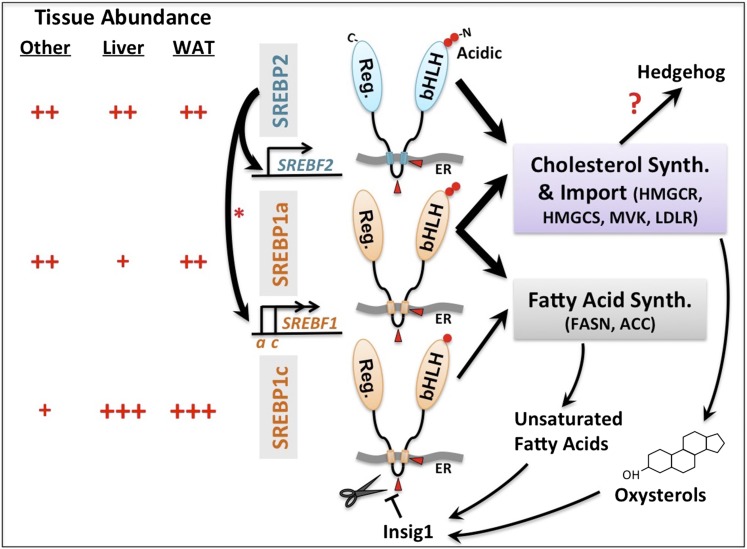
Sterol regulatory element-binding proteins (SREBPs, including SREBP1a, SREBP1c, and SREBP2) are basic-helix-loop-helix leucine zipper (bHLH-Zip) transcription factors that regulate the synthesis and cellular uptake of two major building blocks of cell membranes: cholesterol and fatty acids. For cholesterol biosynthesis, SREBPs activate expression of genes such as HMG-CoA reductase (HMGCR), HMG-CoA synthase (HMGCS), and mevalonate kinase (MVK). For cholesterol uptake, SREBPs activate expression of the LDL receptor (LDLR). For fatty acid synthesis, SREBPs activate expression of genes such as fatty acid synthase (FASN) and acetyl-CoA carboxylase (ACC). Because of the essential role of cholesterol and triglycerides for all cells, it is perhaps no surprise that SREBPs are tightly regulated, exhibiting extensive crosstalk and some redundancy. It has long been viewed that SREBP2 is primarily responsible for activation of genes involved in cholesterol synthesis, as opposed to fatty acid synthesis. In contrast, SREBP1c functions primarily in the liver to drive fatty acid synthesis, while SREBP1a can drive both pathways in all tissues. However, understanding the tissue-specific and differential genetic requirements for each SREBP has proven elusive. In this issue of the Journal of Lipid Research, Vergnes et al. (1) from the laboratory of Karen Reue confirm the role of SREBP2 in regulating cholesterol biosynthesis genes, but discover that SREBP2 has an unanticipated function in survival and limb patterning during development and that it also possesses an important role for fatty acid synthesis via compensatory upregulation of SREBP1 genes (Fig. 1).
Fig. 1.
Regulation of cholesterol and fatty acid bioavailability in cells by SREBPs. SREBPs are conserved bHLH transcription factors localized to the endoplasmic reticulum (ER) via transmembrane domains. Low levels of oxysterols and/or unsaturated fatty acids relieve Insig1-mediated ER-retention, causing SREBP movement to the golgi, proteolytic processing at two cleavage sites (red arrow heads), and nuclear localization. SREBP2 is broadly expressed in embryonic and adult mice and is a potent activator of genes involved in cholesterol synthesis and import, while SREBP1a and SREBP1c play bigger roles in driving expression of genes involved in fatty acid synthesis. Differences in the length and composition of the N-terminal acidic domain (red circles) of each SREBP likely account for differences in their ability to drive transcriptional activation of target genes. Vergnes et al. (1) reveal how loss of Srebp2 (Srebf2) in the mouse greatly compromises expression of both Srebp1a and Srebp1c in the liver (asterisk), which are cotranscribed from the same locus (Srebf1) and differ in their 5′ exons, causing defects in both hepatic cholesterol and fatty acid synthesis. Vergnes et al. also describe how defects in cholesterol synthesis caused by Srebp2 deficiency may negatively impact Hh signaling in developing limb buds of the embryo.
Vergnes et al. use a gene trap allele of murine Srebf2 to examine deficiency of Srebp2 in embryos and adult animals at various dosages through an ingenious conditional allele. This conditional allele is a genetic modification of Srebf2 through a gene trap, which is an insertional mutation, usually in an intron, that contains a strong splice acceptor and polyadenylation signal that disrupts splicing, resulting in a severely truncated protein product. The novelty of this gene trap, within the first intron of Srebf2, is that it enables the conversion of a null allele to a hypomorphic (reduced function) allele through Cre-Lox recombination. This is due to the nature of this specific gene trap, in which the splice acceptor is flanked by loxP sites, followed by the β-GEO fusion protein/reporter. Srebf2 splicing with this gene trap produces β-GEO instead of Srebp2 protein. Removal of the splice acceptor with Cre recombinase largely inactivates the function of the gene trap, after which the remaining β-GEO sequence partially disrupts expression of Srebf2, likely through modest effects on splicing or transcriptional activation. Because Srebp2 autoregulates its own expression in a positive feedback loop (2), an effective gene trap is vital to generating Srebp2 depleted animals by combining one hypomorphic (hyp) Cre-recombined allele and one null “fully-trapped” allele. Vergnes et al. use this allele to generate Srebp2 deficient (Srebf2−/−) and Srebp2 depleted (Srebf2–/hyp) mice. This gene trap is also likely advantageous in that no region of the Srebf2 locus has been deleted to generate the null (or hypomorphic) alleles, and thus regulatory sequences and other important genes are less likely to be affected unintentionally. This is especially important because engineered mutations could affect the Srebf2-embedded microRNA gene, miR-33, which represses Srebp1 and cellular cholesterol efflux 33 (3).
Using the fully trapped null allele, Vergnes et al. document the lethal embryonic phenotype caused by loss of Srebp2, importantly, with no effect on miR-33 expression caused by a lack of Srebp2. The critical importance of Srebf2 is perhaps not too surprising given the embryonic lethal phenotype of engineered Srebf2−/− mouse knockouts that was noted, although not described, more than 18 years ago (4). This early lethality is likely due to the widespread expression of Srebp2 in the developing embryo described by Vergnes et al. and its known role in cholesterol synthesis. However, the cause of embryonic lethality remains unknown along with a gender dimorphism where ~50% female Srebf2–/hyp mice died between 8 and 12 weeks of age.
Surprisingly, the effect of Srebp2 deficiency on fatty acid synthesis genes was just as severe as the effect on cholesterol biosynthesis genes. In whole Srebf2−/− embryos, transcript levels of Srebp1a and Srebp1c (from the Srebpf1 locus) were dramatically reduced, which may be responsible for depletion of fatty acid synthesis genes. This effect was also observed in the liver of a sole surviving Srebf2−/− adult animal. This trend of reduced expression of Srebp1a and 1c was also encountered in adult Srebf2–/hyp animals, albeit less severe. Unlike effects in the liver caused by Srebp2 deficiency, in inguinal subcutaneous white adipose tissue (iWAT), Srebp1a levels were significantly increased, indicating a tissue-specific compensatory feedback. This may also explain why triglyceride levels are not reduced in white adipose tissue in Srebf2−/− or Srebf2–/hyp animals. However, white adipose tissue weight was dramatically reduced in Srebf2−/− animals and also moderately reduced in Srebf2–/hyp animals, so the observed compensation may be difficult to parse given the obvious defect in WAT formation. Srebp2 mediated regulation of Srebp1c has been documented, but here Vergnes et al. may have evidence for regulation of both Srebp1a and Srebp1c, and while they have differing transcriptional start sites, these genes may share sterol-sensitive cis regulatory elements. In essence, the liver is highly dependent on Srebp2, which provides positive feedback to stimulate expression of Srebp1a and Srebp1c, whereas other tissues such as WAT may exhibit more plasticity. It may be informative in the future to analyze Srebp1a and 1c expression in other tissues in Srebf2−/− or Srebf2–/hyp animals in order to understand the range of these compensatory adaptations. Given that whole embryo cholesterol and triglyceride levels appear normal in Srebf2−/− mice, it is reasonable to propose that compensation may be provided by maternally-supplied HDL and LDL, via placental transport, and/or by other undefined basal transcriptional activation of cholesterol synthesis genes. Regardless, the findings here are a testament to the robustness of cholesterol and fatty acid synthesis, which can maintain reasonable levels of these essential molecules in the near absence of three master regulators (SREBPs).
Finally, Vergnes et al. found limb patterning defects in Srebf2−/− embryos that may reflect altered signaling of the critical “morphogen” Hedgehog (Hh). Hh proteins are secreted signaling proteins that are unusual in that they undergo posttranslational modification in the form of cholesterylation and palmitylation (5, 6), which have been reported to modulate Hh diffusion throughout tissues during development (forming a so-called morphogen gradient) (7). While whole embryo cholesterol levels were not changed in Srebf2−/− embryos, cholesterol-modified Sonic hedgehog protein (Shh) may be locally deficient in Srebf2−/− limb buds, suggesting a positive role for this posttranslational modification. Even though the role of cholesterol-modified Hh proteins remains unclear, Vergnes et al. report that Srebp2 depletion blocks the expression of key Shh targets (Ptch1, Gli1, Bmp4, Grem1) critical to limb bud patterning, with some effects recapitulated by sterol depletion in cultured wild-type fibroblasts. Hh defects may thus be implicated in the pathogenesis of the gross anatomical abnormalities observed in Srebf2−/− limb buds, although bizarrely, the sole surviving Srebf2−/− mouse possessed normal limbs. Alternatively, other effects of Srebp2 on limb bud development may occur through cholesterol-independent function of Srebp2, perhaps through effects on autophagy genes (8). Further studies are required to explore Hh cholesterylation in Srebf2−/− embryos. Taken together, the elegant studies by Vergnes et al. greatly advance our understanding of the pleomorphic effects of SREBP signaling in embryonic lipid homeostasis and development.
REFERENCES
- 1.Vergnes L., Chin R. G., de Aguiar Vallim T., Fong L. G., Osborne T. F., Young S. G., and Reue K.. 2016. SREBP-2-deficient and hypomorphic mice reveal roles for SREBP-2 in embryonic development and SREBP-1c expression. J. Lipid Res. 56: 410–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sato R., Inoue J., Kawabe Y., Kodama T., Takano T., and Maeda M.. 1996. Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J. Biol. Chem. 271: 26461–26464. [DOI] [PubMed] [Google Scholar]
- 3.Horie T., Baba O., Kuwabara Y., Nakao T., Nishiga M., Usami S., Izuhara M., Nakazeki F., Ide Y., Koyama S., et al. 2013. MicroRNA-33 regulates sterol regulatory element-binding protein 1 expression in mice. Nat. Commun. 4: 2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimano H., Shimomura I., Hammer R. E., Herz J., Goldstein J. L., Brown M. S., and Horton J. D.. 1997. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J. Clin. Invest. 100: 2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porter J. A., Young K. E., and Beachy P. A.. 1996. Cholesterol modification of hedgehog signaling proteins in animal development. Science. 274: 255–259. [DOI] [PubMed] [Google Scholar]
- 6.Pepinsky R. B., Zeng C., Wen D., Rayhorn P., Baker D. P., Williams K. P., Bixler S. A., Ambrose C. M., Garber E. A., Miatkowski K., et al. 1998. Identification of a palmitic acid-modified form of human Sonic hedgehog. J. Biol. Chem. 273: 14037–14045. [DOI] [PubMed] [Google Scholar]
- 7.Guerrero I., and Chiang C.. 2007. A conserved mechanism of Hedgehog gradient formation by lipid modifications. Trends Cell Biol. 17: 1–5. [DOI] [PubMed] [Google Scholar]
- 8.Seo Y. K., Jeon T. I., Chong H. K., Beisinger J., Xie X., and Osborne T. F.. 2011. Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metab. 13: 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]