Abstract
Phospholipids (PLs), one of the lipid categories, are not only the primary building blocks of cellular membranes, but also can be split to produce products that function as second messengers in signal transduction and play a pivotal role in numerous cellular processes, including cell growth, survival, and motility. Here, we present an integrated novel method that combines a fast and robust TMS-diazomethane-based phosphate derivatization and isotopic labeling strategy, which enables simultaneous profiling and relative quantification of PLs from biological samples. Our results showed that phosphate methylation allows fast and sensitive identification of the six major PL classes, including their lysophospholipid counterparts, under positive ionization mode. The isotopic labeling of endogenous PLs was achieved by deuterated diazomethane, which was generated through acid-catalyzed hydrogen/deuterium (H/D) exchange and methanolysis of TMS-diazomethane during the process of phosphate derivatization. The measured H/D ratios of unlabeled and labeled PLs, which were mixed in known proportions, indicated that the isotopic labeling strategy is capable of providing relative quantitation with adequate accuracy, reproducibility, and a coefficient of variation of 9.1%, on average. This novel method offers unique advantages over existing approaches and presents a powerful tool for research of PL metabolism and signaling.
Keywords: lysophospholipid, lipidomics, mass spectrometry, chemical synthesis, trimethylsilyl-diazomethane, deuterated diazomethane, hydrogen/deuterium exchange
Phospholipids (PLs) have diverse and critical roles in cellular metabolism and function, such as cell growth, survival, and motility (1). First, PLs are the primary building blocks of cellular membranes, which hold the living matter within each cell and also give definition, shape, and protection to many of the organelles within cells. Second, some types of PLs can be split to produce products that function as second messengers in signal transduction (2). Moreover, aberrant PL metabolism has been implicated in numerous human diseases, such as cancer, neurological disorders, diabetes, and others (3, 4). Consequently, analysis of PL profiles has been suggested for early clinical diagnosis (e.g., for the characterization of tumors and other diseases), as research on disease-related PL metabolism has made significant progress over the past decade (5, 6).
Phospholipidomics involves identifying and quantifying the PL species present in biological samples. The conventional MS-based method offers sensitive and high throughput analysis of PLs in both positive and negative ionization mode. The positive or negative ionization will occur at either the phosphate group or on the head group of PLs. Among PLs, phosphatidylcholine (PC) and SM are easily positively charged and detected in the positive mode, whereas phosphatidylethanolamine (PE), phosphatidylserine (PS), phosphatidylglycerol (PG), phosphatidylinositol (PI), and phosphatidic acid (PA) tend to form negatively charged ions and are detected in negative mode. The PE, PS, PG, and PI can also be detected in positive mode by forming as ammonium adducts, but with less ionization efficiency. Previously, Clark et al. (7) showed that TMS-diazomethane could be used to methylate the phosphate groups of phosphoinositides, thus increasing the detection sensitivity of phosphoinositides. In their discussion, they also mentioned the potential of this method for the detection of the other PL species. However, in practice, whether phosphate methylation by TMS-diazomethane is feasible for the detection of these PL classes, as well as their lysophospholipid (LPL) counterparts, remains unclear. For instance, whether the TMS-diazomethane is suitable for the methylation of all these PLs or LPLs, and whether the derivatization efficiency is affected by the chemical properties of individual PL or LPL species, such as FA chain length and double bond numbers, are still unknown. Moreover, identification of PLs and LPLs using ESI-MS/MS after derivatization also needs to be elaborated. Recently, a strategy based on in-solution concomitant quaternization of glycerophospholipid amino groups and methylation of phosphate groups via reaction with diazomethane was reported to be able to enhance the detection sensitivity of some PL subclasses (8). However, the strategy just covers the four subclasses of the PLs, and the preproduction of diazomethane with specific apparatus is required.
In addition, relative quantification of PL species between different samples will be helpful to understand the dynamic change and functions of PLs in biological systems with different conditions. The quantitative analysis of PLs is usually achieved by the inclusion of multiple internal standards (ISDs) and run separately. An alternative strategy for quantification of PLs would be to incorporate a given heavy nuclei (e.g., 2H, 13C, or 15N) into endogenous PLs by metabolic or chemical labeling. Each population of PLs is reacted with a reagent that differs only in its isotopic composition, thereby creating “heavy” and “light” versions of derivatized PLs, which are distinguishable by MS. Using such a strategy, run-to-run, or even scan-to-scan, variance in lipid ion abundance can be avoided because different samples are ionized simultaneously after separate derivatization and mixing. Moreover, the relative abundance of PLs from different biological samples can be directly determined and compared from the relative MS signal intensities. To date, there are several approaches that have been developed for the relative quantitation of PLs using isobaric mass stable isotope labeling derivatization reagents. For example, Berry et al. (11) have reported the isobaric tags for relative and absolute quantitation (9, 10) and isotope-labeled 4-(dimethylamino)benzoic acid N-hydroxysuccinimide ester-based derivatization labeling of primary amino groups of PE species for their quantitative analysis in biological mixtures. Recently, Nie et al. (12) also demonstrated isobaric mass stable isotope-labeled S,S′-dimethylthiobutanoylhydroxysucccinimide ester reagents for characterization and multiplexed quantification of aminophospholipids in biological samples. The advantages of these strategies are that the use of MS2 for quantitation reduces complexity and increases precision, and more than two samples can be analyzed simultaneously. However, they are limited to the relative quantitation of aminophospholipids. Previously, we have reported a strategy based on phosphate methylation and isotopic labeling for the relative quantitation of phosphoinositides from two different samples (13). This method involves isotopic labeling of endogenous phosphoinositides with deuterated diazomethane (CD2N2) for quantitation of phosphoinositides. CD2N2 was generated in situ through acid-catalyzed hydrogen/deuterium (H/D) exchange and methanolysis of TMS-diazomethane. However, whether such a strategy could be adapted directly for the relative quantitation of other major PLs and LPLs remains unclear, due to the high complexity of PLs and LPLs in the biological sample.
In this study, we present an integrated method that combines a fast and robust phosphate derivatization and isotopic labeling strategy using TMS-diazomethane with shotgun lipidomics technique that enables simultaneous profiling and relative quantification of PLs.
EXPERIMENTAL
Chemicals and reagents
TMS-diazomethane [IUPAC name: (diazomethyl)-trimethylsilane] (2 M in hexanes), deuterium oxide, deuterium-labeled methanol (MeOD or methanol-D4), and deuterium chloride (DCl, 20 wt% in deuterium oxide) were purchased from Acros Organics. Acetic acid and hydrochloric acid (HCl) of analytical grade were purchased from Fisher Scientific. Chloroform (SpS grade), methanol (MeOH, SpS grade), water (H2O, ultra-gradient grade), and methyl tert-butyl ether (MTBE, ultra-gradient grade) were from J. T. Bakert. Synthetic PC 17:0/14:1, PE 17:0/14:1, PG 17:0/14:1, PS 17:0/14:1, PA 17:0/14:1, PI 17:0/14:1, lyso (L)PC 17:1, LPE 17:1, LPG 17:1, LPS 17:1, LPA 17:1, and LPI 17:1 were purchased from Avanti Polar Lipids. All the reagents used for cell culture were from Invitrogen unless stated otherwise. All other reagents were from Sigma-Aldrich unless specified.
Cell lines and animals
The prostate cancer cell line, PC3, was obtained from ATCC. The PC3 cells were cultured in DMEM medium with 10% (v/v) FBS and 1% (v/v) penicillin/streptomycin and maintained at 37°C in a 5% CO2 incubator. Wild-type C57BL/6J mice (male, 12 weeks of age) were used in the study.
Lipid extraction
The lipid extraction was performed using 2 ml polypropylene safe-lock tubes. The lipid extraction was performed as described previously with some modifications (14). Briefly, cells were collected after digestion with trypsin and subjected to cell counting assays using the Neubauer plate. Prior to lipid extraction, ∼5 × 105 cells were centrifuged (13,000 g, 5 min, 4°C) and the pellets were washed two times with 1 ml ice-cold PBS. The supernatants were then discarded and the pellet was spiked with 0.4 nmol each of internal standards, including PC 17:0/14:1, PE 17:0/14:1, PG 17:0/14:1, PS 17:0/14:1, PA 17:0/14:1, PI 17:0/14:1, LPC 17:1, LPE 17:1, LPG 17:1, LPS 17:1, LPA 17:1, and LPI 17:1. The pellet was resuspended in1.3 ml MTBE/MeOH (10:3, v/v) for 1 h at room temperature in a shaker. Phase separation was induced by adding 0.25 ml of deionized H2O. After 10 min of incubation at room temperature, the sample was centrifuged at 1,000 g for 10 min. The upper (organic) phase was collected, and the lower phase was re-extracted with the upper phase of a solvent mixture (1.55 ml), whose composition was equivalent to the expected composition of the upper phase (obtained by mixing MTBE-MeOH-H2O (10:3:2.5, v/v/v) and collecting the upper phase. Combined organic phases were dried in a vacuum centrifuge.
For the liver sample, a powder sample (approximately 25 mg) from an individual liver sample of a mouse was weighed and further homogenized in 0.5 ml of ice-cold diluted (0.1×) PBS with a Branson Sonifier S-450 digital ultrasonic cell disruptor/homogenizer. A protein assay on each individual homogenate was performed with a Pierce BCA protein assay kit (Pierce, Rockford, IL).Then, a similar procedure was employed for the lipid extraction from liver samples (∼0.5 mg protein).
Derivatization and stable isotope labeling of lipids
A washing step was performed before derivatization. Briefly, the extracted lipids were resuspended in 682.5 μl MTBE/MeOH (MeOD, in the case of isotopic labeling)/2 N HCl (DCl, in the case of isotopic labeling) (500:150:32.5, v/v/v). Phase separation was induced by adding 250 μl of 0.1 N HCl (DCl, in the case of isotopic labeling). This step was completed within 5 min. The upper organic phase was immediately transferred to a clean tube and rewashed with 500 μl of the lower phase from a two-phase solvent system (MTBE/MeOH or MeOD/0.01 N HCl or DCl, 100:30:25, v/v). After a brief vortex and centrifugation (6,500 g, 2 min at 4°C), the upper phase (lipid extracts) was transferred into another clean tube for derivatization.
Fifty microliters of 2 M TMS-diazomethane in hexane was added into the lipid extract (∼0.5 ml) and incubated for 20 min at room temperature. This reaction was terminated by adding 3 μl glacial acetic acid, with a visible color change. In the case of labeling, the sample solution was washed twice with 500 μl of the lower phase of the two-phase solvent system (MTBE/MeOH/H2O, 100:30:25, v/v). After centrifugation (1,500 g, 3 min), the upper phase was collected and dried in a SpeedVac and redissolved in 80 μl chloroform/MeOH (1:1, v/v) with 5 mM ammonium acetate before mass spectrometric analysis.
Mass spectrometric analysis
Mass spectrometric analyses were performed on the TSQ Vantages instrument (Thermo Fisher Scientific, Bremen), equipped with a robotic nanoflow ion source, Triversa Nanomate® (Advion Biosciences), using chips with 5.5 μm spraying nozzles. The ion source was controlled using the ChipSoftTM software (Advion Biosciences), with the following settings: ionization voltage was set to 1.25 kV, gas backpressure to 0.5 psi in the positive ion mode, 1 μl air gap before chip, and prewetting one time was enabled and vent headspace was disabled. The ion transfer capillary temperature was set to 190°C and s-lens RF amplitude was set to 217V. All MS or MS/MS analyses were operated under Xcalibur software. The Q1 mass range was set to m/z 400–1,200. Precursor ion scan (PIS) or neutral loss scan (NLS) analysis was carried out at a CE of 40 eV, using a dwelling time of 500 ms at a step size of 0.2 Da at unit resolution of the Q1 quadrupole. A 2 min period of signal averaging from 0.5 s per scan was employed for each PIS or NLS spectrum.
Quantitative monitoring of changes in lipid profile
For relative quantitation of PLs, the peak intensities of unlabeled lipid molecular species were extracted and ratiometrically compared with the corresponding labeled ones as Clabeled/Cunlabeled = Ilabeled/Iunlabeled, where Clabeled and Cunlabeled represent the contents of individual species in the labeled and unlabeled lipid extracts, respectively, while Ilabeled and Iunlabeled represent the height of the peak of the labeled and unlabeled PL species after 13C correction, respectively. The results are presented as mean ± SD.
Due to the overlapping nature of the pseudomolecular ion peak of the species of interest with the M+2 isotope peak from another more unsaturated species that has a 2 Da lower mass (e.g., the 13C isotope effect of PC 16:0/18:2 on PC 16:0/18:1), a 13C correction was required for quantification of the PLs. Moreover, due to a mass difference of 2 Da between the 1H- and D-labeled isotopic pairs for PL species, as well as their LPL counterparts, there would be an overlap between the unlabeled ion peak of the species of interest with the M+2 isotope peak and the labeled one (e.g., the 13C isotope effect of unlabeled PE 16:0/18:0 on labeled PE 16:0/18:0). Therefore, a two-step 13C isotope correction was carried out for quantification of the PLs. Briefly, for the quantification of the unlabeled species, the isobaric overlap can be corrected by multiplying the monoisotopic peak intensity by a factor as follows (15): Z = 1-(IM-2/IM)0.0112m(m-1)/2, where Z is a 13C isotope correction factor, m is the total carbon number in the molecular species with lower molecular mass, and IM-2 and IM are peak intensities of ions at m/z of M-2 and M, respectively.
For the quantification of the labeled species, a 13C isotope correction was carried out by multiplying the monoisotopic peak intensity by a modified factor as follows: Z1 = 1-[(IlM-2 + IunlM)/IlM]0.0112m(m-1)/2, where Z1 is a modified 13C isotope correction factor, m is the total carbon number of the molecular species with lower molecular mass, IlM-2, IunlM, and IlM are peak intensities of ions at m/z of labeled (M-2), unlabeled M, and labeled M, respectively.
Safety considerations
The TMS-diazomethane is considered a less toxic alternative to diazomethane, but it should still be handled with care, as inhalation of TMS-diazomethane may cause lung damage or central nervous system depression. Therefore, TMS-diazomethane was handled with the appropriate safety procedures in place, i.e., inside a fume hood, with adequate personal safety equipment. Excess TMS-diazomethane is to be neutralized using acetic acid. Care must be taken if handled in large volumes, as it is highly volatile and produces nitrogen gas.
RESULTS AND DISCUSSION
Phosphate methylation enables simultaneous detection of PLs and LPLs under positive ionization mode
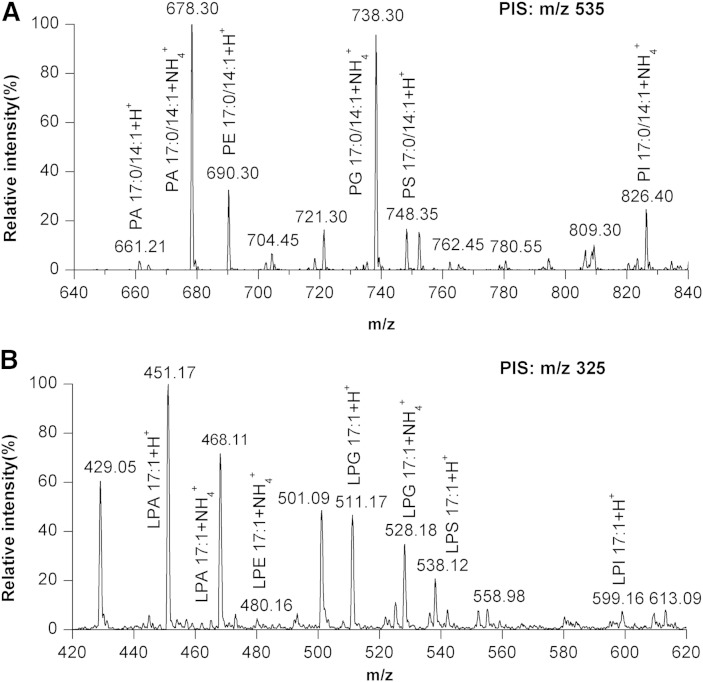
To test whether phosphate methylation by TMS-diazomethane was feasible for the detection of the six major PL classes, as well as their LPL counterparts, a standard mixture containing 1.2 nmol each of 17:0/14:1-PC, -PE, -PS, -PA, -PG, and -PI, as well as 17:1-LPC, -LPE, -LPS, -LPA, -LPG, and -LPI, was subjected to derivatization by TMS-diazomethane as described in the Experimental section (see scheme in supplementary Fig. 1). Our results show that phosphate methylation enables detection of the six major PL classes, as well as their LPL counterparts, under positive ionization mode (Fig. 1). However, the PA, PG, and PI classes preferred to be ionized by forming as ammonium adducts, whereas their LPL counterparts tended to be detected in the form of H+ adducts, though they also can be adducted with ammonium ions. Importantly, the PA, PG, and PI species can be detected readily with high signal response after derivatization, which facilitates the detection of these PLs under positive mode.
Fig. 1.
Detection of the major PL classes, as well as LPLs, under positive ionization mode. A: 17:0/14:1-PA, -PE, -PG, -PS, and -PI were simultaneously detected by PIS at m/z 535 that corresponds to [DG 17:0/14:1+H-H2O]+. B: 17:1-LPA, -LPE, -LPG, -LPS, and -LPI were simultaneously detected by PIS at m/z 325 that corresponds to the [MG 17:1+H-H2O]+.
To examine the overall derivatization efficiency, we prepared a PL and LPL mixture containing the six major PL classes and their LPL counterparts from liver tissue. This mixture was divided into two parts and one of the two parts was subjected to derivatization. After derivatization, the derivatized solution and the one that was not derivatized were analyzed by ESI-MS/MS, respectively. The derivatization efficiency was found to be good enough (more than 93%) for the PC class and significant enhancement in the signal intensity of the PC species was also achieved by derivatization (supplementary Fig. 2). Similar results for other PL classes were obtained from the MS analysis (data not shown). Importantly, the mole percent of the different PC species calculated from the mass spectra obtained by PIS at m/z 184 or 198, respectively, is virtually equal within experimental errors after correction for differential 13C distribution. These results confirmed that the derivatization efficiency was effective enough and not affected by the FA chain length or saturation. Moreover, it also suggested that the ionization efficiencies of individual derivatized molecular species of PLs or LPLs are similar within each class. In addition, our results also reveal that most of the SMs (approximately 80%) can be successfully derivatized by our protocol (supplementary Fig. 3). The derivatized SMs tend to be ionized as proton adducts ([M+H]+) rather than lithium adducts, even in the presence of a high concentration of LiOH in the analyzing solution. Thus, the NLS of 217(213+CH2), corresponding to methylated phosphocholine plus methyl aldehyde from lithiated SMs, is unsuitable to be used for the characterization of the individual derivatized SMs. Instead, the PIS of m/z 198 was employed for the profiling of SMs. Therefore, caution should be required in the calculation of PC abundance, as the isotopes of SMs would interfere with it. However, the signal intensity of the PC species is much larger than that of the SM species after derivatization (supplementary Fig. 3C). Therefore, such interference would be negligible, in practice, or can be corrected by 13C isotopic correction, as described previously.
Characterization of PL and LPL species by ESI-MS/MS
After derivatization, the PL and LPL species were subjected to ESI-MS/MS analysis in the product-ion mode. MS/MS analysis of protonated or ammonium adducted methylated PL species revealed that PL species yielded a fragment ion corresponding to [DG+H-H2O]+. For example, collision-induced dissociation of the methylated PA 17:0/14:1 (m/z 678, ammonium adducted), PC 17:0/14:1 (m/z 718, protonated), PG 17:0/14:1(m/z 738, ammonium adducted), and PS 17:0/14:1 (m/z 748, protonated) can lead to the detection of the same fragment ion at m/z 535, which corresponds to [DG 17:0/14:1+H-H2O]+ (supplementary Fig. 4). In other words, the [DG+H-H2O]+ fragment ion corresponded to the neutral loss of 143 Da (126 Da+NH3), 193 Da, 203 Da (186+NH3), and 213 Da from the polar group of the PA, PC, PG, and PS species in protonated or ammonium adducted form, respectively. These observations indicate that NLS of 143 Da, 193 Da, 203 Da, and 213 Da from the derivatized PA, PC, PG, and PS species could be used for identification and quantification of PA, PC, PG, and PS species after derivatization, respectively. The PE 17:0/14:1 (m/z 690, protonated) or PI 17:0/14:1 (m/z 826, ammonium adducted) species also produced the same fragment ion at m/z 535, thus NLS of 155 Da and 291 Da (274 Da+NH3) were employed for the identification and quantitation of PE and PI species after derivatization, respectively. However, the predominant fragment ion peak in the MS/MS spectrum of PC 17:0/14:1 is not the [DG 17:0/14:1+H-H2O]+, but the m/z 198, which corresponded to the headgroup of methylated phosphocholine (supplementary Fig. 4B). Therefore, PIS of m/z 198 was chosen for the identification and quantification of PC species after derivatization. In a similar way, the 17:1-LPE, -LPA, -LPG, -LPS, and -LPI produced the predominant fragment ion at m/z 325, corresponding to the [MG 17:1+H-H2O]+ (supplementary Fig. 5), and LPC 17:1 produced prime fragment ion at m/z 198 (data not shown). However, as mentioned above, the LPL counterparts of PA, PG, and PI favored the formation of protonated ions. Therefore, the NLS of 126 Da, 155 Da, 186 Da, 213 Da, and 274 Da were used finally for the identification and quantification of derivatized LPA, LPE, LPG, LPS, and LPI species.
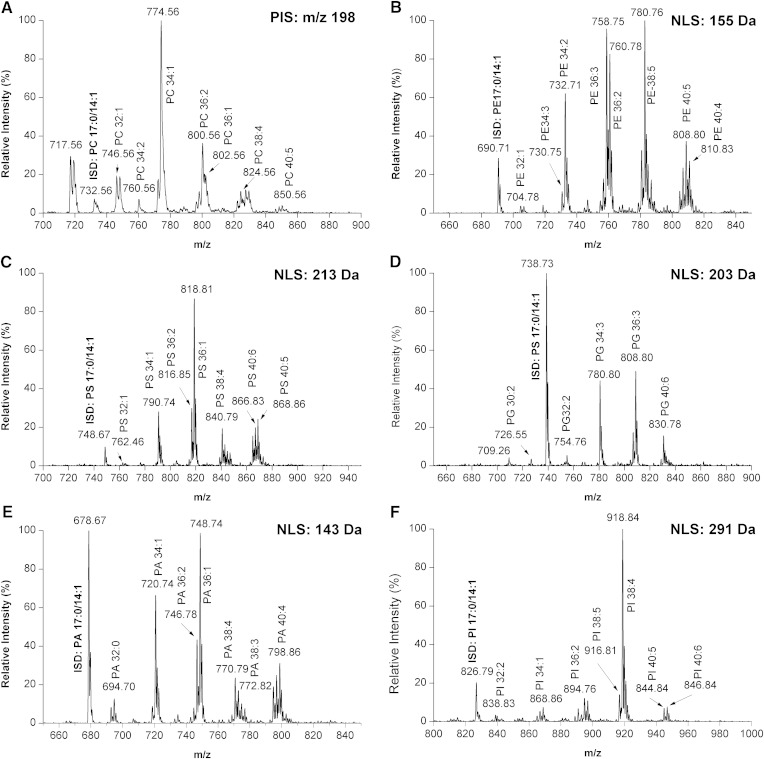
To test whether NLS or PIS was feasible for the identification and quantitation of PLs and LPLs from biological samples, we prepared a cellular PL mixture extracted from a PC3 prostatic cancer cell line spiked with ISDs of PLs and LPLs. This mixture was derivatized, and the derivatized solution was analyzed by the NLS- or PIS-based ESI-MS/MS mentioned above. Our results showed that phosphate methylation enables rapid detection of the six major PLs (Fig. 2) and their LPL counterparts (supplementary Fig. 6) under positive ionization mode. Importantly, the ISDs for the PLs were readily detected under our conditions. In addition, as mentioned above, the derivatization efficiency and ionization efficiency were not affected by the FA chain length or saturation of individual molecular species. Therefore, the different derivatized PL and LPL species can be quantified by PIS- or NLS-based ESI-MS/MS with one corresponding ISD after correction for differential 13C isotope distribution and under a diluted lipid concentration to avoid lipid aggregation, as previously described (15).
Fig. 2.
Profiling of the six major PLs by PIS or NLS under positive ionization mode. A: PIS of m/z 198 for the identification and quantification of PC species after derivatization. B–F: NLS of 143 Da, 155 Da, 203 Da, 213 Da, and 291 Da were chosen for identification and quantification of PA, PE, PG, PS, and PI species after derivatization.
Isotopic labeling of PLs through phosphate methylation by CD2N2 generated in situ
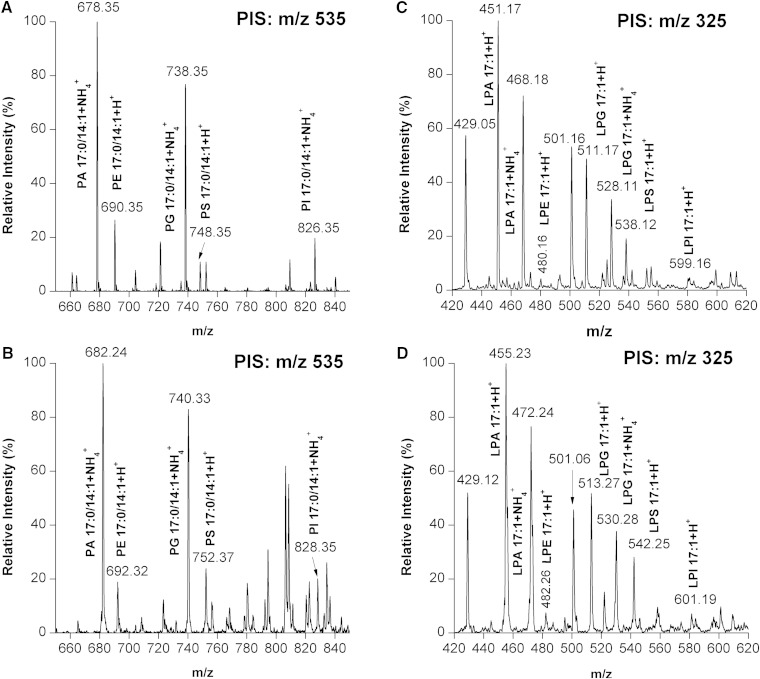
We tested and optimized the strategy based on phosphate methylation and isotopic labeling for the relative quantitation of PLs and their LPL counterparts. It is reasonable to assume that when CD2N2 is utilized instead of a light reagent, a mass increase should be detected by the mass analyzer. Such an increase should be directly proportional to the amount of available phosphate or carboxylic acid groups in the PL species (supplementary Fig. 7). It means that the larger the number of phosphate or carboxylic acid groups in a PL species, the larger the mass difference between the 1H- and the D-labeled isotopic pair that can be identified. To test our hypothesis, a PL and LPL standard mixture, containing 17:0/14:1-PC, -PE, -PS, -PA, -PG, and -PI, as well as 17:1-LPC, -LPE, -LPA, -LPG, and -LPI, was subjected to our stable isotopic labeling analysis. A rapid and efficient phosphate methylation and isotopic labeling of PE, PG, PS, PA, or PI was achieved (Fig. 3). The mass difference between the 1H- and the D-labeled isotopic pairs of PE, PG, PS, PA, and PI were 2, 2, 4, 4, and 2 Da, respectively. No unlabeled PLs were detected in the isotopically labeled samples, demonstrating that the process of generating CD2N2 and phosphate methylation of PLs was achieved close to completion. These values matched the theoretical values calculated from the number of available phosphate or carboxylic acid groups in each PL class. For example, the PE species has only one phosphate group, whereas the PS species has one phosphate group and one carboxylic acid group. Therefore, PE and PS can be labeled with one and two -CHD2 groups, respectively, which leads to a mass difference between the 1H- and the D-labeled isotopic pairs of PtdEtn and PtdSer with 2 and 4 Da, respectively. Similarly, rapid and efficient phosphate methylation and isotopic labeling of LPLs were achieved and the mass differences between the 1H- and the D-labeled isotopic pairs were matched to the theoretical values (Fig. 3).
Fig. 3.
Labeling efficiency of PLs and LPLs by diazomethane generated in situ. A, B: The 1H- and D-labeled 17:0/14:1-PE, -PG, -PS, -PA, and -PI were detected by PIS at m/z 535 that corresponds to [DG 17:0/14:1+H-H2O]+. C, D: The 1H- and D-labeled 17:1-LPA, -LPE, -LPG, -LPS, and -LPI were detected by PIS at m/z 325 that corresponds to the [MG 17:1+H-H2O]+. No unlabeled PLs and LPLs were detected in the isotopically labeled samples, demonstrating the labeling was close to complete.
However, similar to our previous study, we found that the isotopic cluster of D-methylated PLs and LPLs shows the presence of one less abundant peak that is 1 mass unit smaller than the monoisotopic peak for the D-methylated PLs and LPLs. This is partly due to a small percentage of 1H atoms in the D-marked MeOD reagent used in our experiments, as well as due to the effects of back-exchange during the generation of D-diazomethane. To minimize such effects, a prederivatization wash, as described in the Experimental section, was employed, which could reduce the concentration of hydrogen atoms in the environment. However, this could lead to the production of a small proportion of -CD3 group in the resulting methyl ester due to the H/D exchange of acidic protons from the phosphate group with deuterium atoms in the environment. Previous studies had shown that two of the protons in the resulting methyl ester are derived from the diazomethane and the other one is the “donated” acidic proton from the carboxylic acid (phosphate, in our case). Therefore, the final concentration of the DCl in the prederivatization wash solution was controlled to be less than 0.1 N and the process was completed within 5 min. Under such optimal conditions, minimal peaks that were 1 mass unit smaller or larger than the monoisotopic peak for the D-methylated PL species and their LPL counterparts were found (Fig. 3C). The concentration of DCl in derivative reagents and the reaction time were also investigated to optimize the derivatization of PL species and their LPL counterparts. The PL and LPL derivatives from the various reaction conditions showed no significant differences from the ESI-MS or MS/MS spectra (data not shown).
Taken together, our results show that the use of TMS-diazomethane, with MeOD/DCl as a D reservoir, is feasible for isotopic labeling of PL and LPL species, which could then be employed for relative quantitation of PL and LPL species. In addition, our results also indicate that isotopic labeling enables simultaneous fractionation of PL species from two different samples to reduce the sample complexity, as the isotopic pairs were coeluted from the column. For instance, the direct detection of PLs from the lipid droplet sample by direct infusion ESI-MS was hindered by the overabundant amount of TGs in the sample (supplementary Fig. 8A). However, the PLs were readily detected after prefractionation by a hydrophilic interaction liquid chromatography-based chromatography, which separates the PLs from the TGs in the lipid droplet sample (supplementary Fig. 8B). Moreover, the isotopic pair of PCs was coeluted from the column and the profiles of unlabeled or labeled PLs were not affected by the prefractionation.
Relative quantitation of PL and LPL species by ESI-MS/MS
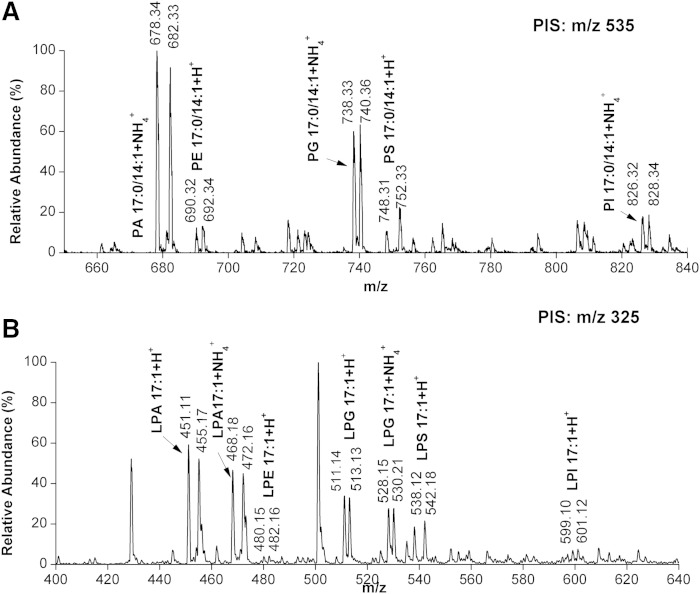
To characterize the accuracy of relative quantitation by phosphate methylation with CD2N2, the PL and LPL standards mixture was divided into two aliquots and separately labeled with either 1H- or D-labeled diazomethane, as described in the Experimental section. The two product solutions were subsequently mixed to simulate relative quantification. This mixture was subjected to [DG+H-H2O]+-specific PIS-based ESI-MS/MS analysis. The peak intensities of unlabeled PL or LPL standards were extracted from the [DG+H-H2O]+-specific PIS spectrum and compared ratiometrically to the corresponding labeled ones for the relative quantitation of labeled and unlabeled lipid species. The signal intensity of the isotopic pair of the PL and LPL standards was found to be in good agreement with the theoretically expected ratios (Fig. 4). The ratios of peak intensities were calculated for each pair of heavy- and light-labeled PL or LPL standards after 13C correction from four runs and the average ratios are displayed in Table 1. The average experimental ratios for the PL and LPL standards were found to be in good agreement with the theoretically expected ratios, with an average coefficient of variation of 9.1%.
Fig. 4.
PIS spectra of the 1H- and D-labeled PL and LPL standards at 1:1 ratios. A: The 1H- and D-labeled 17:0/14:1-PE, -PG, -PS, -PA, and -PI at 1:1 ratios were detected by PIS at m/z 535 that corresponds to [DG 17:0/14:1+H-H2O]+. B: The 1H- and D-labeled 17:1-LPA, -LPE, -LPG, -LPS, and -LPI at 1:1 ratios were detected by PIS at m/z 325 that corresponds to the [MG 17:1+H-H2O]+. The signal intensity of the isotopic pair of the PL and LPL standards was found to be in good agreement with the theoretically expected ratios.
TABLE 1.
Consistency and variability of the isotopic labeling procedure
| Structure | Meastured H/D Ratiosa | Standard Deviation | Coefficient of Variation (%) |
| PC 17:0/14:1 | 1.12 | 0.109 | 8.8 |
| PE 17:0/14:1 | 0.94 | 0.120 | 12.7 |
| PS 17:0/14:1 | 1.02 | 0.061 | 6.0 |
| PG 17:0/14:1 | 0.99 | 0.126 | 12.6 |
| PA 17:0/14:1 | 1.02 | 0.101 | 9.9 |
| PI 17:0/14:1 | 1.06 | 0.148 | 13.9 |
| LPC 17:1 | 1.09 | 0.116 | 10.6 |
| LPE 17:1 | 0.90 | 0.096 | 10.6 |
| LPS 17:1 | 1.09 | 0.065 | 6.0 |
| LPG 17:1 | 0.99 | 0.042 | 4.2 |
| LPA 17:1 | 1.13 | 0.033 | 2.9 |
| LPI 17:1 | 1.07 | 0.119 | 11.2 |
| Average | 1.04 | — | 9.1 |
PL and LPL mixtures were prepared with an expected proportion of H/D-methylated PLs and LPLs at 1:1. The H/D ratios were measured and compared with the theoretical values.
Averaged from three replicates.
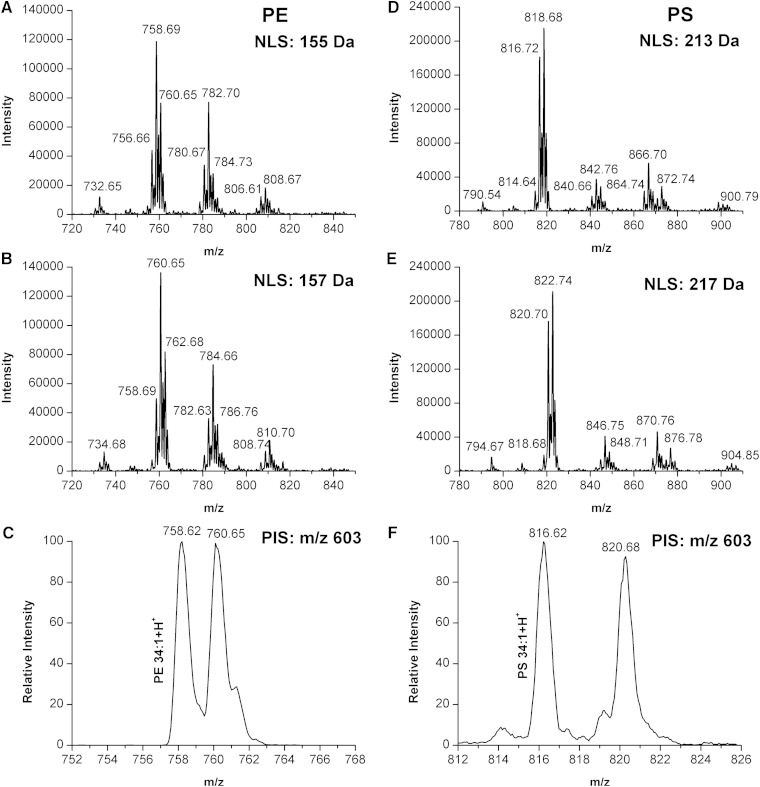
The aim of our experiments was to develop a rapid procedure to measure differences and compare the relative amounts of specific PL species between two populations of PLs extracted from two different samples utilizing stable isotopic labeling. In practice, these two samples could be obtained, for example, from cells, tissues, or body fluid at two different conditions. To further characterize the analytical properties and the accuracy of relative quantitation of PL species from real samples by phosphate methylation with CD2N2, the prostatic cancer cell cultures were divided into several aliquots before PL extraction and separated into the following proportions of cell volume: 3:1, 1:1, and 1:3 and separate labeling with either 1H- or D-labeled diazomethane. The mixture of 1H- or D-labeled PLs at different ratios was subjected to NLS- or PIS-based ESI-MS/MS analysis and the distribution of the H/D ratios was examined. Our results showed that the observed H/D ratios were consistent with the theoretical ratios of PC, PE, PS, PG, PI, and PA present in the mixture. Example mass spectra are shown for PE and PS populations, one with a ratio at 1:1 (Fig. 5) and the other with a ratio at 1:3 (supplementary Fig. 9). These spectra illustrate that the difference in signal intensity of the PE or PS species between the heavy and light forms was found to be in good agreement with the theoretically expected ratios.
Fig. 5.
NLS- or PIS-based ESI-MS/MS analysis for the mixture of 1H- and D-labeled PLs at 1:1 ratios. A: NLS of 155 Da for identification and quantification of 1H-labeled PE species from the mixture of 1H- and D-labeled PLs at 1:1 ratios. B: NLS of 157 Da for identification and quantification of D-labeled PE species from the mixture of 1H- and D-labeled PLs at 1:1 ratios. C: PIS of m/z 603 for identification and quantification of 1H- and D-labeled PE 34:1 from the mixture of 1H- and D-labeled PLs at 1:1 ratios. D: NLS of 213 Da for identification and quantification of 1H-labeled PS species from the mixture of 1H- and D-labeled PLs at 1:1 ratios. E: NLS of 217 Da for identification and quantification of 1D-labeled PS species from the mixture of 1H- and D-labeled PLs at 1:1 ratios. F: PIS of m/z 603 for identification and quantification of 1H- and D-labeled PS 34:1 from the mixture of 1H- and D-labeled PLs at 1:1 ratios. The expected 1:1 intensity ratios and the 2 or 4 Da shift for PE and PS, respectively, are also evident. The monoisotopic peak was employed for quantification.
Potential pitfalls in quantitation of PLs
Although high accuracy for quantitation of PLs is possible under optimal conditions, there are several situations where identification and quantitation of PLs could be compromised. First, unlike the isobaric mass tag labeling strategies mentioned above, in which MS2 space was used to reduce complexity and increase precision, our labeling strategy is not a “mass-balanced” method and can be analyzed only in MS1. Therefore, caution should be used with 13C correction for quantification of PLs and LPLs, as there is only a 2 Da difference between the 1H- and the D-labeled isotopic pairs for PC, PE, PG, and PI species, as well as their LPL counterparts. However, such potential quantitation error can be resolved by careful correction, as described in the Experimental section, or by combining our method with the appropriate chromatographic separation methods. Second, for quantification of PS species, caution is required because parts of the free hydroxyls in the head group of PG species are methylated during the phosphate methylation process (defined here as over-methylated PG), which would lead to an overlap between the D-labeled PS species and the unlabeled, but over-methylated, PGs that have the same fatty acyl composition. For instance, there would be an overlap between the D-labeled PS 31:1 (752 Da, 748+2D) and the unlabeled, but over-methylated, 31:1 PG (752 Da, 738+CH2) in the DAG+-specific PIS spectrum (Fig. 3). However, such overlap can be dissolved by addition of LiOH instead of NH4Ac, as the PG species prefer to adduct with Li+ rather than NH4+ in the presence of LiOH. Third, the plasmalogens would be hydrolyzed upon methylation due to the presence of HCl in our experimental conditions, as the vinyl ether motif within these compounds is acid labile (supplementary Fig. 10). Therefore, caution is required for profiling or quantitative applications of this protocol. However, a further optimization of our protocol, such as finding an alternative acid for HCl or decreasing the concentration of HCl, may resolve such a problem.
CONCLUSIONS
The results presented here demonstrate the feasibility of rapid profiling and relative quantitation of PLs and LPLs from biological samples on the basis of phosphate methylation and isotopic labeling by chemical derivatization with diazomethane. Our novel assay is not only rapid, simple, and sensitive, but also offers several important advantages for PL measurement. First, phosphate methylation allows fast and sensitive identification of the six major PL classes, including their LPL counterparts, under positive ionization mode from biological samples, which excludes the need to switch between positive and negative mode. Importantly, the PA, PG, and PI species can be detected readily with high signal response after derivatization, which facilitates the detection of these PLs under positive mode. Second, isotopic labeling-based quantitation of PLs also minimizes matrix effects or run-to-run differences that could potentially affect results. Moreover, isotopic labeling enables lipid fractionation to reduce sample complexity, as the isotopic pairs were coeluted from the column. Therefore, our methods would present a powerful tool for research of PL metabolism and signaling.
Supplementary Material
Footnotes
Abbreviations:
- CD2N2
- deuterated diazomethane
- DCl
- deuterium chloride
- H/D
- hydrogen/deuterium
- HCl
- hydrochloric acid
- H2O
- water
- ISD
- internal standard
- LPL
- lysophospholipid
- MeOD
- deuterium-labeled methanol
- MeOH
- methanol
- MTBE
- methyl tert-butyl ether
- NLS
- neutral loss scan
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PIS
- precursor ion scan
- PL
- phospholipid
- PS
- phosphatidylserine.
This work was supported by grants from the National Basic Research Program of China (Grants 2010CB833703, 2012CB966803, and 2011CB915501), the National Natural Science Foundation of China (Grants 90919047, 81028009, and 31100614), and Novo Nordisk-CAS Research Foundation NNCAS-2011-1. The authors declare no competing financial interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Escribá P. V., González-Ros J. M., Goñi F. M., Kinnunen P. K., Vigh L., Sánchez-Magraner L., Fernández A. M., Busquets X., Horváth I., and Barceló-Coblijn G.. 2008. Membranes: a meeting point for lipids, proteins and therapies. J. Cell. Mol. Med. 12: 829–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishizuka Y. 1992. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science. 258: 607–614. [DOI] [PubMed] [Google Scholar]
- 3.Dória M. L., Cotrim Z., Macedo B., Simoes C., Domingues P., Helguero L., and Domingues M. R.. 2012. Lipidomic approach to identify patterns in phospholipid profiles and define class differences in mammary epithelial and breast cancer cells. Breast Cancer Res. Treat. 133: 635–648. [DOI] [PubMed] [Google Scholar]
- 4.Hannun Y. A., and Obeid L. M.. 2008. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9: 139–150. [DOI] [PubMed] [Google Scholar]
- 5.Wymann M. P., and Schneiter R.. 2008. Lipid signalling in disease. Nat. Rev. Mol. Cell Biol. 9: 162–176. [DOI] [PubMed] [Google Scholar]
- 6.Fernandis A. Z., and Wenk M. R.. 2009. Lipid-based biomarkers for cancer. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2830–2835. [DOI] [PubMed] [Google Scholar]
- 7.Clark J., Anderson K. E., Juvin V., Smith T. S., Karpe F., Wakelam M. J., Stephens L. R., and Hawkins P. T.. 2011. Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat. Methods. 8: 267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wasslen K. V., Canez C. R., Lee H., Manthorpe J. M., and Smith J. C.. 2014. Trimethylation enhancement using diazomethane (TrEnDi) II: rapid in-solution concomitant quaternization of glycerophospholipid amino groups and methylation of phosphate groups via reaction with diazomethane significantly enhances sensitivity in mass spectrometry analyses via a fixed, permanent positive charge. Anal. Chem. 86: 9523–9532. [DOI] [PubMed] [Google Scholar]
- 9.Berry K. A., and Murphy R. C.. 2005. Analysis of cell membrane aminophospholipids as isotope-tagged derivatives. J. Lipid Res. 46: 1038–1046. [DOI] [PubMed] [Google Scholar]
- 10.Zemski Berry K. A., and Murphy R. C.. 2006. Analysis of polyunsaturated aminophospholipid molecular species using isotope-tagged derivatives and tandem mass spectrometry/mass spectrometry/mass spectrometry. Anal. Biochem. 349: 118–128. [DOI] [PubMed] [Google Scholar]
- 11.Zemski Berry K. A., Turner W. W., VanNieuwenhze M. S., and Murphy R. C.. 2009. Stable isotope labeled 4-(dimethylamino)benzoic acid derivatives of glycerophosphoethanolamine lipids. Anal. Chem. 81: 6633–6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nie S., Pham H. T., Blanksby S. J., and Reid G. E.. 2015. Photoinduced intermolecular cross-linking of gas phase triacylglycerol lipid ions. Eur. J. Mass Spectrom. (Chichester, Eng.). 21: 287–296. [DOI] [PubMed] [Google Scholar]
- 13.Cai T., Shu Q., Hou J., Liu P., Niu L., Guo X., Liu C. C., and Yang F.. 2015. Profiling and relative quantitation of phosphoinositides by multiple precursor ion scanning based on phosphate methylation and isotopic labeling. Anal. Chem. 87: 513–521. [DOI] [PubMed] [Google Scholar]
- 14.Matyash V., Liebisch G., Kurzchalia T. V., Shevchenko A., and Schwudke D.. 2008. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 49: 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han X., and Gross R. W.. 2005. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 24: 367–412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.