLH transcription factor HLH-30 activates the expression of fasting-induced lipases (32). The induction of HLH-30 itself is dependent on the cellular nutrient sensor complex target-of-rapamycin (TOR), whose protein partners Raptor/DAF-15 and Rictor/LPO-6/RICT-1 have been previously shown to regulate body fat (44-46).
In an independent line of investigation, the lipid droplet lipase atgl-1 was found to be transcriptionally induced by serotonin signaling, a conserved neuroendocrine regulator that induces fat loss (47). An RNAi-mediated screen for regulators of atgl-1 induction by serotonin reveals a similar transcriptional regulatory paradigm. The bHLH transcription factor HLH-11, whose cis-regulatory motif is over-represented in the atgl-1 promoter (43), is required to repress ATGL expression in the fed state (S. Srinivasan unpublished observations), whereas the nuclear receptor NHR-76, bearing distant similarity to the mammalian RXR family of nuclear receptors, is required to activate serotonin-dependent ATGL transcription (47). Thus, transcriptional regulation of lipases in C. elegans may be achieved by a generalizable strategy: repression by bHLH-like transcription factors in the basal state followed by recruitment of activating transcriptional regulators in the induced state.
Nuclear Receptors
Nuclear receptors are proteins that regulate gene expression by binding cis-regulatory DNA motifs of target genes upon activation by physiological or environmental ligands. This diverse class of regulatory receptors has the capacity to form complexes with other nuclear receptors, transcription factors and co-regulator proteins, and can have a tremendous impact on shifting organismal metabolic state. Not unexpectedly, some mammalian nuclear receptors are targets for classes of drugs that alter lipid metabolism (48). Because of the immense complexity of nuclear receptor signaling, many nuclear receptors genes remain functionally un-annotated. The C. elegans genome contains 284 nuclear receptor genes (49). In contrast the human genome contains 48 nuclear receptor genes, suggesting a great expansion of the nuclear receptor family in C. elegans. Work from several laboratories has shown functional conservation of individual C. elegans nuclear receptors with respect to lipid metabolism even though strong sequence homology with the mammalian genes is not observed in the majority of cases. The first C. elegans nuclear receptor to be examined in detail was DAF-12, distantly related to the mammalian vitamin D (VDR) and liver X (LXR) receptors. daf-12 mutants were originally isolated in a screen for genes that control an alternative developmental fate called dauer, that is triggered by poor environmental conditions such as lack of food and crowding (50). DAF-12 signaling has been studied in detail and is now known to influence lifespan, lipid metabolism and the switch between developmental arrest and reproductive growth (51, 52). Classical genetics, biochemical experiments and comparative metabolomics approaches have been fruitful in identifying endogenous steroid ligands for DAF-12, a class of molecules called dafachronic acids that are derivatives of cholesterol (53-55). The DAF-12 signaling pathway demonstrates the mechanisms by which environmental signals are transduced via an endocrine nuclear receptor signaling pathway to remodel metabolic and organismal fate.
The complexity of nuclear receptor signaling in metabolic biology in C. elegans can be highlighted by additional examples. A nuclear receptor called NHR-49 promotes fatty acid desaturation and β-oxidation via interaction with another nuclear receptor called NHR-80, and represses sphingolipid metabolism via heterodimerizing with NHR-66 (56). The consequence of loss of NHR-49 signaling is an altered lipid profile, deformed mitochondria and a shortened lifespan (57), suggesting that subtle modifications of fat oxidation and lipid composition can have meaningful phenotypic outcomes. NHR-49 is thought to bear distant sequence similarity to the family of mammalian HNF4α nuclear receptors that regulate lipid metabolism in the liver. Another C. elegans member of this family called NHR-8, regulates cholesterol and bile acid homeostasis, and is genetically upstream of the DAF-12 pathway (58). Finally, the nuclear receptor NHR-76, bearing similarity to the mammalian RXR family of receptors, is required for serotonin-mediated transcriptional induction of the ATGL lipase and increased fat loss (47). At a genomic level, analyses of transcriptional regulators of metabolic genes in C. elegans reveals a strong enrichment of nuclear receptors both on the standard lab diet, and when worms are fed alternate bacterial diets with differing metabolite compositions (59, 60). On-going efforts to decipher the role of nuclear receptors, their ligands and co-activators in lipid metabolism underscore their breadth and complexity in C. elegans (61-64).
Neuronal Control of Body Fat
With respect to body fat control, the central nervous system has historically been thought to exert its effects primarily via food intake. However, a strong body of evidence across different species now shows that the central nervous system is a major regulator of body fat stores and energy expenditure, independent of food intake (65-67). Understanding the neuronal, endocrine and molecular mechanisms by which the nervous system regulates distal body fat stores is a major challenge in biology, with important implications for the management of metabolic disorders resulting from increased body fat stores. Evidence from C. elegans that food intake and body fat are independent outcomes of nervous system function will be discussed in the context of serotonin and TGFβ signaling.
Serotonin
A combination of genetic and pharmacological approaches has yielded information on the serotonergic neuroendocrine pathway that controls body fat. Many of the genes identified through unbiased approaches in C. elegans have mammalian counterparts not previously associated with body fat control and may thus represent new genetic targets. The neuromodulator serotonin is a conserved regulator of energy balance and body fat stores. In mice, loss of the 5HT2c receptor, expressed broadly in the central nervous system, leads to adult-onset obesity (68). In humans, increased neural serotonergic signaling via pharmacological intervention decreases body fat and increases energy expenditure (69, 70). In the nematode C. elegans, serotonin controls many food-related behaviors, body fat and energy expenditure (5, 12). Serotonin is synthesized by the conserved rate-limiting enzyme tryptophan hydroxylase (tph-1), which is expressed in only a few neurons and, unlike mammals, is not found in the gut or other fat storage tissues (71). In a single pair of chemosensory neurons called ADF(L/R) that receive and transduce environmental signals, serotonin synthesis is finely-tuned to food availability: short-term food-deprivation decreases tph-1 expression in the ADF neurons, whereas animals re-introduced to food restore tph-1 expression back to that of well-fed animals (72). Because serotonin signaling has also been shown to regulate the enhanced slowing response, a measure of satiety in C. elegans, increased serotonin signaling is generally regarded as a neuromodulator whose endogenous levels correlate with food availability.
Genes regulating many aspects of serotonin-mediated behavior and physiology have been identified, allowing the dissection of genetic pathways that control various serotonin-regulated behaviors (73). As in mammals, increased serotonin signaling via genetic or pharmacological means leads to a potent decrease in body fat in C. elegans (12, 47). A genetic approach was used to first address the question of whether serotonin-mediated body fat loss occurs via direct regulation of energy expenditure, or indirectly via modulation of food intake, locomotion, reproductive rate or other phenotypes. This approach identified a neural serotonin-gated chloride channel called MOD-1 as a key receptor that controls fat loss without altering food intake. In contrast, three GPCRs SER-1, SER-5 and SER-7 modulate serotonin-mediated food intake and pharyngeal activity in distinct sensory and pharyngeal neural circuits, independent of any effects on body fat (12, 72, 74). Transgenic rescue of MOD-1 in the nervous system restores the suppression of serotonin-mediate fat loss seen in mod-1 mutants (47), whereas ectopic expression of MOD-1 in the intestine does not. Additionally, increased synaptic signaling via loss of the selective serotonin reuptake transporter mod-5, a neuronal gene, also decreases body fat (T. Noble and S. Srinivasan, unpublished results). A similar genetic approach was used to conclude that the potent effects of serotonin signaling on body fat are independent of its other physiological effects including locomotion, reproduction and stress response (12).
An RNAi-based suppressor screen for metabolic genes revealed that the mechanism underlying serotonin-mediated fat loss was increased fatty acid utilization and energy expenditure in the peroxisomes and mitochondria. Each enzymatic step of energy production via oxidation of fatty acids emerged from a suppressor screen for metabolic regulators of serotonin-mediated fat loss. These metabolic genes suppressed not only the fat loss induced by serotonin, but also the increased energy expenditure phenotype, providing evidence that serotonin-induced fat loss and energy expenditure are mechanistically coupled (12). A subsequent RNAi screen revealed that the regulatory lipase ATGL-1 is induced in the intestine by the action of serotonin signaling in neurons via the chloride channel MOD-1 (47).
Elucidation of the neural circuit for serotonin-mediated fat loss reveals two interesting features of serotonin signaling. First, serotonin-mediated fat loss requires octopamine (the invertebrate analog of adrenaline) via the biosynthetic enzyme tyramine beta-hydroxylase (tbh-1). Octopamine signaling in turn modulates the expression of the serotonin biosynthesis enzyme tryptophan hydroxylase (tph-1) (47). Together, serotonin and octopamine/adrenergic-like signaling function in a positive regulatory loop to sustain a neural signal for body fat loss (Figure 4). These observations in C. elegans may provide a potential molecular explanation for the long-observed potent effects of combined serotonergic and adrenergic weight loss drugs. Four pairs of neurons emerge as the critical nodes for serotonergic function. The octopamine signaling neurons RIC and AWB provide a permissive signal to maintain serotonin production, and the serotonin signaling neurons ADF and URX provide an instructive signal to stimulate fat loss. Loss of both the serotonergic and octopaminergic arms of the circuit leads to a complete suppression of fat loss, and a dramatic increase in body fat under basal conditions (Figure 4).
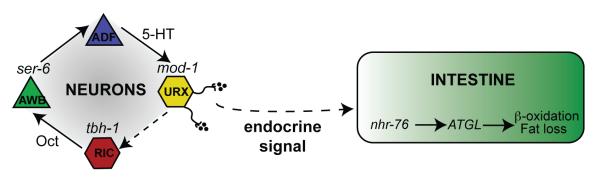
Figure 4. Schematic depiction of the serotonergic neuroendocrine pathway that controls body fat.
The described neural circuit for fat loss functions as a positive regulatory loop in which octopamine/adrenaline-like signaling acts as a permissive cue to maintain serotonin production and function. Serotonin production in the ADF chemosensory neurons activates the serotonergic chloride channel MOD-1 in the URX body cavity neurons to elicit fat loss. Serotonin signaling relays a neuroendocrine instructive signal from the nervous system to the distal intestine via unknown neuroendocrine effectors.
Second, the neural circuitry underlying serotonin-mediated fat loss suggests that rather than serotonin itself, undiscovered downstream neuroendocrine signal(s) are relayed to peripheral metabolic tissues to control fat loss. The inactivation of mod-1 selectively in the URX body cavity neurons prohibits the increased expression of the intestinal atgl-1 lipase during serotonergic activation (47). This result provides evidence for the existence of undiscovered neuroendocrine signals that originate in the nervous system in response to an instructive serotonergic signal. Such signals may be relayed via the coelomic fluid to the intestine where body fat is metabolized. The discovery of new neuroendocrine effectors of body fat loss has the potential to identify potent and conserved regulators of fat loss without the complicating physiological effects of broad serotonergic or octopaminergic signaling.
TGFβ and insulin
TGFβ and insulin are two major neuroendocrine regulators of organismal homeostasis in C. elegans. They have been best-studied in the context of the alternative dauer developmental fate. The signaling pathways for dauer formation downstream of the TGFβ ligand DAF-7 and the insulin receptor DAF-2, have been investigated in depth and are reviewed elsewhere (75). In adults, inactivation of the daf-7 and daf-2 genes causes a dramatic increase in body fat. TGFβ/DAF-7 is expressed in a single pair of sensory neurons called ASI, and regulates body fat via signaling through glutamate receptors in the NSM serotonergic neurons (4). The insulin receptor/DAF-2 is expressed broadly in the nervous system (76, 77). The increased body fat of daf-2 mutants is abrogated by loss of the canonical FOXO transcription factor daf-16. From an RNAi screen, a protein phosphatase called pdp-1 with sequence similarity to the mammalian pyruvate dehydrogenase phosphatase emerged as a link between the insulin and TGFβ signaling pathways (78). However, regulators of insulin-signaling-mediated body fat control distinct from its effects on dauer and lifespan regulation are yet to be identified.
A notable aspect of studies of serotonin and TGFβ signaling in body fat control is that both neuromodulators also function as food sensors in C. elegans. Serotonin functions as an indicator of food presence such that fasted animals restored to food increase serotonin production (72), up-regulate metabolic tone, increase energy expenditure and resume reproductive functions (5, 12). On the other hand, TGFβ/DAF-7 signaling is down-regulated in the absence of food (79), and promotes fat retention (4). Yet, in both cases the regulation of body fat and food intake occur through distinct neural circuits. Together the data suggest that rather food intake alone, the perception of nutrient sufficiency in the nervous system is relayed to maintain metabolic rate and stimulate fat loss. In contrast, the perception of nutrient deprivation down-regulates metabolism and leads to fat retention.
Neuroendocrine Signals
There is strong evidence to show that for serotonin and other neuromodulators that function solely in the nervous system to regulate body fat, endocrine mechanisms must exist to allow communication between neurons and the intestine, where body fat is stored. Unlike mammals, the C. elegans gut is not directly innervated. This anatomical feature serves as an excellent platform for the discovery of neuroendocrine peptides, hormones and other signaling molecules that relay information between the nervous and metabolic tissues. Despite great strides in the discovery of neuroendocrine hormones using biochemical approaches (80-82), much remains to be discovered in the field of neuroendocrine signaling in the context of body fat control. One can envision two models for the regulatory logic underlying neuroendocrine communication in C. elegans. In the first, each fat-regulatory neuron relays its own signal to mobilize or retain fat, and the intestine functions as the locus of control for the sorting and organization of multiple signals received from the nervous system. Other than the body cavity neurons, additional C. elegans sensory neurons are known to secrete neuropeptides in a manner not restricted by synaptic wiring (83, 84). In the second model, the locus of control resides in a small subset of central regulatory neurons. Sensory neurons relay external information to this central hub, which may also access internal nutrient status. A balance of internal and external states would then lead to the release of a few key neuroendocrine signals that are interpreted by the intestine to either mobilize or synthesize fat. Both models are plausible; of the 100+ neuropeptide genes in the C. elegans genome, each of which encodes multiple neuropeptides (85), many modulate body fat (S. Srinivasan unpublished observations).
Summary and Perspectives
Genetic and pharmacological screening approaches suggest that there are many additional regulators of body fat that await further study. Tractable genetic and pharmacological tools allowing tight temporal and reversible control of neuronal activation and inhibition are now available for C. elegans (86), and will facilitate the rapid discovery of key neurons that are instructive regulators of body fat. GFP-based reporters for body fat and its cellular compartmentalization make the study of the cell biology of lipid regulation feasible at a genome-wide scale. The coming years are expected to be an especially informative period in gaining a deeper appreciation of the regulatory logic of body fat control.
Acknowledgements
I thank Tallie Noble for editorial assistance with the figures, and members of the Srinivasan Laboratory for comments on the manuscript. We thank the Cravatt Laboratory at TSRI for assistance with mass spectrometry of C. elegans lipids. Malcolm Wood (Microscopy Core, TSRI) provided the technical skills and expertise to perform and evaluate the electron microscopy images. S.S. is supported by research grants from the National Institute of Diabetes and Digestive and Kidney Diseases, the Ray Thomas Edwards Foundation and the Novartis Advanced Discovery Institute.
Footnotes
Disclosure Statement
The author is not aware of any affiliations, memberships, funding or financial holdings that might be perceived to affect the objectivity of this review.
Literature Cited
- 1.Avery L, Shtonda BB. Food transport in the C. elegans pharynx. J Exp Biol. 2003;206:2441–57. doi: 10.1242/jeb.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schafer WR. Deciphering the neural and molecular mechanisms of C. elegans behavior. Curr Biol. 2005;15:R723–9. doi: 10.1016/j.cub.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 3.Schafer WR, Sanchez BM, Kenyon CJ. Genes affecting sensitivity to serotonin in Caenorhabditis elegans. Genetics. 1996;143:1219–30. doi: 10.1093/genetics/143.3.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greer ER, Perez CL, Van Gilst MR, Lee BH, Ashrafi K. Neural and molecular dissection of a C. elegans sensory circuit that regulates fat and feeding. Cell Metab. 2008;8:118–31. doi: 10.1016/j.cmet.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sze JY, Victor M, Loer C, Shi Y, Ruvkun G. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature. 2000;403:560–4. doi: 10.1038/35000609. [DOI] [PubMed] [Google Scholar]
- 6.Sawin ER, Ranganathan R, Horvitz HR. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26:619–31. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 7.You YJ, Kim J, Raizen DM, Avery L. Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metab. 2008;7:249–57. doi: 10.1016/j.cmet.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barros AG, Liu J, Lemieux GA, Mullaney BC, Ashrafi K. Analyses of C. elegans fat metabolic pathways. Methods Cell Biol. 2012;107:383–407. doi: 10.1016/B978-0-12-394620-1.00013-8. [DOI] [PubMed] [Google Scholar]
- 9.Yen K, Le TT, Bansal A, Narasimhan SD, Cheng JX, Tissenbaum HA. A comparative study of fat storage quantitation in nematode Caenorhabditis elegans using label and label-free methods. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wahlby C, Lee Conery A, Bray MA, Kamentsky L, Larkins-Ford J, et al. High- and low-throughput scoring of fat mass and body fat distribution in C. elegans. Methods. 2014 doi: 10.1016/j.ymeth.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, et al. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013;154:430–41. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srinivasan S, Sadegh L, Elle IC, Christensen AG, Faergeman NJ, Ashrafi K. Serotonin regulates C. elegans fat and feeding through independent molecular mechanisms. Cell Metab. 2008;7:533–44. doi: 10.1016/j.cmet.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang P, Na H, Liu Z, Zhang S, Xue P, et al. Proteomic study and marker protein identification of Caenorhabditis elegans lipid droplets. Mol Cell Proteomics. 2012;11:317–28. doi: 10.1074/mcp.M111.016345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroeder LK, Kremer S, Kramer MJ, Currie E, Kwan E, et al. Function of the Caenorhabditis elegans ABC transporter PGP-2 in the biogenesis of a lysosome-related fat storage organelle. Mol Biol Cell. 2007;18:995–1008. doi: 10.1091/mbc.E06-08-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu N, Zhang SO, Cole RA, McKinney SA, Guo F, et al. The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J Cell Biol. 2012;198:895–911. doi: 10.1083/jcb.201201139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inglis PN, Ou G, Leroux MR, Scholey JM. The sensory cilia of Caenorhabditis elegans. WormBook. 2007:1–22. doi: 10.1895/wormbook.1.126.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beets I, Janssen T, Meelkop E, Temmerman L, Suetens N, et al. Vasopressin/oxytocin-related signaling regulates gustatory associative learning in C. elegans. Science. 2012;338:543–5. doi: 10.1126/science.1226860. [DOI] [PubMed] [Google Scholar]
- 18.Garrison JL, Macosko EZ, Bernstein S, Pokala N, Albrecht DR, Bargmann CI. Oxytocin/vasopressin-related peptides have an ancient role in reproductive behavior. Science. 2012;338:540–3. doi: 10.1126/science.1226201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watts JL. Fat synthesis and adiposity regulation in Caenorhabditis elegans. Trends Endocrinol Metab. 2009;20:58–65. doi: 10.1016/j.tem.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watts JL, Browse J. Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2002:5854–9. doi: 10.1073/pnas.092064799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez CL, Van Gilst MR. A 13C isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans. Cell Metabolism. 2008:266–74. doi: 10.1016/j.cmet.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 22.Vrablik TL, Watts JL. Emerging roles for specific fatty acids in developmental processes. Genes Dev. 2012;26:631–7. doi: 10.1101/gad.190777.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Samuel BS, Breen PC, Ruvkun G. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature. 2014;508:406–10. doi: 10.1038/nature13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kniazeva M, Sieber M, McCauley S, Zhang K, Watts JL, Han M. Suppression of the ELO-2 FA elongation activity results in alterations of the fatty acid composition and multiple physiological defects, including abnormal ultradian rhythms, in Caenorhabditis elegans. Genetics. 2003;163:159–69. doi: 10.1093/genetics/163.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludewig AH, Schroeder FC. Ascaroside signaling in C. elegans. WormBook. 2013:1–22. doi: 10.1895/wormbook.1.155.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang SO, Box AC, Xu N, Le Men J, Yu J, et al. Genetic and dietary regulation of lipid droplet expansion in Caenorhabditis elegans. Proceedings of the National Academy of Sciences. 2010:4640–45. doi: 10.1073/pnas.0912308107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hermann GJ, Schroeder LK, Hieb CA, Kershner AM, Rabbitts BM, et al. Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol Biol Cell. 2005;16:3273–88. doi: 10.1091/mbc.E05-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mak HY. Lipid droplets as fat storage organelles in Caenorhabditis elegans: Thematic Review Series: Lipid Droplet Synthesis and Metabolism: from Yeast to Man. J Lipid Res. 2012;53:28–33. doi: 10.1194/jlr.R021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Rourke EJ, Soukas AA, Carr CE, Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 2009;10:430–5. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marks MS, Heijnen HF, Raposo G. Lysosome-related organelles: unusual compartments become mainstream. Curr Opin Cell Biol. 2013;25:495–505. doi: 10.1016/j.ceb.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding Y, Zhang S, Yang L, Na H, Zhang P, et al. Isolating lipid droplets from multiple species. Nat Protoc. 2013;8:43–51. doi: 10.1038/nprot.2012.142. [DOI] [PubMed] [Google Scholar]
- 32.O'Rourke EJ, Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol. 2013;15:668–76. doi: 10.1038/ncb2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salway JG. Metabolism at a glance. Blackwell Science; 1999. [Google Scholar]
- 34.Lee KZ, Kniazeva M, Han M, Pujol N, Ewbank JJ. The fatty acid synthase fasn-1 acts upstream of WNK and Ste20/GCK-VI kinases to modulate antimicrobial peptide expression in C. elegans epidermis. Virulence. 2010;1:113–22. doi: 10.4161/viru.1.3.10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Paik YK. A potential role for fatty acid biosynthesis genes during molting and cuticle formation in Caenorhabditis elegans. BMB Rep. 2011;44:285–90. doi: 10.5483/BMBRep.2011.44.4.285. [DOI] [PubMed] [Google Scholar]
- 36.Sampath H, Ntambi JM. Polyunsaturated fatty acid regulation of genes of lipid metabolism. Annual Reviews of Nutrition. 2005:317–40. doi: 10.1146/annurev.nutr.25.051804.101917. [DOI] [PubMed] [Google Scholar]
- 37.Coleman RA, Lewin TM, Muoio DM. Physiological and nutritional regulation of enzymes of triacylglycerol synthesis. Annu Rev Nutr. 2000;20:77–103. doi: 10.1146/annurev.nutr.20.1.77. [DOI] [PubMed] [Google Scholar]
- 38.McKay RM, McKay JP, Avery L, Graff JM. C elegans: a model for exploring the genetics of fat storage. Dev Cell. 2003;4:131–42. doi: 10.1016/s1534-5807(02)00411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker AK, Jacobs RL, Watts JL, Rottiers V, Jiang K, et al. A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell. 2011;147:840–52. doi: 10.1016/j.cell.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun ZY, et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700–4. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- 41.Van Gilst MR, Hadjivassiliou H, Yamamoto KR. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc Natl Acad Sci U S A. 2005;102:13496–501. doi: 10.1073/pnas.0506234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jo H, Shim J, Lee JH, Lee J, Kim JB. IRE-1 and HSP-4 contribute to energy homeostasis via fasting-induced lipases in C. elegans. Cell Metab. 2009;9:440–8. doi: 10.1016/j.cmet.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 43.Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, et al. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138:314–27. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- 45.Jones KT, Greer ER, Pearce D, Ashrafi K. Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol. 2009;7:e60. doi: 10.1371/journal.pbio.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes & Development. 2009:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noble T, Stieglitz J, Srinivasan S. An integrated serotonin and octopamine neuronal circuit directs the release of an endocrine signal to control C. elegans body fat. Cell Metab. 2013;18:672–84. doi: 10.1016/j.cmet.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beaven SW, Tontonoz P. Nuclear receptors in lipid metabolism: targeting the heart of dyslipidemia. Annu Rev Med. 2006;57:313–29. doi: 10.1146/annurev.med.57.121304.131428. [DOI] [PubMed] [Google Scholar]
- 49.Taubert S, Ward JD, Yamamoto KR. Nuclear Hormone Receptors in Nematodes: Evolution and Function. Molecular and cellular endocrinology. 2010 doi: 10.1016/j.mce.2010.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–83. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Antebi A, Culotti JG, Hedgecock EM. daf-12 regulates developmental age and the dauer alternative in Caenorhabditis elegans. Development. 1998;125:1191–205. doi: 10.1242/dev.125.7.1191. [DOI] [PubMed] [Google Scholar]
- 52.Ludewig AH, Kober-Eisermann C, Weitzel C, Bethke A, Neubert K, et al. A novel nuclear receptor/coregulator complex controls C. elegans lipid metabolism, larval development, and aging. Genes Dev. 2004;18:2120–33. doi: 10.1101/gad.312604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jeong PY, Jung M, Yim YH, Kim H, Park M, et al. Chemical structure and biological activity of the Caenorhabditis elegans dauer-inducing pheromone. Nature. 2005;433:541–5. doi: 10.1038/nature03201. [DOI] [PubMed] [Google Scholar]
- 54.Mahanti P, Bose N, Bethke A, Judkins JC, Wollam J, et al. Comparative metabolomics reveals endogenous ligands of DAF-12, a nuclear hormone receptor, regulating C. elegans development and lifespan. Cell Metab. 2014;19:73–83. doi: 10.1016/j.cmet.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Motola DL, Cummins CL, Rottiers V, Sharma KK, Li T, et al. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–23. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 56.Pathare PP, Lin A, Bornfeldt KE, Taubert S, Van Gilst MR. Coordinate regulation of lipid metabolism by novel nuclear receptor partnerships. PLoS Genet. 2012;8:e1002645. doi: 10.1371/journal.pgen.1002645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 2005;3:e53. doi: 10.1371/journal.pbio.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Magner DB, Wollam J, Shen Y, Hoppe C, Li D, et al. The NHR-8 nuclear receptor regulates cholesterol and bile acid homeostasis in C. elegans. Cell Metab. 2013;18:212–24. doi: 10.1016/j.cmet.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.MacNeil LT, Watson E, Arda HE, Zhu LJ, Walhout AJ. Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell. 2013;153:240–52. doi: 10.1016/j.cell.2013.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watson E, MacNeil LT, Ritter AD, Yilmaz LS, Rosebrock AP, et al. Interspecies systems biology uncovers metabolites affecting C. elegans gene expression and life history traits. Cell. 2014;156:759–70. doi: 10.1016/j.cell.2014.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watson E, MacNeil LT, Arda HE, Zhu LJ, Walhout AJ. Integration of metabolic and gene regulatory networks modulates the C. elegans dietary response. Cell. 2013;153:253–66. doi: 10.1016/j.cell.2013.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mullaney BC, Blind RD, Lemieux GA, Perez CL, Elle IC, et al. Regulation of C. elegans fat uptake and storage by acyl-CoA synthase-3 is dependent on NR5A family nuclear hormone receptor nhr-25. Cell Metab. 2010;12:398–410. doi: 10.1016/j.cmet.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arda HE, Taubert S, MacNeil LT, Conine CC, Tsuda B, et al. Functional modularity of nuclear hormone receptors in a Caenorhabditis elegans metabolic gene regulatory network. Mol Syst Biol. 2010;6:367. doi: 10.1038/msb.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taubert S, Van Gilst MR, Hansen M, Yamamoto KR. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 2006;20:1137–49. doi: 10.1101/gad.1395406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kong D, Tong Q, Ye C, Koda S, Fuller PM, et al. GABAergic RIP-Cre neurons in the arcuate nucleus selectively regulate energy expenditure. Cell. 2012;151:645–57. doi: 10.1016/j.cell.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu J, Li T, Yang D, Ma R, Moran TH, Smith WW. Synphilin-1 alters metabolic homeostasis in a novel Drosophila obesity model. Int J Obes (Lond) 2012;36:1529–36. doi: 10.1038/ijo.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu M, Sarruf DA, Talukdar S, Sharma S, Li P, et al. Brain PPAR-gamma promotes obesity and is required for the insulin-sensitizing effect of thiazolidinediones. Nat Med. 2011;17:618–22. doi: 10.1038/nm.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nonogaki K, Abdallah L, Goulding EH, Bonasera SJ, Tecott LH. Hyperactivity and reduced energy cost of physical activity in serotonin 5-HT(2C) receptor mutant mice. Diabetes. 2003;52:315–20. doi: 10.2337/diabetes.52.2.315. [DOI] [PubMed] [Google Scholar]
- 69.Chan EW, He Y, Chui CS, Wong AY, Lau WC, Wong IC. Efficacy and safety of lorcaserin in obese adults: a meta-analysis of 1-year randomized controlled trials (RCTs) and narrative review on short-term RCTs. Obes Rev. 2013;14:383–92. doi: 10.1111/obr.12015. [DOI] [PubMed] [Google Scholar]
- 70.Wise SD. Clinical studies with fluoxetine in obesity. Am J Clin Nutr. 1992;55:181S–84S. doi: 10.1093/ajcn/55.1.181s. [DOI] [PubMed] [Google Scholar]
- 71.Gershon MD. 5-Hydroxytryptamine (serotonin) in the gastrointestinal tract. Curr Opin Endocrinol Diabetes Obes. 2013;20:14–21. doi: 10.1097/MED.0b013e32835bc703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cunningham KA, Hua Z, Srinivasan S, Liu J, Lee BH, et al. AMP-activated kinase links serotonergic signaling to glutamate release for regulation of feeding behavior in C. elegans. Cell Metab. 2012;16:113–21. doi: 10.1016/j.cmet.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chase DL, Koelle MR. Biogenic amine neurotransmitters in C. elegans. WormBook. 2007:1–15. doi: 10.1895/wormbook.1.132.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Song BM, Avery L. Serotonin activates overall feeding by activating two separate neural pathways in Caenorhabditis elegans. J Neurosci. 2012;32:1920–31. doi: 10.1523/JNEUROSCI.2064-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu PJ. Dauer. WormBook. 2007:1–19. doi: 10.1895/wormbook.1.144.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kimura KD, Riddle DL, Ruvkun G. The C. elegans DAF-2 insulin-like receptor is abundantly expressed in the nervous system and regulated by nutritional status. Cold Spring Harb Symp Quant Biol. 2011;76:113–20. doi: 10.1101/sqb.2011.76.010660. [DOI] [PubMed] [Google Scholar]
- 77.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–6. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 78.Narasimhan SD, Yen K, Bansal A, Kwon ES, Padmanabhan S, Tissenbaum HA. PDP-1 links the TGF-beta and IIS pathways to regulate longevity, development, and metabolism. PLoS Genet. 2011;7:e1001377. doi: 10.1371/journal.pgen.1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ren P, Lim CS, Johnsen R, Albert PS, Pilgrim D, Riddle DL. Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science. 1996;274:1389–91. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]
- 80.Guillemin R. Endorphins, brain peptides that act like opiates. N Engl J Med. 1977;296:226–8. doi: 10.1056/NEJM197701272960414. [DOI] [PubMed] [Google Scholar]
- 81.Guillemin R. A conversation with Roger Guillemin. Interview by Greg Lemke. Annu Rev Physiol. 2013;75:1–22. doi: 10.1146/annurev-physiol-082712-104641. [DOI] [PubMed] [Google Scholar]
- 82.Yalow RS, Berson SA. Immunoassay of endogenous plasma insulin in man. 1960. Obes Res. 1996;4:583–600. doi: 10.1002/j.1550-8528.1996.tb00274.x. [DOI] [PubMed] [Google Scholar]
- 83.Kao G, Nordenson C, Still M, Ronnlund A, Tuck S, Naredi P. ASNA-1 positively regulates insulin secretion in C. elegans and mammalian cells. Cell. 2007;128:577–87. doi: 10.1016/j.cell.2006.12.031. [DOI] [PubMed] [Google Scholar]
- 84.Lee BH, Ashrafi K. A TRPV channel modulates C. elegans neurosecretion, larval starvation survival, and adult lifespan. PLoS Genet. 2008;4:e1000213. doi: 10.1371/journal.pgen.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li C, Kim K. Neuropeptides. WormBook. 2008:1–36. doi: 10.1895/wormbook.1.142.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pokala N, Liu Q, Gordus A, Bargmann CI. Inducible and titratable silencing of Caenorhabditis elegans neurons in vivo with histamine-gated chloride channels. Proc Natl Acad Sci U S A. 2014;111:2770–5. doi: 10.1073/pnas.1400615111. [DOI] [PMC free article] [PubMed] [Google Scholar]