Abstract
Autosomal dominant hypercholesterolemia (ADH) is a human disorder characterized phenotypically by isolated high-cholesterol levels. Mutations in the low density lipoprotein receptor (LDLR), APOB, and proprotein convertase subtilisin/kexin type 9 (PCSK9) genes are well known to be associated with the disease. To characterize the genetic background associated with ADH in France, the three ADH-associated genes were sequenced in a cohort of 120 children and 109 adult patients. Fifty-one percent of the cohort had a possible deleterious variant in LDLR, 3.1% in APOB, and 1.7% in PCSK9. We identified 18 new variants in LDLR and 2 in PCSK9. Three LDLR variants, including two newly identified, were studied by minigene reporter assay confirming the predicted effects on splicing. Additionally, as recently an in-frame deletion in the APOE gene was found to be linked to ADH, the sequencing of this latter gene was performed in patients without a deleterious variant in the three former genes. An APOE variant was identified in three patients with isolated severe hypercholesterolemia giving a frequency of 1.3% in the cohort. Therefore, even though LDLR mutations are the major cause of ADH with a large mutation spectrum, APOE variants were found to be significantly associated with the disease. Furthermore, using structural analysis and modeling, the identified APOE sequence changes were predicted to impact protein function.
Keywords: familial hypercholesterolemia, apolipoproteins, apolipoprotein E, cholesterol, low density lipoprotein, phenotype/genotype correlation
The clinical phenotype of autosomal dominant hypercholesterolemia (ADH) is characterized by an increase of plasma low density lipoprotein-cholesterol (LDL-C) levels, enhanced tendon xanthomas, and a premature risk of CVD. ADH is a frequent, inherited human disorder with a heterozygous prevalence of 1 in 500, albeit for a few European populations this prevalence has been reported to be 1 in 200 (1). This disease usually results from mutations in genes of the low density lipoprotein receptor (LDLR), apoB-100 (APOB), and proprotein convertase subtilisin/kexin type 9 (PCSK9) (2, 3). An in-frame deletion in apoE gene (APOE) has been described in two unrelated European families affected with ADH (4, 5), thereby adding a fourth gene associated with the disease. Mutations in APOE have been found previously in dominant familial dysbetalipoproteinemia (FD) and familial combined hyperlipidemia (FCHL), both diseases being characterized by a high risk of CVD and by markedly elevated cholesterol and TG levels (6, 7).
The severity of the phenotype in ADH varies genetically with the type of mutation and the gene affected (8, 9). LDLR is the gene most frequently associated with ADH and is also the best characterized. It is responsible for the disease called familial hypercholesterolemia (FH) (9–11). With the exception of a few founder populations, the spectrum of LDLR causing FH mutations is large. To date, more than 1,600 variants have been identified [Leiden Open Variation Database (LOVD) of ADH, http://www.ucl.ac.uk/ldlr/; and Human Gene Mutation Database, http://www.hgmd.cf.ac.uk/ac/]. France is among the most heterogeneous in the world: recent studies have identified almost 400 mutations in more than 1,000 affected subjects (9, 12).
In contrast, only a few mutations implicated in ADH were identified in the APOB gene, causing the familial defective apoB-100 (13, 14). Eight pathogenic APOB variants were localized within or adjacent to the LDLR binding domain, with the p.Arg3527Gln being the most common variant in Europe (15–17). Very recently, three novel probable disease-causing alterations found outside the LDL binding domain were reported (18, 19). It is interesting to note that the apoB-defective patients are usually less affected with hypercholesterolemia, having plasma cholesterol levels lower than those encountered in FH patients (9, 13).
The PCSK9 gene, the third gene known to be associated with ADH, encodes a protein that has been shown to degrade the LDLR. ADH due to “gain of function” PCSK9 mutations is uncommon, occurring in less than 1% of cases (9–11). Note that PCSK9 inhibitors are a promising new class of drugs for LDL-C reduction (20, 21).
In the lipid-free state, the apoE protein exhibits two distinct structural domains: an N-terminal domain (residues 1–191), which includes the LDLR binding region, and a C-terminal domain (residues 216–299), which contains the major lipid binding region (22). The apoE is polymorphic with three major isoforms that differ in cysteine and arginine content at positions 130 and 176 (note that without the peptide signal, these positions are 112 and 158, respectively). The E3 isoform, with Cys130 and Arg176, is the most common form and hence is identified as the wild type. The E4 isoform (Arg130/Arg176) is present in almost 15% of the population and has been associated with hypercholesterolemia and with an increased CVD risk (23, 24). The E2 isoform (Cys130/Cys176) affects almost 6% of the population and is associated with low cholesterol and hypertriglyceridemia in heterozygotes and type III hyperlipoproteinemia in homozygotes (23, 24).
As FH responds well to drug treatment, early diagnosis is beneficial for the reduction of atherosclerosis and CVD risks. Diagnostic tools have been developed using different criteria (25–28). Here, a cohort of 229 ADH French patients, from an unpublished population and from unrelated families, was analyzed with global genetic testing, including LDLR, a fragment of exon 26 of APOB, PCSK9, and APOE analysis. This study explores the genetic background of ADH in France and, for the first time, the frequency of APOE variants in patients with a clinical diagnosis of ADH and the specific phenotypes according to the altered genes.
MATERIALS AND METHODS
Patients
Patients with an ADH phenotype were selected, without any genetic testing, from unrelated families (n = 229) residing in France and attending clinics in various parts of the country from September 2010 to April 2014. Informed consent was obtained from all patients regarding genetic testing analysis and participation in the study. It should be noted that diabetes mellitus, hypothyroidism, and increased body mass index were not exclusion criteria in the study; at this stage, patients were solely selected on the basis of an ADH phenotype.
The Dutch Lipid Clinic Network criteria were used to score all the ADH patients of more than 17 years old (n = 109), as recommended by the New French Society of Atherosclerosis. The criteria introduce a point system. In summary, a level of LDL-C from 1.55 to 1.99 g/l (4.0–4.9 mM) gives 1 point, 3 points from 2.00 to 2.49 g/l (5.0–6.4 mM), 5 points from 2.50 to 3.30 g/l (6.5–8.4 mM), and 8 points with a level more than 3.30 g/l (8.5 mM). Concerning the familial history: one parent of first degree with hypercholesterolemia (LDL-C >2g/l or 5.0 mM) gives 2 points and 1 point for first-degree familial antecedent of CVD (i.e., in the case of premature CVD, before the age of 55 and 65 years for the father and the mother of the proband, respectively). We added 2 points to the patient’s score in the case of personal CVD and 6 points in presence of xanthomas. ADH is diagnosed as “definite” (patient’s score >8 points), “probable” (6–8 points), and “possible” (3–5 points). The patients with score <3 points were ruled out of the study.
The child cohort (n = 120, ADH patients <17 years) was scored with a specific scoring developed by clinicians (29). A comparison between the LDL-C levels before and after a 3-month dietary period was used to score the child. The diet consisted in reducing saturated fat and cholesterol-rich foods. For example, as indicated in Benlian et al. (29), to a child with one parent with statin treatment, a definite score was attributed, if the LDL-C was greater than 2.10 g/l and remained above 1.40 g/l after the diet, and a probable score if the LDL-C remained at a level between 1.70 and 2.09 g/l, without any decreasing after the diet.
Study design
All 229 patients were analyzed by sequencing of the entire LDLR gene, a fragment of exon 26 of APOB, and a fragment of exon 4 of APOE for isoform identification. The negative patients were analyzed by the multiplex ligation-dependent probe amplification (MLPA) technique to detect large rearrangements within LDLR. The PCSK9 gene was further studied for patients with no identified genetic variation in loci LDLR and APOB. The entire PCSK9 gene was sequenced in negative patients with a severe phenotype (“probable” or “definite” score), and, in the case of moderate phenotypes, only the presence of functional polymorphisms p.Arg46Leu (rs11591147), p.Ala443Thr (rs28362263), and p.Glu670Gly (rs505151) was analyzed. All the negative patients after LDLR, APOB, and PCSK9 screening were analyzed by sequencing the entire APOE gene. Where a new variant or a variant of uncertain clinical significance (VUS) was found during the LDLR and APOB screening, the analysis was completed by PCSK9 and APOE sequencing and MLPA analysis in the LDLR locus. The entire LDLR, PCSK9, and APOE sequencing included the promoter region and intronic boundaries (50 to 100 flanking base pairs in both sides). The 3′ untranslated regions were not entirely analyzed as the sequencing stopped around 100 base pairs after the termination codon.
Molecular analysis
Genomic DNA was extracted from peripheral blood leukocytes by using a phenol/chloroform extraction. Genotyping for the LDLR, a fragment of exon 26 of APOB, PCSK9, and APOE was carried out using primers and conditions as described in the supplementary data (supplementary Table 1). Sequencing analysis of PCR products was performed by the Sanger method using ABI 3500xL DX sequencer and the Big Dye Terminator v.3.1 cycle sequencing kit (Applied Biosystems). The MLPA was carried out to detect rearrangements within the LDLR in accordance with the manufacturer’s protocol (kit P062; MRC-Holland, The Netherlands). In all cases, the presence of a mutation was confirmed by an independent PCR amplification followed by sequencing or by a second MLPA analysis. The sequence variants of this study were designated according to the Human Genome Variation Society guidelines (http://www.hgvs.org).
Bioinformatic tools and molecular modeling
The programs SIFT (30) and PolyPhen2 (31) were used to predict possible impact of genetic variations on the structure and function of altered proteins. For splicing predictions, we used four algorithms to computationally score 5′ and 3′ splice sites. The four algorithms were based on different concepts [Neural Network Splice Prediction (NNSP), MaxEntScan (MES), Splice Site Finder Like (SSFL), and Human Splicing Finder (HSF)] included in the ALAMUT (Interactive Biosoftware v2.4.0), and the default parameter settings were used.
The prediction of changes in thermal protein stability for each observed APOE variant was obtained from the CUPSAT web server (32). The functional/structural consequences of the APOE amino acid changes were estimated using the NMR structure of the full-length isoform E3 [pdb id 2L7B (33)].
Splicing reporter minigene assay
The genomic variations identified as having a possible effect on splicing in the bioinformatics predictions (10–20% minimum score change) were analyzed by performing a splicing reporter minigene assay. Amplicons were designed that included the exons in question with a variable length of flanking 5′ and 3′ intronic bases (see primers in supplementary Table 1) and were amplified from DNA of the heterozygous patients. Wild-type and mutated PCR products were inserted in the NdeI restriction site of the pTB minigene vector (34) using In-Fusion HD Cloning Plus CE (Clontech Laboratories). The pTB minigene is composed of 4 exons and 3 introns, and the PCR products were inserted in the NdeI restriction site of the third intron. The sequence of the constructs was verified with vector-specific sequence primers surrounding the inserted amplicon. Transfection in HeLa cells using the FuGENE HD transfection reagent (Promega, The Netherlands), RT-PCR procedures, and analysis have been previously described (35, 36). RT-PCR products were analyzed by agarose gel electrophoresis and direct Sanger sequencing using pTB exonic primers, in exons 2–3 and exon 4 of the pTB vector (supplementary Figure 2).
RESULTS
Global molecular analysis of ADH in France
In this cohort of 229 patients, 105 (46%), 77 (34%), and 47 (20%) were diagnosed with definite, probable, and possible ADH, respectively. A likely pathogenic variant or a VUS was found in the four studied genes in 85 patients (81%) with a definite score, in 36 (47%) with a probable score, and in 9 (19%) with a possible score. LDL-C levels, first-degree antecedent of hypercholesterolemia, and personal or familial history of CVD for all 229 patients are given in the supplementary data (supplementary Table 2).
The relative contribution of the four examined genes to ADH in the cohort is shown in supplementary Figure 1. All the identified variants with in silico analysis results, frequency data, and references are presented in the supplementary data (supplementary Table 3).
We identified, in 116 patients of the cohort (50.6%), 91 different probable damaging variants or VUSs in LDLR, spanning the entire length of the gene: 13 patients with a major rearrangement (11%), 15 patients with a small deletion/insertion (13%), and 88 patients with a single sequence variant including 59 with a missense (51%), 17 with a nonsense substitution (15%), and 12 patients with a variant that could modify the natural splice site (10%).
A variant in APOB was identified in eight different ADH patients. The most frequent variant was the p.(Arg3527Gln) (5/8) and two patients carried another likely pathogenic variant, p.(Glu3527Trp) and p.(Arg3558Cys), all already described in ADH. The last APOB variant, the p.(Gln3432Glu), was identified in a patient with a pathogenic variant in LDLR and was described as a benign polymorphism (supplementary Table 3).
With regard to PCSK9, the p.Ser127Arg was present in two unrelated patients with “definite” ADH score (2/4). It is the most frequent PCSK9 mutation causing ADH in France (9). The two other identified variants, p.(Leu41Gln) and p.(Gly516Val), have not been described before (Table 1).
TABLE 1.
List of new variants, possibly disease causing, identified in LDLR and PCSK9
| Gene | Exon | DNA Change | Protein | Type | Predicted Effect | PolyPhen2 Prediction | SIFT Prediction | Species Conservation | Patient Score | Variant Group |
| LDLR OMIM#606945 | 2 | c.178C>T | p.(Gln60*) | NS | Truncated | Definite | A | |||
| 4 | c.680_682delinsCA | p.(Asp227Alafs*38) | Indel | Truncated | Definite | A | ||||
| 4 | c.686A>T | p.(?) | Splice | Crypt splice | Definite | A | ||||
| 5–8 | c.695-?_1186+?del | p.(?) | LR | Truncated | Definite | A | ||||
| 6 | c.905G>T | p.(Cys302Phe) | MS | Damaging | NT | High | Definite | B | ||
| 7 | c.974G>T | p.(Cys325Phe) | MS | Damaging | NT | High | Probable | B | ||
| 8 | c.1118del | p.(Gly373Valfs*40) | Indel | Truncated | Definite | A | ||||
| 9 | c.1201C>T | p.(Leu401Phe) | MS | Damaging | NT | High | Definite | B | ||
| 10 | c.1438G>A | p.(Ala480Thr) | MS | Damaging | NT | High | Probable | B | ||
| 12 | c.1730G>A | p.(Trp577*) | NS | Truncated | Definite | A | ||||
| 14 | c.2106G>A | p.(Met702Ile) | MS | Benign | Tolerated | Low | Possible | VUS | ||
| 15 | c.2167G>T | p.(Glu723*) | NS | Truncated | Definite | A | ||||
| 15 | c.2211G>T | p.(Arg737Ser) | MS | Benign | Tolerated | Low | Probable | VUS | ||
| 15 | c.2229_2234dup | p.(Arg744_Pro745dup) | Indel | Elongated | Possible | B | ||||
| 15 | c.2295_2302del | p.(Thr766Serfs*13) | Indel | Truncated | Definite | A | ||||
| 15 | c.2311G>A | p.(?) | Splice | Crypt splice | Definite | A | ||||
| 16–18 | c.2312-?_2583+?dup | p.(?) | LR | Elongated | Definite | A | ||||
| 17 | c.2482T>A | p.(Tyr828Asn) | MS | Damaging | NT | High | Definite | B | ||
| PCSK9 OMIM#607786 | 1 | c.122T>A | p.(Leu41Gln) | MS | Damaging | Tolerated | Medium | Probable | VUS | |
| 10 | c.1547G>T | p.(Gly516Val) | MS | Damaging | NT | High | Probable | B |
Missense variants (MS), nonsense mutations (NS), small deletion/insertion (indel), and large rearrangements (LR) identified in LDLR and PCSK9. The effect of the variants at the protein level was predicted by SIFT (30) and PolyPhen2 (31). The term “Truncated” means truncated peptide; “Crypt splice,” cryptic splice site creation; and “Elongated,” elongated peptide. NT in SIFT prediction is for “not tolerated.” The level of conservation between species was indicated using SIFT data: highly conserved (High) = none or 1 other amino acid found at this position, medium conserved (Medium) = 2 to 3 amino acids, and lowly conserved (Low) stands for >3 amino acids. Patient score was defined according to the Dutch score: definite >8 points, probable = 6–8 points, and possible = 3–5 points. Variant group A = null allele; group B = defective allele. The nucleotide and protein sequence variants were designated according to the Human Genome Variation Society guidelines (http://www.hgvs.org).
The complete APOE was analyzed in the 106 patients for whom no change was predicted to be functionally damaging in LDLR, APOB, and PCSK9. We found that four patients exhibited a change in APOE (1.7%) (Table 2). Considering that one APOE mutation was detected for a patient suffering from mixed dyslipidemia and not from isolated hypercholesterolemia, the frequency of APOE variant in our ADH cohort was recalculated to be 1.3% (3/228).
TABLE 2.
Genotype/phenotype correlations in patients with a variant in APOE gene
| Patient #ID | Gender | Age (years) | Genotype | Variant Group | LDL-C(g/l) | HDL (g/l) | TGs (g/l) | Predicted Protein Effect | Familial Segregation | Statin Treatment | APOE Isoform |
| #1 | M | 9 | p.[(Arg163Cys)] ; | B | 4.15 | 0.56 | 0.77 | Probably damaging | Yes | Yes | E3E3 |
| [(Arg163Cys)] | |||||||||||
| #1b | F | 29 | p.[(Arg163Cys)] ; [(=)] | B | 2.70 | 0.43 | 1.00 | Probably damaging | Mother of patient #1 | No | E3E3 |
| #2 | F | 19 | p.[(Leu167del)] ; | B | 2.87 | ND | <2.00 | Truncated peptide | ND | Yes | E3E3 |
| [(=)] | |||||||||||
| #3 | F | 6 | p.[Leu46Pro ]; | VUS | 1.97 | 0.70 | 0.85 | Structure destabilizing | Mother carrying mutation with isolated hypercholesterolemia | No | E3E4 |
| [(=)] | |||||||||||
| #4 | M | 43 | p.[(Gly145Asp)] ; | B | >2.00 | ND | >2.00 | Discordance between predictions | Three children carrying no mutation (without mixed dyslipidemia) | Yes | E2E2 |
| [(=)] |
F, female; M, male; ND, not determined. Serum lipids levels: LDL-C, HDL, and TGs were measured without any medication or treatment. No familial or personal CVD antecedents were known for all the patients. The effect on protein of the APOE variants was predicted by SIFT (30) and PolyPhen2 (31) and completed by structural consequence estimation via the CUPSAT web server (32). Variant group B = defective allele. Additional information can be found in supplementary Tables 1 and 3.
Variants functionality and new mutations identified in ADH patients
The variants found in LDLR, APOB, PCSK9, or APOE genes could be separated into four functional groups according to the ADH identification in LOVD, the in silico and in vitro analysis, and/or familial segregation studies. Group A pathogenic variants (n = 41) included major rearrangements, nonsense substitutions, frame-shift insertion/deletion and variants causing splice site abolition, positions ±1 or 2 within the consensus splice site, and other positions tested in vitro. Fifty-five patients carried one group A variant, all found in the LDLR gene, and certainly explained by the observed phenotype. Among group A, 10 variants in LDLR have never been reported before (see Table 1).
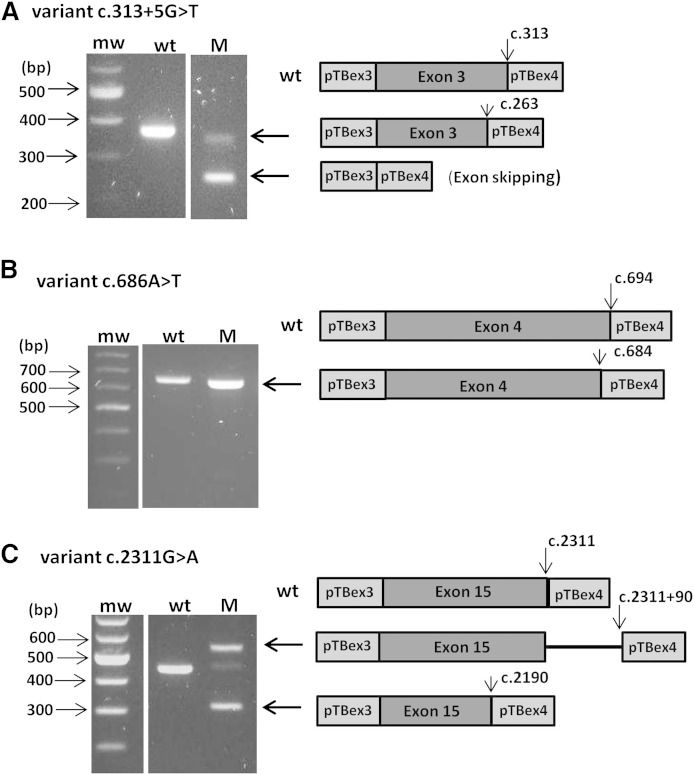
The deleterious effect predicted on splicing of three LDLR variants was confirmed by the splicing reporter minigene assay (Table 3). These three variants were therefore assigned to group A. The first variant, c.313+5G>T, was previously described in Maroccan FH patients (http://www.ucl.ac.uk/ldlr/) and predicted as deleterious via the in silico study. The minigene assay showed de facto the complete abolition of the splice site (Fig. 1A). This variant was found here in a North African patient with a “probable” ADH score. The second variant, a new substitution c.686A>T in exon 4 of LDLR, was prognosticated in silico to be benign by PolyPhen2 and SIFT but foreseen to modify the natural splice site (Table 3). The minigene assay confirmed the creation of a cryptic site in position 684, the natural splice site in 694 being completely abolished (Fig. 1B). The last variant, another new LDLR variant c.2311G>A changing the last nucleotide of the exon 15, was forecasted to cause probable alternative splicing. Indeed, the complete abolition of the natural splice site and the creation of two cryptic sites in the positions 2311+90 and 2190 were revealed by the minigene assay (Fig. 1C).
TABLE 3.
LDLR splicing mutations: in silico analysis and splicing reporter minigene assay
| Mutation | In Silico Analysis | Minigene Assay | |||
| SSFL [0–100] WT→ Mutant | MES [0–12] WT→ Mutant | NNSP [0–1] WT→ Mutant | HSF [0–100] WT→ Mutant | Observed Consequence | |
| Donor | |||||
| 313+5G>T | 75 → − (Abolition) | 9.9 → 1.8 (−81.5%) | 0.95 → − (Abolition) | 82 → 70 (−14.9%) | Complete abolition of the physiological splice site and use of the cryptic splice site at c.263 |
| c.686A>T | − → 79 (Physiological splice site at c.694: 73) | − → 7.2 (Physiological splice site at c.694: 7.6) | − → 0.9 (Physiological splice site at c.694: 0.9) | − → 84 (Physiological splice site at c.694: 82) | Exclusive use of the cryptic splice site at c.684 |
| c.2311G>A | 82.5 → 70 (−14.7%) | 9 →1.6 (−82%) | 1 → − (Abolition) | 85 →74 (−12.5%) | Complete abolition of the physiological splice site and exclusive use of two cryptic splice sites at c.2311+90 and at c.2190 |
Four algorithms for computational scoring of 5′ (donor) and 3′ (acceptor) splice sites based on different concepts: NNSP, MES, SSFL, and HSF, all included in the ALAMUT (Interactive Biosoftware v2.4.0). “_” indicates a value below the threshold. Modifications occurred only on donor splice site in all the studied variants. Value variations of natural splice site are indicated for c.313+5G>T and c.2311G>A variants and force decreasing in brackets. Value variations of the cryptic splice site at c.684 are indicated for the variant c.686A>T and values of the natural splice site in brackets.
Fig. 1.
Differences in RT-PCR transcripts produced by splicing reporter minigene assays with the constructions containing the three LDLR variants c.313+5G>T (A), c.686A>T (B), and c.2311G>A (C). RT-PCR fragments produced in the minigene assay and separated by agarose gel electrophoresis are shown at the left, and schematic representation of fragments corresponding with agarose bands are shown at the right, with flanking exons 3 and 4 of pTB vector in light gray, denoted pTBex3 and pTBex4, respectively, and the inserted LDLR exon in dark gray. The natural splicing sites in wild-type constructs are indicated by an arrow on the schematic representation, as well as the revealed cryptic slice sites in variant constructs, that is, c.263 in c.313+5G>T (A), c.684 in variant c.686A>T (B), and both c.2311+90 and c.2190 in c.2311G>A (C). When the exon was skipped, the RT-PCR product gave only the 248 bp transcript corresponding to the flanking exons 3 and 4 of the pTB vector. M, variant construct; mw, molecular weight marker; wt, wild-type construct.
Group B likely pathogenic variants included missense substitutions with a probable functional effect (n = 49) and in-frame insertion/deletion (n = 3). Sixty-nine ADH patients presented one variant of group B in LDLR, APOB, PCSK9, or APOE.
PolyPhen2 and SIFT programs were used to predict the pathogenicity of the missense changes. Forty-one missense substitutions, prognosticated to be deleterious by the two programs and identified in LOVD of ADH mutations in LDLR, APOB, and PCSK9, belong to group B. Two of variants identified in APOE, namely the p.(Gly145Asp) and the p.(Arg163Cys), were reported in FD or FCHL patients (6, 7), so these variants were classified in group B too. Six newly identified changes, predicted as deleterious, were also classified in group B because they occurred in a highly conserved position among species: five in LDLR and one in PCSK9 (Table 1). The new in-frame duplication p.(Arg744_Pro745dup) in LDLR was identified in two French families: in a mother and her daughter both with a definite score and in a second family, in which just one child with a possible ADH was analyzed and carried this variant.
Group C included all variants that may not cause the disease (n = 8), such as the benign polymorphisms p.(Glu277Lys), p.(Asp342Asn) in LDLR, p.(Gln3432Glu) in APOB, and p.(Glu670Gly) in PCSK9. The benign silent or intronic variants with no consequence for splice site were also categorized in group C. We also classified two intronic and three silent mutations found in LDLR as benign variants, according to the in silico analysis as it did not suggest that the splicing reporter minigene assay was necessary. These variants were the c.1706-10G>A, c.941-4G>A, c.1167G>A, c.1920C>T, and c.1977C>A (see supplementary Table 4). Note that the c.1167G>A had never been described before in LOVD.
The group of VUSs included missense substitutions with possible functional effects or other intronic or exonic changes with possible effects on transcription or causing alternative splicing (n = 5). Nine ADH patients carried at least one VUS; of these, patients five had the p.(Thr726Ile) in LDLR. The pathogenicity of this variant is controversial (37). This variant was found in two patients with a pathogenic variant in LDLR and in three other patients with a probable or possible score of ADH.
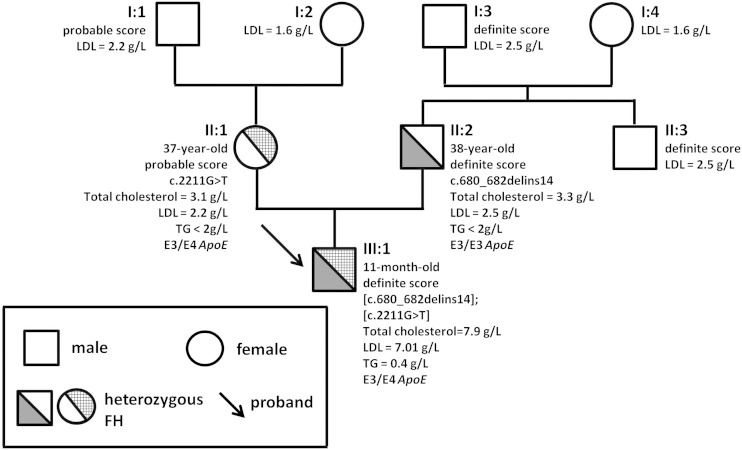
Among the likely pathogenic VUSs, the p.Leu46Pro in APOE and the newly identified p.(Leu41Gln) in PCSK9 were predicted to be tolerated by SIFT and possibly or probably damaging by PolyPhen2. The p.Leu46Pro in APOE could affect the protein structure as explained below. The p.(Leu41Gln) was carried by a 61-year-old woman with 2.5 g/l of LDL-C level and premature CVD. This patient also had the variant p.(Asp342Asn) in LDLR (rs139361635), which is considered to have no effect on the protein (supplementary Table 3). Unfortunately, as she did not know her family, we were unable to investigate further. Another new variant in LDLR, the p.(Arg737Ser), was predicted to be benign by PolyPhen2 and SIFT. However, this substitution was found in a 1-year-old baby with an LDL-C level of 7.01 g/l that could be a compound heterozygous form of FH. He inherited the p.(Arg737Ser) and a deletion causing a heterozygous frame shift from his mother and his father, respectively (Fig. 2). Antecedents of hypercholesterolemia were known in the mother’s and the father’s families. No deleterious effect on splicing was predicted by in silico analysis for this variant (supplementary Table 4). Further genetic studies on the complete family or in vitro analysis are required to confirm the pathogenicity of these variants.
Fig. 2.
Pedigree and genetic analysis in a family with a new identified variant in LDLR. The arrow indicates the proband: a compound heterozygous baby carrying a mutation (c.680_682delins14) and a VUS [c.2211G>T; p.(Arg737Ser)] in LDLR. Circles represent females and squares males. Filled symbols indicate heterozygous subjects with FH. When available, score and clinical and genetic data are indicated next to symbol. LDL, serum LDL cholesterol level; TG, serum triglycerides level. The age of the tested patients at the time of study is also given.
APOE variants and structural analysis
The four variants present in APOE were as follows: the p.(Arg163Cys) previously described in type III dyslipidemia (38) (patients #1 and #1b); the p.(Leu167del), a 3 bp in-frame deletion described in ADH disease (4, 5) (patient #2); the rare polymorphism p.Leu46Pro (rs769452) (patient #3); and the p.(Gly145Asp) described in FD patients (6) (patient #4) (Table 2). Three of these patients (#1 to #3) are suffering from a severe form of hypercholesterolemia, and the last patient (#4) has a severe form of mixed dyslipidemia. The latter was included in the cohort as suffering with severe hypercholesterolemia treated by statins, and his daughter presented a possible score for hypercholesterolemia. Biological and clinical information and familial antecedents on the four patients with a variant in APOE are summarized in Table 2. The mother of patient #1 was also added (identified as patient #1b), in order to examine the homozygous and heterozygous status of the p.(Arg163Cys). More precisely, patient #1, a 9-year-old boy, carried the homozygous p.(Arg163Cys) and had an LDL-C level of 4.15 g/l ; his mother (patient #1b) is suffering from hypercholesterolemia with a “definite” score (LDL-C = 2.7 g/l). Patient #2, a 19-year-old patient with a “probable” ADH score, carried the p.(Leu167del) and a silent variant c.1920C>T in LDLR predicted to not affect the protein or the splicing (supplementary Table 4). Patient #3 was a 6-year-old girl with a blood LDL-C level of 1.97 g/l and a clear history of FH from her mother, who carried this heterozygous variant too. Patient #4 was suffering with a severe form of mixed dyslipidemia and carried the E2E2 genotype that is known to increase TG levels.
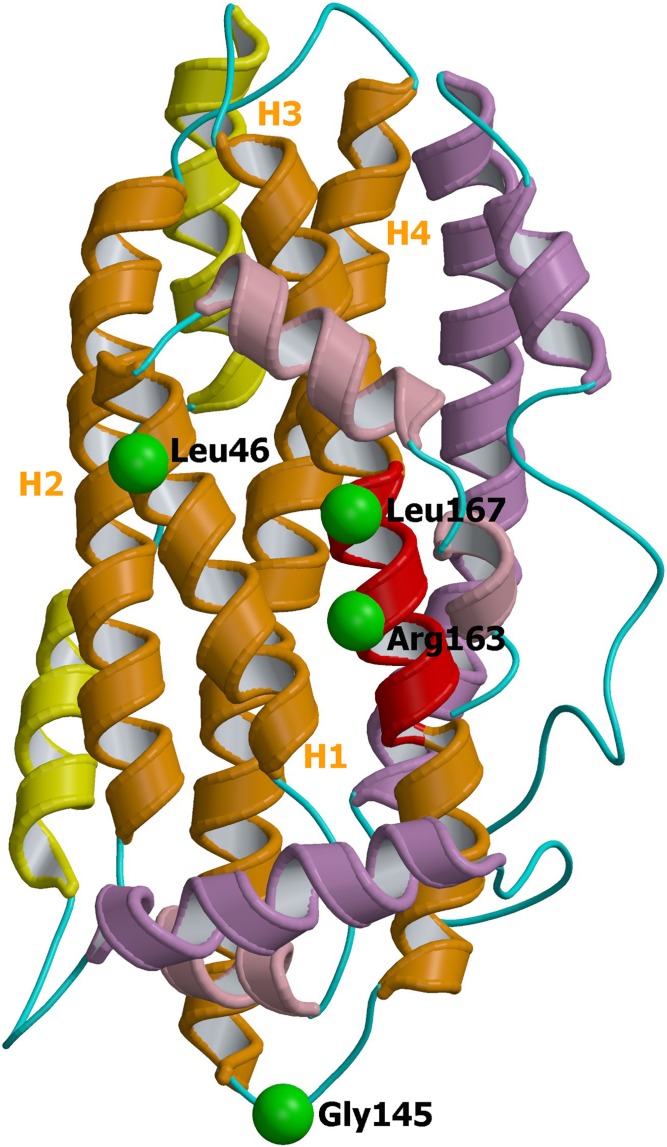
Modeling analysis based on the NMR structure of the full-length apoE isoform E3 (33) was used for interpreting the effect of the four identified mutations. The mutations are all situated in the N-terminal domain (Fig. 3). Leu46 is located in the first α-helix H1 of N-terminal core domain, and hence the p.Leu46Pro variant inserts a proline residue into an α-helix, a situation that is generally known to be a destabilizing structural feature (39). Accordingly, the p.Leu46Pro mutation was predicted to be destabilizing by the CUPSAT program, with a large change in free folding energy of 1.4 kcal/mol. Indeed, very recently the variant Leu46Pro of apoE isoform E4 was experimentally found to be thermodynamically destabilized, more prone to proteolysis, and affected in its lipid binding properties (40). Gly145 is located within a loop region connecting helices H3 and H4; this residue being solvent-accessible, the CUPSAT program predicts a stabilizing mutation for the variant p.(Gly145Asp). Contrariwise, Arg163 and Leu167 are in the middle of the α-helix H4, precisely in the putative LDLR binding site (region 158–168). Therefore, p.(Arg163Cys) and p.(Leu167del) mutants are thought to impair LDLR binding properties of the apoE protein.
Fig. 3.
Ribbon representation of apoE (pdb id 2L7B). Mutated residues encountered in the present study are represented and labeled for their localization on the 3D protein structure. The additional N-terminal helices are colored in salmon, the N-terminal domain in orange, the hinge domain (i.e., the region between the two functionally distinct domains) in yellow, and the C-terminal domain in purple. The putative LDLR binding region (residues 158–168) is shown in red. The four α-helices of the N-terminal domain are labeled as H1 to H4. The image was obtained using consecutively MolScript (47) and Raster3D (48) programs.
DISCUSSION
A probable damaging variant or a VUS was identified in 56.8% of our ADH French cohort. Changes in LDLR remained the main cause of ADH and represented 89% of the detected variants. Eighteen new possibly damaging variants were added to the last study of Marduel et al. (9), in which 175 novel mutational events had been reported from a large French cohort of ADH patients. We identified 88 different variants in LDLR, in addition to the 391 from the previous French study (9) and confirm that France has among the most heterogeneous LDLR variant spectrum in the world. Furthermore, the large number of new variants, especially in the LDLR gene, which appears particularly permissive to mutational events, has to be considered for the molecular diagnosis strategy.
Among the new changes identified in LDLR and PCSK9, group A and B variants could be classified as null or defective alleles and so likely linked to the disease. We further experimentally established by splicing reporter minigene assays the splice site alteration induced by three LDLR variants, namely c.2311G>A, c.313+5G>T, and c.686A>T. The p.(Arg737Ser) in LDLR and the p.(Leu41Gln) in PCSK9 could be classified as likely pathogenic VUSs and required further familial genetic studies or in vitro analysis to confirm the pathogenicity. Finally, one new exonic substitution, p.(Met702Ile), and one silent variant, c.1167G>A, in LDLR could be considered to have no effect on the protein according to the in silico analysis.
The relatively high percentage of ADH patients with no changes in the four considered genes (43.2% of the cohort) suggests the existence of other causative genes and/or polygenic causes. Recently, sequence variants in STAP1, encoding signal transducing adaptor family member 1, has been found in ADH patients, with a frequency of 1.2% (41). Another locus, called HCHOLA4, mapped at 16q22.1 region, was linked to ADH, but the causative gene has not been identified yet (42). Thus, other ADH disease-causative genes probably remain to be discovered even if the main causative genes are known (3). However, the distribution of the three common apoE isoforms could explain the cases with moderate hypercholesterolemia. Indeed, 32% of the cohort exhibited the E4 isoform, and this proportion was even greater in patients without any pathogenic variant detected (42%), while the E4 frequency in the European population is about 15% (24).
Interestingly, four unrelated patients bear a change in APOE (Table 2). Patient #4, suffering from a mixed dyslipidemia, carried the p.(Gly145Asp) in association with the E2E2 genotype. This genotype is known to increase TG levels due to a defect in the IDL catabolism. It is established that the function of apoE protein is intimately linked to its conformational properties, so that structural perturbations brought by a single-site mutation may be accompanied by a dysfunction associated with pathogenesis (22, 43). It was difficult to evaluate the structural impact of the p.(Gly145Asp) change, but, taking into account the key role played by the charge-charge interactions in LDLR binding (44), this variant, by modifying the net charge of the apoE protein toward acidic, could reduce the affinity for LDLR. Furthermore, the homozygous E2 isoform was demonstrated to be severely defective in in vitro LDLR binding activity compared with the E3 isoform (45). The severe mixed dyslipidemia observed in patient #4 could be explained by a quite complete apoE deficiency of the mutated allele.
Concerning the other apoE variants, the p.Leu46Pro was described as a VUS (rs769452) without clinical information. This APOE variant was predicted to have a destabilizing effect on the protein structure here, and this has been confirmed experimentally recently for the E4 isoform (40); our patient carried precisely the E4 isoform (patient #3). The p.(Leu167del) found in patient #2 was previously described in ADH patients (4, 5), and our in silico structural analyses suggest that this apoE variant will have impaired LDLR binding properties. The binding defect of p.(Leu167del) was, however, not demonstrated (4).
The p.(Arg163Cys) variant has previously been described in a patient with FD, who in addition bore a deleterious mutation also in APOE (38). Contrariwise, in our study, patient #1 carried only the homozygous C163 change in APOE and no other mutation. This boy and his mother are suffering an isolated severe hypercholesterolemia. Unfortunately, we cannot obtain any information about the father. As both patients came from the island of Martinique, the homozygous status could be explained by a founder effect or unknown consanguinity. The in silico structural analysis suggests that the p.(Arg163Cys) will have impaired LDLR binding properties as well.
Even if experimental investigations are needed to confirm the effects of mutations, the variants p.(Leu167del), p.(Arg163Cys), and possibly the VUS p.Leu46Pro are thought to be responsible for the observed ADH phenotype. As the TG levels were found unaffected (Table 2), the observed phenotype of patients #1, #2, and #3 could be explained by a partial apoE deficiency, contrary to patient #4. Even the homozygous form Cys163 appears to retain usual properties toward the VLDL and IDL remnants in plasma, considering the normal TG level observed in patient #1.
From our data, it appears that the homozygous form C163 in APOE is less severe than a homozygous FH form and looks like the APOB homozygous form, at least with respect to the LDL-C levels (46). Interestingly, the statin treatment was associated with good outcome in patient #1, reducing the LDL-C level from 4.15 to 2.70 g/l. Interestingly, we did not notice any notable CVD history for the four families with APOE changes. This observation has to be confirmed by a precise outcome as our data suggest that the APOE modifications in ADH could lead to less atherosclerosis than LDLR or PCSK9 changes.
In conclusion, changes in LDLR remain the main cause of ADH accounting for 89% of the mutated ADH patients in this study. We identified 18 new possible damaging variants in LDLR and 2 in PCSK9. This study shows further that the variants in APOE are not an insignificant cause of ADH with a frequency of 1.3% of altered patients in our cohort. Finally, two of the three variants found in patients with isolated hypercholesterolemia were located in the LDLR binding domain of the apoE protein.
Supplementary Material
Acknowledgments
The authors would like to thank the Laboratoire Commun de Biologie et Génétiques Moléculaires (LCBGM) at St. Antoine Hospital, Paris, for performing the molecular tests. The authors are indebted to all patients and clinicians for participating in this study. The authors thank Drs. N. Nunan and A. Wohlkönig for critical reading of the manuscript.
Footnotes
Abbreviations:
- ADH
- autosomal dominant hypercholesterolemia
- FD
- dominant familial dysbetalipoproteinemia
- FH
- familial hypercholesterolemia
- HSF
- Human Splicing Finder
- LDL-C
- low density lipoprotein-cholesterol
- LDLR
- low density lipoprotein receptor
- LOVD
- Leiden Open Variation Database
- MES
- MaxEntScan
- MLPA
- multiplex ligation-dependent probe amplification
- NNSP
- Neural Network Splice Prediction
- PCSK9
- proprotein convertase subtilisin/kexin type 9
- SSFL
- Splice Site Finder Like
- TC
- total cholesterol
- VUS
- variant of unknown clinical significance.
This work was supported by the Biology-Imaging District of East-Hospitals Group of AP-HP (Assistance Publique-Hôpitaux de Paris). R. Wintjens is Research Associate at the National Fund for Scientific Research FNRS-FRS (Belgium).
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Nordestgaard B. G., Chapman M. J., Humphries S. E., Ginsberg H. N., Masana L., Descamps O. S., Wiklund O., Hegele R. A., Raal F. J., Defesche J. C., et al.; European Atherosclerosis Society Consensus Panel. 2013. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European Atherosclerosis Society. Eur. Heart J. 34: 3478–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varret M., Abifadel M., Rabès J. P., and Boileau C.. 2008. Genetic heterogeneity of autosomal dominant hypercholesterolemia. Clin. Genet. 73: 1–13. [DOI] [PubMed] [Google Scholar]
- 3.Futema M., Plagnol V., Li K., Whittall R. A., Neil H. A. W., Seed M., Bertolini S., Calandra S., Descamps O. S., Graham C. A., et al.; Simon Broome Consortium; UK10K Consortium. 2014. Whole exome sequencing of familial hypercholesterolaemia patients negative for LDLR/APOB/PCSK9 mutations. J. Med. Genet. 51: 537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marduel M., Ouguerram K., Serre V., Bonnefont-Rousselot D., Marques-Pinheiro A., Berge K. E., Devillers M., Luc G., Lecerf J.-M., Tosolini L., et al. 2013. Description of a large family with autosomal dominant hypercholesterolemia associated with the APOE p.Leu167del mutation. Hum. Mutat. 34: 83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Awan Z., Choi H. Y., Stitziel N., Ruel I., Bamimore M. A., Husa R., Gagnon M. H., Wang R. H., Peloso G. M., Hegele R. A., et al. 2013. APOE p.Leu167del mutation in familial hypercholesterolemia. Atherosclerosis. 231: 218–222. [DOI] [PubMed] [Google Scholar]
- 6.Miller D. B., Hegele R. A., Wolfe B. M., and Huff M. W.. 1995. Identification, molecular characterization, and cellular studies of an apolipoprotein E mutant (E1) in three unrelated families with hyperlipidemia. J. Clin. Endocrinol. Metab. 80: 807–813. [DOI] [PubMed] [Google Scholar]
- 7.Solanas-Barca M., de Castro-Orós I., Mateo-Gallego R., Cofán M., Plana N., Puzo J., Burillo E., Martín-Fuentes P., Ros E., Masana L., et al. 2012. Apolipoprotein E gene mutations in subjects with mixed hyperlipidemia and a clinical diagnosis of familial combined hyperlipidemia. Atherosclerosis. 222: 449–455. [DOI] [PubMed] [Google Scholar]
- 8.Guardamagna O., Restagno G., Rolfo E., Pederiva C., Martini S., Abello F., Baracco V., Pisciotta L., Pino E., Calandra S., et al. 2009. The type of LDLR gene mutation predicts cardiovascular risk in children with familial hypercholesterolemia. J. Pediatr. 155: 199–204. [DOI] [PubMed] [Google Scholar]
- 9.Marduel M., Carrié A., Sassolas A., Devillers M., Carreau V., Di Filippo M., Erlich D., Abifadel M., Marques-Pinheiro A., Munnich A., et al.; French ADH Research Network. 2010. Molecular spectrum of autosomal dominant hypercholesterolemia in France. Hum. Mutat. 31: E1811–E1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Graaf A., Avis H. J., Kusters D. M., Vissers M. N., Hutten B. A., Defesche J. C., Huijgen R., Fouchier S. W., Wijburg F. A., Kastelein J. J., et al. 2011. Molecular basis of autosomal dominant hypercholesterolemia: assessment in a large cohort of hypercholesterolemic children. Circulation. 123: 1167–1173. [DOI] [PubMed] [Google Scholar]
- 11.Medeiros A. M., Alves A. C., and Bourbon M.. 2015. Mutational analysis of a cohort with clinical diagnosis of familial hypercholesterolemia: considerations for genetic diagnosis improvement. Genet. Med. Epub ahead of print. May 28, 2015; doi:10.1038/gim.2015.71. [DOI] [PubMed] [Google Scholar]
- 12.Amsellem S., Briffaut D., Carrié A., Rabès J. P., Girardet J. P., Fredenrich A., Moulin P., Krempf M., Reznik Y., Vialettes B., et al. 2002. Intronic mutations outside of Alu-repeat-rich domains of the LDL receptor gene are a cause of familial hypercholesterolemia. Hum. Genet. 111: 501–510. [DOI] [PubMed] [Google Scholar]
- 13.Whitfield A. J., Barrett P. H., van Bockxmeer F. M., and Burnett J. R.. 2004. Lipid disorders and mutations in the APOB gene. Clin. Chem. 50: 1725–1732. [DOI] [PubMed] [Google Scholar]
- 14.Borén J., Ekström U., Agren B., Nilsson-Ehle P., and Innerarity T. L.. 2001. The molecular mechanism for the genetic disorder familial defective apolipoprotein B100. J. Biol. Chem. 276: 9214–9218. [DOI] [PubMed] [Google Scholar]
- 15.Gaffney D., Reid J. M., Cameron I. M., Vass K., Caslake M. J., Shepherd J., and Packard C. J.. 1995. Independent mutations at codon 3500 of the apolipoprotein B gene are associated with hyperlipidemia. Arterioscler. Thromb. Vasc. Biol. 15: 1025–1029. [DOI] [PubMed] [Google Scholar]
- 16.Pullinger C. R., Gaffney D., Gutierrez M. M., Malloy M. J., Schumaker V. N., Packard C. J., and Kane J. P.. 1999. The apolipoprotein B R3531C mutation. Characteristics of 24 subjects from 9 kindreds. J. Lipid Res. 40: 318–327. [PubMed] [Google Scholar]
- 17.Motazacker M. M., Pirruccello J., Huijgen R., Do R., Gabriel S., Peter J., Kuivenhoven J. A., Defesche J. C., Kastelein J. J., Hovingh G. K., et al. 2012. Advances in genetics show the need for extending screening strategies for autosomal dominant hypercholesterolaemia. Eur. Heart J. 33: 1360–1366. [DOI] [PubMed] [Google Scholar]
- 18.Thomas E. R., Atanur S. S., Norsworthy P. J., Encheva V., Snijders A. P., Game L., Vandrovcova J., Siddiq A., Seed M., Soutar A. K., et al. 2013. Identification and biochemical analysis of a novel APOB mutation that causes autosomal dominant hypercholesterolemia. Mol. Genet. Genomic Med. 1: 155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alves A. C., Etxebarria A., Soutar A. K., Martin C., and Bourbon M.. 2014. Novel functional APOB mutations outside LDL-binding region causing familial hypercholesterolaemia. Hum. Mol. Genet. 23: 1817–1828. [DOI] [PubMed] [Google Scholar]
- 20.Farnier M. 2014. PCSK9: from discovery to therapeutic applications. Arch. Cardiovasc. Dis. 107: 58–66. [DOI] [PubMed] [Google Scholar]
- 21.Shimada Y. J., and Cannon C. P.. 2015. PCSK9 (proprotein convertase subtilisin/kexin type 9) inhibitors: past, present, and the future. Eur. Heart J. 36: 2415–2424. [DOI] [PubMed] [Google Scholar]
- 22.Hatters D. M., Peters-Libeu C. A., and Weisgraber K. H.. 2006. Apolipoprotein E structure: insights into function. Trends Biochem. Sci. 31: 445–454. [DOI] [PubMed] [Google Scholar]
- 23.Eichner J. E., Dunn S. T., Perveau G., Thompson D. M., Stewart K. E., and Stroehla B. C.. 2002. Apolipoprotein E polymorphism and cardiovascular disease: a HuGE review. Am. J. Epidemiol. 155: 487–495. [DOI] [PubMed] [Google Scholar]
- 24.Khan T. A., Shah T., Prieto D., Zhang W., Price J., Fowkes G. R., Cooper J., Talmud P. J., Humphries S. E., Sundstrom J., et al. 2013. Apolipoprotein E genotype, cardiovascular biomarkers and risk of stroke: systematic review and meta-analysis of 14,015 stroke cases and pooled analysis of primary biomarker data from up to 60,883 individuals. Int. J. Epidemiol. 42: 475–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams R. R., Hunt S. C., Schumacher M. C., Hegele R. A., Leppert M. F., Ludwig E. H., and Hopkins P. N.. 1993. Diagnosing heterozygous familial hypercholesterolemia using new practical criteria validated by molecular genetics. Am. J. Cardiol. 72: 171–176. [DOI] [PubMed] [Google Scholar]
- 26.Scientific Steering Committee on behalf of the Simon Broome Register Group. 1991. Risk of fatal coronary heart disease in familial hypercholesterolaemia. BMJ. 303: 893–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scientific Steering Committee on behalf of the Simon Broome Register Group. 1999. Mortality in treated heterozygous familial hypercholesterolaemia: implications for clinical management. Atherosclerosis. 142: 105–112. [PubMed] [Google Scholar]
- 28.Defesche J. C. 2000. Familial hypercholesterolemia. In Lipids and Vascular Disease. D. J. Betteridge, editor. Dunitz, London. 65–76. [Google Scholar]
- 29.Benlian P., Turquet A., Carrat F., Amsellem S., Sanchez L., Briffaut D., and Girardet J. P.. 2009. Diagnosis scoring for clinical identification of children with heterozygous familial hypercholesterolemia. J. Pediatr. Gastroenterol. Nutr. 48: 456–463. [DOI] [PubMed] [Google Scholar]
- 30.Kumar P., Hinokoff S., and Ng P. C.. 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 31.Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., Bork P., Kondrashov A. S., and Sunyaev S. R.. 2010. A method and server for predicting damaging missense mutations. Nat. Methods. 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parthiban V., Gromiha M. M., and Schomburg D.. 2006. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 34: W239–W242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J., Li Q., and Wang J.. 2011. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc. Natl. Acad. Sci. USA. 108: 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baralle D., and Baralle M.. 2005. Splicing in action: assessing disease causing sequence changes. J. Med. Genet. 42: 737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crehalet H., Latour P., Bonnet V., Attarian S., Labauge P., Bonello N., Bernard R., Millat G., Rousson R., and Bozon D.. 2010. U1 snRNA mis-binding: a new cause of CMT1B. Neurogenetics. 11: 13–19. [DOI] [PubMed] [Google Scholar]
- 36.Millat G., Lafont E., Nony S., Rouvet I., and Bozon D.. 2015. Functional characterization of putative novel splicing mutations in the cardiomyopathy-causing genes. DNA Cell Biol. 34: 489–496. [DOI] [PubMed] [Google Scholar]
- 37.Graham C. A., McIlhatton B. P., Kirk C. W., Beattie E. D., Lyttle K., Hart P., Neely R. D., Young I. S., and Nicholls D. P.. 2005. Genetic screening protocol for familial hypercholesterolemia which includes splicing defects gives an improved mutation detection rate. Atherosclerosis. 182: 331–340. [DOI] [PubMed] [Google Scholar]
- 38.Lohse P., Mann W. A., Stein E. A., and Brewer H. B. Jr.. 1991. Apolipoprotein E-4Philadelphia (Glu13→Lys, Arg145→Cys). Homozygosity for two rare point mutations in the apolipoprotein E gene combined with severe type III hyperlipoproteinemia. J. Biol. Chem. 266: 10479–10484. [PubMed] [Google Scholar]
- 39.Barlow D. J., and Thornton J. M.. 1988. Helix geometry in proteins. J. Mol. Biol. 201: 601–619. [DOI] [PubMed] [Google Scholar]
- 40.Argyri L., Dafnis I., Theodossiou T. A., Gantz D., Stratikos E., and Chroni A.. 2014. Molecular basis for increased risk for late-onset Alzheimer disease due to the naturally occurring L28P mutation in apolipoprotein E4. J. Biol. Chem. 289: 12931–12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fouchier S. W., Dallinga-Thie G. M., Meijers J. C., Zelcer N., Kastelein J. J., Defesche J. C., and Hovingh G. K.. 2014. Mutations in STAP1 are associated with autosomal dominant hypercholesterolemia. Circ. Res. 115: 552–555. [DOI] [PubMed] [Google Scholar]
- 42.Marques-Pinheiro A., Marduel M., Rabès J. P., Devillers M., Villéger L., Allard D., Weissenbach J., Guerin M., Zair Y., Erlich D., et al.; French Research Network on ADH. 2010. A fourth locus for autosomal dominant hypercholesterolemia maps at 16q22.1. Eur. J. Hum. Genet. 18: 1236–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahley R. W., Weisgraber K. H., and Huang Y.. 2009. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer’s disease to AIDS. J. Lipid Res. 50: S183–S188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sjouke B., Kusters D. M., Kindt I., Besseling J., Defesche J. C., Sijbrands E. J., Roeters van Lennep J. E., Stalenhoef A. F., Wiegman A., de Graaf J., et al. 2014. Homozygous autosomal dominant hypercholesterolaemia in the Netherlands: prevalence, genotype-phenotype relationship, and clinical outcome. Eur. Heart J. 36: 560–565. [DOI] [PubMed] [Google Scholar]
- 45.Zaiou M., Arnold K. S., Newhouse Y. M., Innerarity T. L., Weisgraber K. H., Segall M. L., Phillips M. C., and Lund-Katz S.. 2000. Apolipoprotein E-low density lipoprotein receptor interaction: influences of basic residue and amphipathic alpha-helix organization in the ligand. J. Lipid Res. 41: 1087–1095. [PubMed] [Google Scholar]
- 46.Weisgraber K. H., Innerarity T. L., and Mahley R. W.. 1982. Abnormal lipoprotein receptor-binding activity of the human E apoprotein due to cysteine-arginine interchange at a single site. J. Biol. Chem. 257: 2518–2521. [PubMed] [Google Scholar]
- 47.Kraulis P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24: 946–950. [Google Scholar]
- 48.Merritt E. A., and Bacon D. J.. 1997. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277: 505–524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.