Abstract
Long-chain acyl-CoA synthetase 1 (ACSL1) converts free fatty acids into acyl-CoAs. Mouse studies have revealed that ACSL1 channels acyl-CoAs to β-oxidation, thereby reducing glucose utilization, and is required for diabetes-accelerated atherosclerosis. The role of ACSL1 in humans is unknown. We therefore examined common variants in the human ACSL1 locus by genetic association studies for fasting glucose, diabetes status, and preclinical atherosclerosis by using the MAGIC and DIAGRAM consortia; followed by analyses in participants from the Multi-Ethnic Study of Atherosclerosis, the Penn-T2D consortium, and a meta-analysis of subclinical atherosclerosis in African Americans; and finally, expression quantitative trait locus analysis and identification of DNase I hypersensitive sites (DHS). The results show that three SNPs in ACSL1 (rs7681334, rs735949, and rs4862423) are associated with fasting glucose or diabetes status in these large (>200,000 subjects) data sets. Furthermore, rs4862423 is associated with subclinical atherosclerosis and coincides with a DHS highly accessible in human heart. SNP rs735949 is in strong linkage disequilibrium with rs745805, significantly associated with ACSL1 levels in skin, suggesting tissue-specific regulatory mechanisms. This study provides evidence in humans of ACSL1 SNPs associated with fasting glucose, diabetes, and subclinical atherosclerosis and suggests links among these traits and acyl-CoA synthesis.
Keywords: endocrine disorders, fatty acid, fatty acid/metabolism, genetics, genomics
The enzyme long-chain acyl-CoA synthetase 1 (ACSL1), which catalyzes conversion of free long-chain fatty acids into their acyl-CoA derivatives, has emerged as a metabolic rheostat in mouse skeletal muscle, heart, and adipose tissue (1–3). Thus, mice deficient in ACSL1 in skeletal muscle exhibit a marked reduction in fatty acid utilization through β-oxidation and a concomitant increase in glucose utilization during fasting, and they are hypoglycemic during endurance training (4). In the heart, which relies heavily on fatty acids as an energy source, ACSL1 deficiency results in reduced fatty acid oxidation and increased glucose utilization (1, 2). Mice with adipose tissue-selective ACSL1 deficiency have reduced blood glucose levels during cold exposure (3). Furthermore, an inducible whole-body ACSL1-deficient mouse model exhibits lower blood glucose levels than matched controls (1). Thus, mouse studies have revealed a clear relationship between reduced blood glucose levels and reduced ACSL1 activity in several tissues due to metabolic flexibility in these tissues.
ACSL1 is ubiquitously expressed, with high levels in typical insulin target tissues, such as skeletal muscle, liver, and adipose tissue (5). ACSL1 is also expressed in myeloid cells, particularly in myeloid cells stimulated with inflammatory molecules, such as Toll-like receptor (TLR) 4 ligands, Gram-negative bacteria, TLR1/2 ligands, TLR3 ligands, TNF-α, and IFN-γ (6–8). Both in mouse and human macrophages, plasma membrane-associated ACSL1 levels increase after TLR4 and IFN-γ stimulation (6, 9) and also increase in human macrophages exposed to increased metabolic activation by insulin, elevated glucose, and palmitate (10). When these cells are exposed to metabolic or inflammatory activation, ACSL1 is required for effective phospholipid replenishment, neutral lipid accumulation, and early atherosclerosis in the context of diabetes (6, 7, 11). Mice deficient in ACSL1 selectively in myeloid cells or endothelial cells do not exhibit reduced blood glucose levels (6, 12).
Whereas mouse studies indicate that ACSL1 might play a critical role in maintaining glucose homeostasis and atherosclerosis, the role of ACSL1 in humans is unknown. Because of the studies discussed above, we were particularly interested in the relationship between human ACSL1 SNPs with variation in fasting glucose, diabetes, and risk of atherosclerosis. We interrogated published [Meta-Analyses of Glucose and Insulin-Related Traits Consortium (MAGIC) and Diabetes Genetics Replication and Meta-Analysis (DIAGRAM) consortium] genome-wide association study (GWAS) scans of fasting glucose (13, 14) and diabetes status (15) with respect to common [minor allele frequency (MAF) >5%] SNPs in the ACSL1 locus (4q35.1). MAGIC contains >46,000 healthy nondiabetic individuals of European descent, while DIAGRAM has >56,000 individuals of primarily European descent (>12,000 with type 2 diabetes). For the common ACSL1 SNPs identified through the MAGIC and DIAGRAM consortia, we conducted follow-up analyses of participants with and without diabetes from the Multi-Ethnic Study of Atherosclerosis (MESA) and in >200,000 subjects with and without type 2 diabetes included in the Penn-T2D consortium. The analyses in MESA allowed us to expand discoveries from the published GWAS effort to examine evidence of association a) across race/ethnic groups and b) with subclinical atherosclerosis traits, and the analysis in the Penn-T2D consortium allowed us to expand our analysis to a much larger sample size. The association between ACSL1 SNPs and coronary artery calcification (CAC) was validated in African Americans (16). Furthermore, expression quantitative trait locus (eQTL) analysis was used to investigate if the identified ACSL1 SNPs were associated with ACSL1 levels in skin, lymphocytes, and adipose tissue, and finally, DNase I hypersensitive sites (DHSs) coinciding with these SNPs were analyzed in several fetal and adult human tissues.
Together, our results demonstrate that three SNPs in the ACSL1 gene are associated with markers of fasting glucose or diabetes status and that one of these SNPs is also significantly associated with subclinical atherosclerosis. This represents the first indication of ACSL1 contributing to the regulation of fasting glucose and risk of diabetes and subclinical atherosclerosis in humans.
MATERIALS AND METHODS
Candidate association analyses from large-scale consortia
To perform association analysis of common variants within the ACSL1 gene region, we examined published GWAS and custom-array results from the MAGIC and DIAGRAM consortia. We report the SNP demonstrating the strongest association with the analyzed traits for each study.
MAGIC.
MAGIC published a GWAS of fasting glucose and fasting insulin in up to 46,196 nondiabetic participants of European descent in 2010 (13). A subsequent effort from MAGIC performed a GWAS of the same traits with adjustment for BMI in an expanded sample of up to 51,750 nondiabetic participants of European descent (14).
DIAGRAM.
The DIAGRAM consortium performed a GWAS of type 2 diabetes in 12,171 cases and 56,862 controls comprising the DIAGRAM v3 GWAS (15). The SNPs identified through GWAS contributed to the design of the Metabochip, a custom genotyping array used for genetic association analysis of type 2 diabetes in an expanded sample of (primarily European descent) 34,840 cases and 114,981 controls.
Genetic association analysis of ACSL1 variants in MAGIC and DIAGRAM.
We focused on ACSL1 variants within 50 kb of the ACSL1 gene, using LocusZoom (17) to identify SNPs within the region from the published GWAS and custom genotyping array results. We then conducted candidate association analyses for each of the four selected published data sets, the original MAGIC GWAS (13), the MAGIC BMI-adjusted GWAS (14), the DIAGRAM v3 GWAS (15), and the DIAGRAM Metabochip (15). For each lookup, we applied a Bonferroni correction for the number of SNPs identified within the target region. Here, we report those SNPs reaching the specified Bonferroni threshold for at least one of the published data sets.
Analysis of ACSL1 SNPs in MESA
MESA.
The MESA is a longitudinal cohort study of subclinical cardiovascular disease and risk factors in 6,814 men and women 45 to 84 years of age, initially free of overt disease, that predict progression to disease or progression of subclinical disease (18). The MESA Family Study (MESAFS) recruited 1,595 African American and Hispanic family members 45 to 84 years of age specifically for genetic analysis, and the MESA Air Pollution Study recruited an additional 257 participants (19). MESA participants had detailed medical histories and examinations for anthropometry, blood pressure, and vascular imaging [to obtain measures of subclinical atherosclerosis of CAC and common (cIMT) and internal carotid intima-media thickness (iIMT)] (20). Glucose concentrations were determined from fasting samples, and diabetes status was defined using the 2003 American Diabetes Association fasting criteria (fasting glucose ≥126 mg/dl) (21) and/or diabetes treatment. Characterization of MESA subjects included in the present study is included in the supplementary Materials and supplementary Tables 1 and 2. MESA participants were genotyped using the Affymetrix Human SNP array 6.0 (22). Imputation was conducted with IMPUTE v2 using Phase 1 v3 of the cosmopolitan 1,000 genomes reference set (23), followed by robust quality control (24).
Genetic association analysis in MESA.
MESA participants were stratified by race/ethnic group using an unrelated subset of individuals from MESA and from MESAFS, by selecting, at most, one individual from each pedigree, based on inferred relationships in KING (25). Linear regression of quantitative phenotypes or logistic regression of dichotomous phenotypes was performed under an additive 1 d.f. genetic dosage model in R. For phenotypes with a substantial familial component, we performed analysis using linear mixed-effects models for quantitative traits, or generalized estimating equations for dichotomous traits, to account for familial relationships as implemented in the package R/GWAF (26). A basic model including age, gender, study site, and principal components of ancestry was used. Fixed effect meta-analysis was performed to combine race/ethnic specific results using METAL (27).
For each of the three SNPs identified in consortium studies (Table 1), primary analyses in MESA focused on the phenotype analyzed in discovery of each SNP, including glucose (log-transformed) and diabetes status. Secondary analysis examined subclinical atherosclerosis (cIMT, iIMT, CAC presence/absence, and Agatston score for those with CAC >0).
TABLE 1.
Summary of SNPs identified in the ACSL1 gene region from MAGIC and DIAGRAM
| Data | Trait | Number of ACSL1 Region SNPsa | SNP ID | HG 19 Position | Effect/ Other Allele | Consortia Results | MESA Resultsb | ||||||
| EAF | Beta | SE | P | EAFc | Beta | SE | P | ||||||
| MAGIC GWAS | Fasting glucose in nondiabetics | 173 | rs7681334 | 185,710,859 | A/G | 0.455 | 0.016 | 0.004 | 8.4E-6 | 0.496/ 0.452/ 0.644/ 0.928 | 0.003 | 0.002 | 0.177 |
| MAGIC BMI-adjusted GWAS | Fasting glucose in nondiabetics | 183 | rs4862423 | 185,726,548 | T/C | 0.350 | 0.017 | 0.003 | 4.6E-7 | 0.407/ 0.268/ 0.502/ 0.661 | 0.004 | 0.002 | 0.037 |
| Data | Trait | Number of SNPsa | SNP ID | HG 19 Position | Effect/ Other Allele | EAFd | Odds Ratio | 95% CI | P | EAFc | Odds Ratio | 95% CI | P |
| DIAGRAM v3 GWAS | Diabetes status | 173 | rs735949 | 185,716,232 | T/C | 0.825 | 1.11 | (1.05, 1.17) | 7.8E-5 | 0.853/ 0.948/ 0.909/ 0.9993 | 1.04 | (0.870, 1.25) | 0.716 |
| DIAGRAM Metabochip | Diabetes status | 9 | 1.09 | (1.05, 1.12) | 3.7E-6 |
CEU, northern Caucasian Europeans (from Utah); CI, confidence interval; EAF, effect allele frequency; HGQ1219, human genome version 19.
The ACSL1 gene region was defined as the genomic region containing the ACSL1 gene itself ±50 kb.
Results for MESA are presented based on combined meta-analysis across race/ethnic groups.
EAF for MESA shown separately for Caucasian/African American/Hispanic/Chinese.
EAF reported for HapMap CEU because this information is not included in the DIAGRAM summary files.
Penn-T2D consortium.
The Penn-T2D consortium used publicly available DIAGRAM meta-analyses data, updated by adding additional T2D GWAS/Metabochip studies that have not previously contributed data to the DIAGRAM consortium (findings from these analyses are currently submitted for publication). The Penn-T2D consortium consists of data from 56,241 type 2 diabetes cases and 187,815 controls.
Replication and validation in MESA and Penn-T2D for SNPs identified in consortia studies
For each of the three ACSL1 SNPs identified in MAGIC and DIAGRAM associated with fasting glucose and diabetes (Table 1), analyses were focused on replicating and validating association with the corresponding trait analyzed in discovery of each SNP in MESA and the Penn-T2D consortium. Statistical significance for replication of the three SNPs was determined at a Bonferroni-corrected threshold of α= 0.05/3 = 0.017, whereas α= 0.05 was considered as nominal evidence of replication. For secondary analysis of subclinical atherosclerosis traits in MESA, we report findings as significant if they reach a Bonferroni-corrected threshold of α= 0.05/12 = 0.0042, corresponding to three SNPs and four traits, or 12 tests.
Meta-analysis of CAC in African Americans
This study amassed data on 5,823 African Americans from eight US studies in CAC, examining various definitions of CAC: ln[CAC+1]; CAC present/absent; ln(CAC) where CAC >0; and BLOM transformed CAC, defined by Gomez et al. (28). Results from the eight studies were meta-analyzed in METAL (16). For this validation study, we used the results from subjects with CAC >0 on 2,520 subjects. This cohort included African Americans from MESA (29% of subjects). Statistical significance for replication of the three SNPs was determined at a threshold of α= 0.05/3 = 0.017 after Bonferroni correction.
CARDIoGRAMplusC4D consortium
In order to investigate association of the three identified ACSL1 SNPs with cardiovascular events (myocardial infarction, symptomatic coronary events, or coronary stenosis), we took advantage of the CARDIoGRAMplusC4D consortium. CARDIoGRAM GWAS is a meta-analysis of 22 GWASs in participants of European descent involving 22,233 cases and 64,762 controls, and C4D GWAS is a meta-analysis of GWASs in participants of European and South Asian descent involving 15,420 coronary heart disease cases and 15,062 controls. GWAS data from both of these consortia contain nonoverlapping participants (29). We used this publicly available data and conducted a fixed-effects meta-analysis yielding a sample size of 37,653 coronary heart disease cases and 80,182 controls.
Multiple Tissue Human Expression Resource analysis
The Multiple Tissue Human Expression Resource (MuTHER) consortium data (http://www.muther.ac.uk) were used to investigate if the three identified ACSL1 SNPs were associated with ACSL1 levels, representing an eQTL analysis. MuTHER contains genome-wide expression (Illumina HT-12v3 array) profiles on lymphocytes, subcutaneous fat, and skin biopsies from ∼856 twins from the TwinsUK BioResource. MuTHER was interrogated for changes in probe ILMN_1684585 (ACSL1).
Identification of chromatin accessibility and transcription factor binding motifs
Chromatin accessibility data were obtained from public repositories for human cell lines [ENCODE Consortium (30)] as well as fetal and adult tissues and primary cell lines [Roadmap Epigenomics Consortium (31) and unpublished observations] and intersected with the three SNPs examined by this work. Transcription factor binding motif locations were determined as described by Vierstra et al. (32).
Human study approvals
Each study obtained approval from their respective institutional review board and the ethics committee of each participating institution, including the University of Alabama at Birmingham, Washington University, University of Mississippi Medical Center, University of Minnesota, Northwestern University, Kaiser Permanente (Oakland, CA), University of Washington, Columbia University, Johns Hopkins School of Medicine, University of California Los Angeles School of Medicine, Wake Forest University School of Medicine, University of Michigan Health Sciences and Behavioral Sciences, the Cedars-Sinai Medical Center, the University of Virginia, and the University of Pennsylvania, and informed consent from participants. All methodology was compliant with the principles set forth in the Declaration of Helsinki and Title 45, US Code of Federal Regulations, Protection of Human Subjects.
RESULTS
ACSL1 association analysis in published results from MAGIC
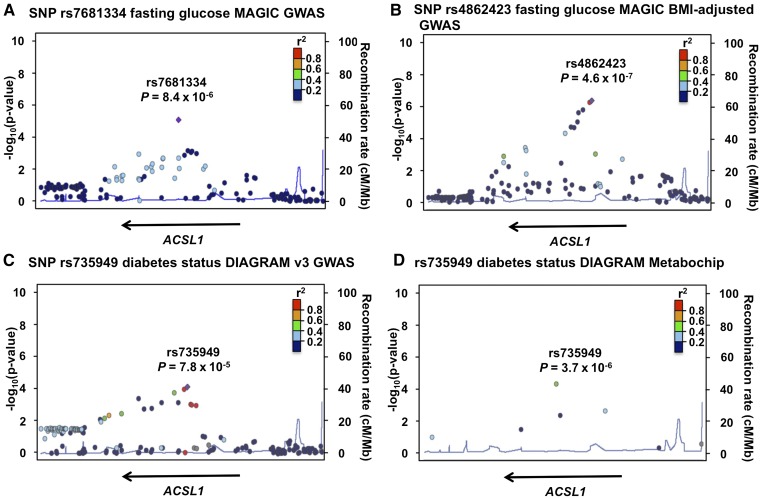
After Bonferroni correction (significance threshold α= 0.05/173 = 2.9 × 10−4) for 173 SNPs within ACSL1 in the MAGIC GWAS, we identified rs7681334 as significantly associated with fasting glucose (MAF = 0.40, P = 8.4 × 10−6; Fig. 1A and Table 1). After Bonferroni correction (significance threshold of α= 0.05/183 = 2.7 × 10−4) for 183 SNPs within ACSL1 in the MAGIC BMI-adjusted GWAS, we identified rs4862423 in addition to rs7681334 as significantly associated with fasting glucose (MAF = 0.43, P = 4.6 × 10−7; Fig. 1B and Table 1).
Fig. 1.
Regional association plots for statistically significant SNPs identified through candidate association studies in ACSL1 in published results from genetic consortia. SNP rs7681334 association with fasting glucose in the original MAGIC GWAS (A), SNP rs4862423 association with fasting glucose in the MAGIC BMI-adjusted GWAS (B), SNP rs735949 association with diabetes status in DIAGRAM v3 GWAS (C), and SNP rs735949 association with diabetes status in DIAGRAM Metabochip (D). The plots are generated using LocusZoom with 1000 Genomes CEU as the reference for calculating linkage disequilibrium.
ACSL1 association analysis in published results from DIAGRAM
We identified rs735949 as significantly associated with diabetes status (MAF = 0.06, P = 7.8 × 10−5) after Bonferroni correction for 173 ACSL1 SNPs in the DIAGRAM v3 GWAS. We also identified rs735949 as significantly associated with diabetes status (MAF = 0.06, P = 3.7 × 10−6) after correction for 9 ACSL1 SNPs in the DIAGRAM Metabochip data (Fig. 1C, D and Table 1).
Replication of ACSL1 results identified from consortia studies in MESA
In combined meta-analysis across race/ethnic groups for nondiabetic participants from MESA, we identified nominal (but not Bonferroni-corrected) evidence of replication for the association of ACSL1 rs4862423 with BMI-adjusted fasting glucose; further, the observed direction of effect was consistent with that seen in MAGIC (Table 1; P = 0.037). The effect allele (T/C) frequency was 0.350 in MAGIC European Caucasians and 0.407 in MESA Caucasian participants. The observed effect size and strength of association for rs4862423 was most notable in MESA African Americans without diabetes compared with other race/ethnic groups (P = 0.033; supplementary Table 3). We did not observe additional race/ethnic specific associations for fasting glucose-related SNPs (supplementary Tables 3 and 4). We did not observe statistically significant evidence of replication for rs7681334 with fasting glucose or for rs735949 with diabetes status (Table 1 and supplementary Table 5). Thus, these results demonstrate that the T allele in rs4862423 (T/C) associates significantly with increased fasting blood glucose levels in MAGIC, with suggestive evidence of replication in MESA.
Replication and validation of ACSL1 SNP associations in Penn-T2D meta-analyses
In a second study [Penn-T2D meta-analyses (Penn-T2D Meta) data], which is larger than either of the two consortia used for discovery analysis and the MESA replication study, we were able to confirm the association of ACSL1 rs735949 with diabetes status. Furthermore, rs7681334, which was associated with fasting blood glucose in MAGIC (Table 1) was also significantly associated with diabetes status in the Penn-T2D Meta (Table 2). The observed direction of effect was consistent with that seen in MAGIC and DIAGRAM. ACSL1 rs4862423, which was associated with fasting glucose in the BMI-adjusted MAGIC with suggestive evidence of replication in MESA (Table 1), showed a suggestive association with diabetes status (P = 0.065) in the Penn-T2D Meta for which the direction of effect was consistent with that seen with fasting glucose in MAGIC and MESA. Thus, the association of ACSL1 rs735949 and rs7681334 with fasting glucose and diabetes status is replicated in large cohorts. The association of rs4862423 with fasting glucose was significant in the BMI-adjusted MAGIC in nondiabetic subjects, yet failed to reach significant association with diabetes status in the Penn-T2D Meta.
TABLE 2.
Summary of association with type 2 diabetes status of the three identified SNPs in the ACSL1 gene region from Penn-T2D Meta
| SNP ID | Effect Allele | Noneffect Allele | Beta | SE | P | N Case | N Control |
| rs7681334 | A | G | 0.031 | 0.009 | 6.56E-04 | 56,241 | 187,815 |
| rs735949 | T | C | 0.069 | 0.014 | 1.02E-06 | 56,241 | 187,815 |
| rs4862423 | T | C | 0.024 | 0.013 | 6.51E-02 | 20,516 | 66,774 |
ACSL1 SNP association with subclinical atherosclerosis in MESA
For subclinical atherosclerosis traits, we observed association of ACSL1 rs7681334 with CAC Agatston score (among participants with CAC >0) in combined meta-analysis across race/ethnic groups (Table 3; P = 0.003). The association was stronger in participants without diabetes (Table 3; P = 0.001). We further observed association of rs4862423 with CAC in participants without diabetes (Table 3; P = 0.002). Race/ethnic stratification provides consistent directions of effect for both of these ACSL1 SNPs for Caucasian, African American, and Hispanic participants, but not Chinese participants (supplementary Tables 6 and 7). In addition, both SNPs showed stronger effects on CAC in nondiabetics compared with those with diabetes (Cochran’s Q test for heterogeneity, rs7681334, P = 0.12; rs4862423, P = 0.04).
TABLE 3.
Summary of statistically significant genetic association results for Agatston calcium score in MESA
| Group | SNP ID | HG 19 Position | Effect/Other Allele | EAFa | N | Beta | SE | P |
| All participants | rs7681334 | 185,710,859 | A/G | 0.497/0.450/ 0.647/0.944 | 3,852 | −0.126 | 0.043 | 0.003 |
| Participants without diabetes | rs7681334 | 185,710,859 | A/G | 0.496/0.452/ 0.644/0.928 | 3,172 | −0.157 | 0.046 | 0.001 |
| Participants without diabetes | rs4862423 | 185,726,548 | T/C | 0.407/0.270/ 0.499/0.658 | 3,172 | −0.150 | 0.048 | 0.002 |
Results are presented based on the basic model of genetic association including adjustment for age, gender, study site, and principal components of ancestry.
EAF for MESA shown separately for Caucasian/African American/Hispanic/Chinese participants.
CAC results in meta-analysis of African Americans
The association of rs4862423 with CAC observed in MESA African Americans among those with CAC scores >0 was significant in the expanded sample (Table 4). However, the association of rs7681334 with CAC in MESA was not observed in the African American sample. Furthermore, an association between rs735949 and CAC was observed in the African American sample, but not in MESA (Tables 3 and 4). Thus, the association of rs4862423 with subclinical atherosclerosis was consistent between the two studies. The association of rs735949 and rs7681334 with CAC appeared to be influenced by other factors.
TABLE 4.
Summary of results on genetic association results for Agatston calcium score in meta-analysis of CAC among African Americans
| SNP ID | HG 19 Position | Effect/Other Allele | EAF | N | Beta | SE | P |
| rs4862423 | 185,726,548 | T/C | 0.2448 | 2,523 | −0.1757 | 0.0611 | 0.004 |
| rs735949 | 185,716,232 | T/C | 0.954 | 2,523 | −0.343 | 0.1322 | 0.009 |
| rs7681334 | 185,710,859 | A/G | 0.4555 | 2,523 | −0.0066 | 0.0538 | 0.912 |
Analysis of association of ACSL1 SNPs in the CARDIoGRAMplusC4D consortium
In addition to testing ACSL1 SNPs for association with diabetes and subclinical atherosclerosis, we determined whether the ACSL1 SNPs modified risk for clinical end points (coronary heart disease). In the CARDIoGRAMplusC4D consortium data, there were no significant associations of ACSL1 SNPs rs7681334, rs735949, or rs4862423 with coronary heart disease (supplementary Table 8).
Chromatin accessibility at the ACSL1 locus
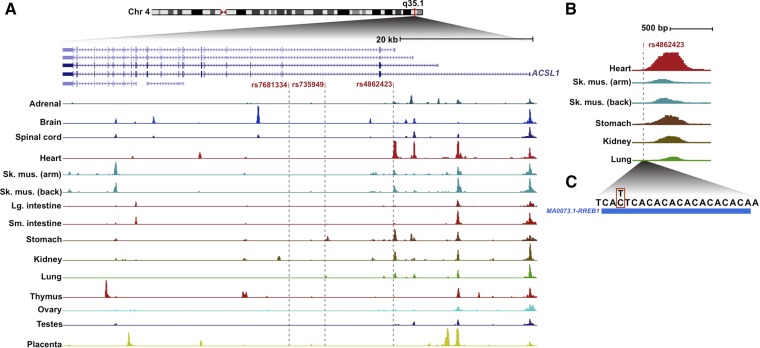
The three trait-associated ACSL1 SNPs are located within the first (rs4862423) and second (rs7681334, rs735949) introns of the ACSL1 gene. Noncoding GWAS SNPs are concentrated in regulatory DNA elements, where they may modulate transcription factor binding (33). Thus, we asked whether these ACSL1 polymorphisms coincided with DHSs, which generically indicate transcription factor binding at active regulatory elements (30). ACSL1 utilizes alternative promoters, which appear to have different levels of tissue selectivity (Fig. 2A). SNP rs4862423 is located in intron 1 of ACSL1, a region that is highly accessible to DNase I in fetal heart tissue, and to a lesser extent in other fetal tissues (Fig. 2B) as well as adult heart, mucosal (gastric and small bowel), pancreatic, and skeletal muscle tissue (data not shown). This site does not appear to be active in the hepatocyte cell line HepG2 but is accessible in multiple cultured epithelial and skeletal muscle cell lines (data not shown). Furthermore, rs4862423 coincides with a recognition site for RREB1 (ras responsive element binding protein 1; Fig. 2C). Although rs735949 coincides with a DHS in primary skin melanocytes (data not shown), we did not observe regulatory element activity coinciding with rs7681334 in any of the cell lines and tissues for which DHS data are available.
Fig. 2.
Chromatin accessibility at the ACSL1 locus. A: Normalized densities of DNase I cleavages are shown for 14 human fetal tissues, with the positions of three GWAS SNPs indicated by gray vertical lines. B: Localization of rs4862423 within a DNase I accessible region with the highest activity in fetal heart. C: Position of SNP (red box) within a binding motif of RREB1 (blue box).
eQTL analysis
ACSL1 SNPs were analyzed for evidence of eQTLs using the MuTHER consortium data on three tissues: subcutaneous fat, lymphocytes, and skin. There were no significant associations for any of the three SNPs with probe ILMN_1684585 (ACSL1). P values for rs7681334 were 0.45 in fat, 0.59 in lymphocytes, and 0.76 in skin. The corresponding P values for rs735949 were 0.13, 0.56, and 0.054, and for rs4862423, they were 0.35, 0.73, and 0.68 (supplementary Table 9). The most significant SNP associated with probe ILMN_1684585 in skin biopsies was rs745805 (P = 0.0014), which is in high linkage disequilibrium with rs735949 (r2 = 0.832 in 1000G pilot 1 CEU; supplementary Table 9). These results suggest that rs735949 might be associated with ACSL1 expression levels in some tissues.
DISCUSSION
The role of ACSL1 in humans is unknown, although mouse studies have shown that this enzyme plays an important role as a metabolic rheostat mechanism in several tissues, and hence, that loss of ACSL1 results in reduced blood glucose levels due to reduced fatty acid oxidation under conditions in which excess energy is needed. We demonstrate here that specific SNPs in the ACSL1 gene are significantly associated with fasting glucose (rs7681334 and rs4862423) or with diabetes status (rs735949) in the MAGIC and DIAGRAM consortia. We also report evidence of replication of the association of rs4862423 with fasting glucose in nondiabetic participants from MESA, and association of both rs7681334 and rs735949 with diabetes status in Penn-T2D Meta. Although the Penn-based consortium includes the DIAGRAM data, it contains twice the number of cases included in DIAGRAM. Together, these four studies make a strong case for significant association of the three ACSL1 SNPs with fasting glucose and diabetes. Furthermore, whereas MAGIC and DIAGRAM included subjects primarily of European descent, MESA includes subjects of different race/ethnic groups (mostly Caucasian, African American, and Hispanic participants and a smaller fraction of Chinese participants). The smaller sample size in MESA (7,847 participants) compared with the MAGIC and DIAGRAM consortia and Penn-T2D Meta probably contributed to the lower level of significance and statistical power. It is unlikely that the association of the ACSL1 SNPs with glucose levels is significantly confounded by BMI or adiposity because no nominally statistically significant association (defined as P < 0.05) was observed for any of the three SNPs in a large-scale GWAS meta-analysis of BMI (34).
The present study also demonstrates a significant association of rs4862423 with subclinical atherosclerosis, measured as CAC scores >0 in MESA and in a follow-up meta-analysis study in African Americans. The directionality of the association was different for fasting glucose and CAC. Thus, rs4862423 allele T was associated with increased fasting glucose and decreased CAC. The reason for the difference in directionality is unclear. Analysis of cardiovascular events in CARDIoGRAMplusC4D did not show association with the three ACSL1 SNPs. It is possible that the association occurs only with early atherosclerosis or with calcification, rather than with more advanced lesions and clinical events. In this context, it is interesting that ACSL1 in mice is required for early lesions of atherosclerosis, with a less obvious effect on advanced lesions (6).
All three ACSL1 SNPs are located in intronic regions of ACSL1 (introns 1 and 2), and our studies demonstrate that the two SNPs most clearly associated with fasting glucose and diabetes (rs7681334 and rs735949) do not associate with identified DNase I accessible regions in several cells, fetal or adult tissues most likely to be relevant for these phenotypes. However, we show that rs735949 is associated with a DNase accessible region in primary skin melanocytes. A previous study identified association of rs735949 with islet transcription factors (35). It is possible that rs7681334 and rs735949 coincide with regulatory elements active in other tissues, ages, or disease states relevant to fasting glucose and diabetes traits, or that these SNPs are in linkage disequilibrium with other regulatory variants at this locus. Indeed, rs735949 was in strong linkage disequilibrium with another SNP in the same ACSL1 intron 2, rs745805, which is significantly associated with ACSL1 expression in skin, suggesting links between rs735949 and ACSL1 expression in a tissue-selective manner. However, rs745805 is not located within a region of appreciable DNase I hypersensitivity, nor does it coincide with a transcription factor recognition sequence (data not shown). Thus, the ACSL1 SNPs investigated in regions that do not coincide with DHSs may be in linkage disequilibrium with transcription factor binding-altering SNPs within regulatory elements nearby. Furthermore, eQTL analysis did not reveal significant associations with the SNPs investigated and ACSL1 levels in fat, skin, or lymphocytes. ACSL1 expression is known to be regulated differently in different tissues (7), and the ACSL1 gene contains three alternative exon 1s controlled by different promoters (36). It can therefore not be ruled out that one or several of the SNPs are associated with ACSL1 expression levels in other tissues or in disease states. Furthermore, a long noncoding RNA, SLED1, is located in the vicinity of rs7681334 and rs735949. The putative role of SLED1 is unknown, but it is plausible that these SNPs could affect SLED1.
ACSL1 rs4862423, which is associated with fasting glucose and CAC, coincides with an ACSL1 DHS active in multiple fetal and adult tissues (including heart and pancreas) and is localized within a binding motif for RREB1. RREB1 has recently been implicated by GWAS as a regulatory factor associated with glucose levels (37). Mass spectrometry-based analysis of the human proteome (Proteomics DB) has demonstrated that heart, pancreas, lymphocytes, and colon express RREB1 (38). The role of RREB1 in regulation of blood glucose and atherosclerosis is unknown, but interestingly, RREB1 was recently demonstrated to modify histones by aiding in removal of histone H3 lysine 27 trimethylation (H3K27me3) marks, which often characterize silenced regions, thereby promoting Ucp1 and Cidea expression in brown adipocytes (39). This mechanism of action of RREB1 was shown to be due to recruitment of Jmjd3, an H3K27me3 demethylase. Thus, RREB1 binding to the motif associated with rs4862423 might alter chromatin modification and increase ACSL1 transcription in tissues that express RREB1. This hypothesis remains speculative until tissues expressing RREB1 from subjects with rs4862423 polymorphism have been examined by electrophoretic mobility shift assays for differences in RREB1 binding and ACSL1 expression. SNP rs4862423 is located in intron 1 of human ACSL1. Previous studies have identified other SNPs located in this intron as being associated with metabolic parameters in humans (40, 41): rs6552828 has been associated with gains in maximal O2 uptake after exposure to an exercise program in sedentary adults participating in the Health, Risk Factors, Exercise Training, and Genetics (HERITAGE) Family Study (40), and rs9997745 has been associated with increased metabolic syndrome risk (41). A recent study suggested that rs6552828 (located near rs4862423) might result in differences in ACSL1 expression (42). It is therefore possible that rs4862423 and other polymorphisms in intron 1 of the ACSL1 gene result in altered expression of ACSL1 in specific tissues.
In summary, our study is the largest to date examining ACSL1 SNPs in humans, and the first to examine the association between ACSL1 SNPs, fasting glucose, diabetes, and subclinical atherosclerosis. Our results demonstrate that three SNPs in the ACSL1 gene are associated with markers of fasting glucose or diabetes status and that one of these SNPs is also significantly associated with subclinical atherosclerosis. This represents the first evidence of ACSL1 association with regulation of fasting glucose and risk of diabetes and subclinical atherosclerosis in humans and suggests possible links between these traits and acyl-CoA synthesis.
Supplementary Material
Acknowledgments
We are grateful to Dr. Daniel Rader (Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA) for helpful suggestions and advice. The authors thank the participants of the MESA study, the Coordinating Center, MESA investigators, and study staff for their valuable contributions. The authors from the Candidate-Gene Association Resource (CARe) Consortium wish to acknowledge the support of the NHLBI and the contributions of the research institutions, study investigators, field staff, and study participants in creating this resource for biomedical research. We thank Eric Boerwinkle (Human Genetics Center and Institute of Molecular Medicine and Division of Epidemiology, University of Texas Health Science Center, Houston, TX) and Julie Cunningham (Department of Health Sciences Research, Mayo Clinic College of Medicine, Rochester, MN) for their help with genotyping.
Footnotes
Abbreviations:
- ACSL1
- acyl-CoA synthetase 1
- CAC
- coronary artery calcification
- CEU
- northern Caucasian Europeans (from Utah)
- DHS
- DNase I hypersensitive site
- DIAGRAM
- Diabetes Genetics Replication and Meta-Analysis
- eQTL
- expression quantitative trait locus
- GWAS
- genome-wide association study
- MAGIC
- Meta-Analyses of Glucose and Insulin-Related Traits Consortium
- MAF
- minor allele frequency
- MESA
- Multi-Ethnic Study of Atherosclerosis
- MuTHER
- Multiple Tissue Human Expression Resource
- RREB1
- ras responsive element binding protein 1
- TLR
- Toll-like receptor
The research reported in this publication was supported by the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Multi-Ethnic Study of Atherosclerosis (MESA) and the MESA SHARe project are conducted and supported by the following contracts: N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-001079, and UL1-TR-000040 from the National Heart, Lung, and Blood Institute (NHLBI). MESA Family is conducted and supported in collaboration with MESA investigators; support is provided by the following grants and contracts: R01HL071051, R01HL071205, R01HL071250, R01HL071251, R01HL071252, R01HL071258, and R01HL071259. Funding for MESA SHARe genotyping was provided by NHLBI Contract N02-HL6-4278. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, Clinical and Translational Science Institute Grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease (NIDDK) Diabetes Research Center (DRC) Grant DK063491 to the Southern California Diabetes Endocrinology Research Center. This publication was developed under a STAR research assistance agreement, No. RD831697 (MESA Air), awarded by the US Environmental Protection Agency (EPA). It has not been formally reviewed by the EPA. The views expressed in this document are solely those of the authors, and the EPA does not endorse any products or commercial services mentioned in this publication. K.E.B. is supported by NHLBI Grants R01HL062887, P01HL092969, and R01HL126028, by DP3DK108209 from the NIDDK, and by the DRC at the University of Washington (P30DK017047). A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org. Data on coronary artery disease/myocardial infarction have been contributed by CARDIoGRAMplusC4D investigators and have been downloaded from http://www.cardiogramplusc4d.org. D.S. is supported by NHLBI Grants R01HL111398 and R01HL122843 and by funds awarded by Pfizer. Grant support for the African American meta-analysis is as follows: The NHLBI’s Family Heart Study (FamHS) was supported by National Institutes of Health Grants R01HL087700 and R01HL088215 [Michael A. Province, principal investigator (PI)] from NHLBI, and R01DK8925601 and R01DK075681 (I. B. Borecki, PI) from NIDDK. The following studies have contributed parent study data, ancillary study data, and DNA samples through the Broad Institute (N01-HC-65226): N01-HC-48047, N01-HC-48048, N01-HC-48049, N01-HC-48050, N01-HC-95095, N01-HC-45204, N01-HC-45205, N01-HC-05187, N01-HC-45134, N01-HC-95100, N01-HC-95170, N01-HC-95171, N01-HC-95172, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95168, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95169, and R01HL071205. The Coronary Artery Risk Development in Young Adults (CARDIA) study is funded by contracts N01-HC-95095, N01-HC-48047, N01-HC-48048, N01-HC-48049, N01-HC-48050, N01-HC-45134, N01-HC-05187, N01-HC-45205, and N01-HC-45204 from the NHLBI to the CARDIA investigators. GWAS genotyping and quality control for the CARDIA African Americans was supported by the NHLBI’s CARe Study. Statistical analysis of CARDIA data was supported by Grants R01HL084099 and U01-HG004729 (Myriam Fornage, Center for Human Genetics, University of Texas, Houston, TX). This manuscript has been reviewed by CARDIA for scientific content and consistency of data interpretation with previous CARDIA publications. The MESA Family/Air Studies were conducted and supported by the NHLBI and the EPA in collaboration with MESA Family and MESA Air investigators, respectively. Funding for genotyping was provided by NHLBI Contract N02-HL64278. Genotyping was performed at the Broad Institute of Harvard and Massachusetts Institute of Technology (Boston, MA) and at Affymetrix (Santa Clara, CA) using the Affymetrix Genome-Wide Human SNP Array 6.0. The GeneSTAR Study was supported by the NHLBI through the STAMPEED (R01HL087698) consortium and Grants R01HL58625, R01HL59684, and R01HL071025, as well as a grant from the National Institutes of Health/National Institute of Nursing Research (NR008153). Additional support was provided by a grant from the National Institutes of Health/National Center for Research Resources (M01-RR000052) to the Johns Hopkins General Clinical Research Center. The Genetic Epidemiology Network of Arteriopathy (GENOA) is supported by National Institutes of Health Grants HL085571, HL087660, and HL100245 from NHLBI. M.P.R. is supported by Grants R01DK071224, R01DK090505, U01-HL108636, K24-HL107643 and R01-HL113147. The authors declare no conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Ellis J. M., Mentock S. M., Depetrillo M. A., Koves T. R., Sen S., Watkins S. M., Muoio D. M., Cline G. W., Taegtmeyer H., Shulman G. I., et al. 2011. Mouse cardiac acyl coenzyme a synthetase 1 deficiency impairs fatty acid oxidation and induces cardiac hypertrophy. Mol. Cell. Biol. 31: 1252–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schisler J. C., Grevengoed T. J., Pascual F., Cooper D. E., Ellis J. M., Paul D. S., Willis M. S., Patterson C., Jia W., and Coleman R. A.. 2015. Cardiac energy dependence on glucose increases metabolites related to glutathione and activates metabolic genes controlled by mechanistic target of rapamycin. J. Am. Heart Assoc. 4: e001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellis J. M., Li L. O., Wu P. C., Koves T. R., Ilkayeva O., Stevens R. D., Watkins S. M., Muoio D. M., and Coleman R. A.. 2010. Adipose acyl-CoA synthetase-1 directs fatty acids toward beta-oxidation and is required for cold thermogenesis. Cell Metab. 12: 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L. O., Grevengoed T. J., Paul D. S., Ilkayeva O., Koves T. R., Pascual F., Newgard C. B., Muoio D. M., and Coleman R. A.. 2015. Compartmentalized acyl-CoA metabolism in skeletal muscle regulates systemic glucose homeostasis. Diabetes. 64: 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L. O., Klett E. L., and Coleman R. A.. 2010. Acyl-CoA synthesis, lipid metabolism and lipotoxicity. Biochim. Biophys. Acta. 1801: 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanter J. E., Kramer F., Barnhart S., Averill M. M., Vivekanandan-Giri A., Vickery T., Li L. O., Becker L., Yuan W., Chait A., et al. 2012. Diabetes promotes an inflammatory macrophage phenotype and atherosclerosis through acyl-CoA synthetase 1. Proc. Natl. Acad. Sci. USA. 109: E715–E724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubinow K. B., Wall V. Z., Nelson J., Mar D., Bomsztyk K., Askari B., Lai M. A., Smith K. D., Han M. S., Vivekanandan-Giri A., et al. 2013. Acyl-CoA synthetase 1 is induced by Gram-negative bacteria and lipopolysaccharide and is required for phospholipid turnover in stimulated macrophages. J. Biol. Chem. 288: 9957–9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Y. L., Morales-Rosado J., Ray J., Myers T. G., Kho T., Lu M., and Munford R. S.. 2014. Toll-like receptor agonists promote prolonged triglyceride storage in macrophages. J. Biol. Chem. 289: 3001–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker L., Liu N. C., Averill M. M., Yuan W., Pamir N., Peng Y., Irwin A. D., Fu X., Bornfeldt K. E., and Heinecke J. W.. 2012. Unique proteomic signatures distinguish macrophages and dendritic cells. PLoS One. 7: e33297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kratz M., Coats B. R., Hisert K. B., Hagman D., Mutskov V., Peris E., Schoenfelt K. Q., Kuzma J. N., Larson I., Billing P. S., et al. 2014. Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab. 20: 614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanter J. E., Tang C., Oram J. F., and Bornfeldt K. E.. 2012. Acyl-CoA synthetase 1 is required for oleate and linoleate mediated inhibition of cholesterol efflux through ATP-binding cassette transporter A1 in macrophages. Biochim. Biophys. Acta. 1821: 358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X., Gonzalez O., Shen X., Barnhart S., Kramer F., Kanter J. E., Vivekanandan-Giri A., Tsuchiya K., Handa P., Pennathur S., et al. 2013. Endothelial acyl-CoA synthetase 1 is not required for inflammatory and apoptotic effects of a saturated fatty acid-rich environment. Arterioscler. Thromb. Vasc. Biol. 33: 232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A. U., Wheeler E., Glazer N. L., Bouatia-Naji N., Gloyn A. L., et al. 2010. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 42: 105–116. [Erratum. 2010. Nat Genet. 42: 464.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manning A. K., Hivert M. F., Scott R. A., Grimsby J. L., Bouatia-Naji N., Chen H., Rybin D., Liu C. T., Bielak L. F., Prokopenko I., et al. 2012. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 44: 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris A. P., Voight B. F., Teslovich T. M., Ferreira T., Segre A. V., Steinthorsdottir V., Strawbridge R. J., Khan H., Grallert H., Mahajan A., et al.; Wellcome Trust Case Control Consortium; Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. 2012. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44: 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wojczynski M. K., Li M., Bielak L. F., Kerr K. F., Reiner A. P., Wong N. D., Yanek L. R., Qu L., White C. C., Lange L. A., et al. 2013. Genetics of coronary artery calcification among African Americans, a meta-analysis. BMC Med. Genet. 14: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pruim R. J., Welch R. P., Sanna S., Teslovich T. M., Chines P. S., Gliedt T. P., Boehnke M., Abecasis G. R., and Willer C. J.. 2010. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 26: 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bild D. E., Bluemke D. A., Burke G. L., Detrano R., Diez Roux A. V., Folsom A. R., Greenland P., Jacob D. R. Jr., Kronmal R., Liu K., et al. 2002. Multi-ethnic study of atherosclerosis: objectives and design. Am. J. Epidemiol. 156: 871–881. [DOI] [PubMed] [Google Scholar]
- 19.Kaufman J. D., Adar S. D., Allen R. W., Barr R. G., Budoff M. J., Burke G. L., Casillas A. M., Cohen M. A., Curl C. L., Daviglus M. L., et al. 2012. Prospective study of particulate air pollution exposures, subclinical atherosclerosis, and clinical cardiovascular disease: the Multi-Ethnic Study of Atherosclerosis and Air Pollution (MESA Air). Am. J. Epidemiol. 176: 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Folsom A. R., Kronmal R. A., Detrano R. C., O’Leary D. H., Bild D. E., Bluemke D. A., Budoff M. J., Liu K., Shea S., Szklo M., et al. 2008. Coronary artery calcification compared with carotid intima-media thickness in the prediction of cardiovascular disease incidence: the Multi-Ethnic Study of Atherosclerosis (MESA). Arch. Intern. Med. 168: 1333–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genuth S., Alberti K. G., Bennett P., Buse J., Defronzo R., Kahn R., Kitzmiller J., Knowler W. C., Lebovitz H., Lernmark A., et al.; Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. 2003. Follow-up report on the diagnosis of diabetes mellitus. Diabetes Care. 26: 3160–3167. [DOI] [PubMed] [Google Scholar]
- 22.Manichaikul A., Naj A. C., Herrington D., Post W., Rich S. S., and Rodriguez A.. 2012. Association of SCARB1 variants with subclinical atherosclerosis and incident cardiovascular disease: the Multi-Ethnic Study of Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 32: 1991–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abecasis G. R., Auton A., Brooks L. D., DePristo M. A., Durbin R. M., Handsaker R. E., Kang H. M., Marth G. T., and McVean G. A.; 1000 Genomes Project Consortium. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature. 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y., Willer C. J., Ding J., Scheet P., and Abecasis G. R.. 2010. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 34: 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manichaikul A., Mychaleckyj J. C., Rich S. S., Daly K., Sale M., and Chen W. M.. 2010. Robust relationship inference in genome-wide association studies. Bioinformatics. 26: 2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen M. H., and Yang Q.. 2010. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 26: 580–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willer C. J., Li Y., and Abecasis G. R.. 2010. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 26: 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez F., Wang L., Abel H., Zhang Q., Province M. A., and Borecki I. B.. 2015. Admixture mapping of coronary artery calcification in African Americans from the NHLBI Family Heart Study. BMC Genet. 16: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deloukas P., Kanoni S., Willenborg C., Farrall M., Assimes T. L., Thompson J. R., Ingelsson E., Saleheen D., Erdmann J., Goldstein B. A., et al.; CARDIoGRAMplusC4D Consortium; DIAGRAM Consortium; CARDIOGENICS Consortium; MuTHER Consortium; Wellcome Trust Case Control Consortium. 2013. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 45: 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thurman R. E., Rynes E., Humbert R., Vierstra J., Maurano M. T., Haugen E., Sheffield N. C., Stergachis A. B., Wang H., Vernot B., et al. 2012. The accessible chromatin landscape of the human genome. Nature. 489: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M. J., et al.; Roadmap Epigenomics Consortium. 2015. Integrative analysis of 111 reference human epigenomes. Nature. 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vierstra J., Rynes E., Sandstrom R., Zhang M., Canfield T., Hansen R. S., Stehling-Sun S., Sabo P. J., Byron R., Humbert R., et al. 2014. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science. 346: 1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maurano M. T., Humbert R., Rynes E., Thurman R. E., Haugen E., Wang H., Reynolds A. P., Sandstrom R., Qu H., Brody J., et al. 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science. 337: 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Locke A. E., Kahali B., Berndt S. I., Justice A. E., Pers T. H., Day F. R., Powell C., Vedantam S., Buchkovich M. L., Yang J., et al.; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium. 2015. Genetic studies of body mass index yield new insights for obesity biology. Nature. 518: 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasquali L., Gaulton K. J., Rodriguez-Segui S. A., Mularoni L., Miguel-Escalada I., Akerman I., Tena J. J., Moran I., Gomez-Marin C., van de Bunt M., et al. 2014. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet. 46: 136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki H., Watanabe M., Fujino T., and Yamamoto T.. 1995. Multiple promoters in rat acyl-CoA synthetase gene mediate differential expression of multiple transcripts with 5′-end heterogeneity. J. Biol. Chem. 270: 9676–9682. [DOI] [PubMed] [Google Scholar]
- 37.Mahajan A., Sim X., Ng H. J., Manning A., Rivas M. A., Highland H. M., Locke A. E., Grarup N., Im H. K., Cingolani P., et al.; T2D-GENES consortium and GoT2D consortium. 2015. Identification and functional characterization of G6PC2 coding variants influencing glycemic traits define an effector transcript at the G6PC2-ABCB11 locus. PLoS Genet. 11: e1004876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilhelm M., Schlegl J., Hahne H., Moghaddas Gholami A., Lieberenz M., Savitski M. M., Ziegler E., Butzmann L., Gessulat S., Marx H., et al. 2014. Mass-spectrometry-based draft of the human proteome. Nature. 509: 582–587. [DOI] [PubMed] [Google Scholar]
- 39.Pan D., Huang L., Zhu L. J., Zou T., Ou J., Zhou W., and Wang Y. X.. 2015. Jmjd3-mediated H3K27me3 dynamics orchestrate brown fat development and regulate white fat plasticity. Dev. Cell. 35: 568–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouchard C., Sarzynski M. A., Rice T. K., Kraus W. E., Church T. S., Sung Y. J., Rao D. C., and Rankinen T.. 2011. Genomic predictors of the maximal O2 uptake response to standardized exercise training programs. J. Appl. Physiol. 110: 1160–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phillips C. M., Goumidi L., Bertrais S., Field M. R., Cupples L. A., Ordovas J. M., Defoort C., Lovegrove J. A., Drevon C. A., Gibney M. J., et al. 2010. Gene-nutrient interactions with dietary fat modulate the association between genetic variation of the ACSL1 gene and metabolic syndrome. J. Lipid Res. 51: 1793–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuku N., He Z. H., Sanchis-Gomar F., Pareja-Galeano H., Tian Y., Arai Y., Abe Y., Murakami H., Miyachi M., Zempo H., et al. 2015. Exceptional longevity and muscle and fitness related genotypes: a functional in vitro analysis and case-control association replication study with SNPs THRH rs7832552, IL6 rs1800795, and ACSL1 rs6552828. Front. Aging Neurosci. 7: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.