Abstract
Cerebrotendinous Xanthomatosis (CTX) is a treatable inborn error of metabolism caused by recessive variants in CYP27A1. Clinical presentation varies, but typically includes infant-onset chronic diarrhea, juvenile-onset bilateral cataracts, and later-onset tendinous xanthomas and progressive neurological dysfunction. CYP27A1 plays an essential role in side-chain oxidation of cholesterol necessary for the synthesis of the bile acid, chenodeoxycholic acid, and perturbations in this gene that reduce enzyme activity result in elevations of cholestanol. It is commonly held that CTX is exceedingly rare, but epidemiological studies are lacking. In order to provide an accurate incidence estimate of CTX, we studied the ExAC cohort of ~60,000 unrelated adults from global populations to determine the allele frequency of the 57 variants in CYP27A1 reported pathogenic for CTX. In addition, we conducted bioinformatics analyses on these CTX-causing variants and determined a bioinformatics profile to predict variants that may be pathogenic but have not yet been reported in the CTX patient literature. An additional 29 variants were identified that met bioinformatics criteria for being potentially pathogenic. Incidence was estimated based allele frequencies of pathogenic CTX variants plus those determined to be potentially pathogenic. One variant, p.P384L, previously reported in three unrelated CTX families had an allele frequency ≥ 1% in European, Latino and Asian populations. Three additional mutations had a frequency of ≥ 0.1% in Asian populations. CTX disease incidence was calculated excluding the high frequency p.P384L and separately using a genetic paradigm where this high frequency variant only causes classic CTX when paired in trans with a null variant. These calculations place CTX incidence ranging from 1:134,970 – 1:461,358 in Europeans, 1:263,222 – 1:468,624 in Africans, 1:71,677 – 1:148,914 in Americans, 1:64,267 – 1:64,712 in East Asians and 1:36,072 – 1:75,601 in South Asians. This work indicates CTX is under-diagnosed and improved patient screening is needed as early intervention prevents disease progression.
Keywords: CTX, incidence, bioinformatics, population genetics, IEM, xanthoma, cataract, cholestanol, sterol, bile acids, cholic acid, CDCA
Introduction
Cerebrotendinous Xanthomatosis (CTX, OMIM 213700) is an inborn error of bile acid synthesis caused by recessive mutations in CYP27A1 encoding sterol-27-hydroxylas. The classic clinical presentation includes infantile-onset chronic diarrhea, juvenile-onset bilateral cataracts, with later development of tendinous xanthomas and progressive neurological dysfunction including variably spasticity, ataxia, cognitive dysfunction and dementia. However, clinical manifestations can vary significantly even within families. CYP27A1 plays an essential role in normal cholesterol metabolism and perturbations in this gene result in elevations of cholestanol in tissue and plasma, as well as build-up of toxic bile acid intermediates. Treatment is available for this inborn error of metabolism (IEM) which, if started early in life, can prevent the major clinical problems associated with this disease, reviewed in (Berginer et al., 2015). Thus, the identification of all patients at a young age significantly benefits patients and families.
It is commonly held that CTX is exceedingly rare and while definitive diagnosis can be made with available diagnostic testing of plasma sterols, most often patients who fall into the differential for CTX are not tested for CTX. Given this background and the paucity of epidemiological studies of CTX, we hypothesized that CTX may be under-diagnosed. We sought to provide an accurate disease incidence based on the ExAC cohort, a large cohort of over 60,000 unrelated adults who do not have a diagnosis of CTX and were not ascertained for studies of CTX.
We assessed the previously reported CTX-causing variants bioinformatically. The results of analyzing these known pathogenic variants were then utilized to generate an empirically determined bioinformatic framework for prediction of pathogenicity of other CYP27A1 variants. Few patients with CTX have been reported in the literature and it is most likely that there are pathogenic mutations segregating in the population that have not yet been described. We identified these additional ‘potentially pathogenic’ variants by conducting a multiplex bioinformatic analysis on the variants in the ExAC cohort. The resulting list of pathogenic and ‘potentially pathogenic’ variants was used to calculate CTX disease incidence. Results show this disease is under-diagnosed and raises questions of variable penetrance and possible modified genetic paradigms for CTX as observed in other IEMs such as phenylketonuria (OMIM 261600) and biotinidase deficiency (OMIM 253260) (Mitchell, 1993; Wolf, 1993, 2015).
Methods
Databases and genetic variants
Searches of medical literature and publicly available databases were conducted to identify variants reported as causing CTX. PubMed http://www.ncbi.nlm.nih.gov/pubmed, ClinVar http://www.ncbi.nlm.nih.gov/clinvar and Online Inheritance in Man (OMIM) http://www.omim.org were queried February 2015.
The Exome Aggregation Consortium (ExAC) Cohort was used to obtain allele frequencies of CYP27A1 variants in found in subjects with CTX as well as a source of potentially pathogenic variants segregating in populations that may be predicted damaging to CYP27A1 function (Exome Aggregation Consortium Cohort, Cambridge, MA, http://exac.broadinstitute.org, accessed February, 2015).
Nomenclature for all variants follow HGVS standards and is reported in reference to CYP27A1 NM_000784.3 and NP_000775.1. All genetic alleles studied that were not previously found in ClinVar have been submitted to ClinVar http://www.ncbi.nlm.nih.gov/clinvar/.
Bioinformatic analysis of variant potential pathogenicity
All CTX-causing missense variants were assessed for potential to perturb protein function using CADD 1.0 (Kircher et al., 2014), SIFT (Ng and Henikoff, 2001), PolyPhen2 HDIV (Adzhubei et al., 2010), Genomic Evolutionary Rate Profiling (GERP++) (Cooper et al., 2005a; Davydov et al., 2010a), and PhyloP using custom perl scripts as previously described (Bonnen et al., 2013). Any value residing outside 1.5 times the inter-quartile range was labeled an outlier and removed for calculation of reported mean, standard deviation, boxplots and violin plots. SIFT is based on the assumption that amino acids in a protein sequence that are highly conserved throughout evolution are more functionally impactful and that substitutions in these positions may affect protein function. Using a sequence alignment based approach, SIFT predicts and ranks the effects of all possible substitutions at each position in a protein sequence. PolyPhen-2 predicts the functional significance of an allele change from its individual features by Naïve Bayes classifier trained using supervised machine-learning. PolyPhen-2 HDIV is trained on all damaging alleles with known effects on the molecular function causing human Mendelian diseases, present in the UniProtKB database, together with differences between human proteins and their closely related mammalian homologs, assumed to be non-damaging. GERP++ detects evolutionary constraint while PhyloP considers both conservation and acceleration in the evolutionary rate of substitutions. The phyloP score reported here results from analysis of an alignment of 46 different species. The GERP++ score reported here is based on the alignment of 35 mammalian species.
Incidence estimation
The disease incidence estimates are based on carrier frequencies of variants reported to cause CTX. The first step in incidence estimation was the identification of genetic variants reported as causing CTX. We queried the mutation databases HGMD and ClinVar, as of December 22, 2014. Searches of medical literature identified additional variants pertaining to CTX. All variants were mapped to HG19 and CYP27A1 NM_000784.3 and NP_000775.1. Any mutations not currently present in ClinVar were deposited into ClinVar. To determine carrier allele frequencies we queried a large adult cohort that did not include anyone known to have CTX (Exome Aggregation Consortium (ExAC), Cambridge, MA, http://exac.broadinstitute.org, accessed December, 2014). The total carrier frequency was calculated as the number of individuals carrying a CTX-pathogenic mutation divided by the total number of individuals ascertained.
Disease incidence was calculated based on the total carrier frequency and Hardy-Weinberg principles.
Calculation of incidence of disease while taking into account the possibility of the presence of an allele, A, segregating in the population that only causes disease when present in an individual in combination with specific other alleles (for example nonsense alleles, N) was computed as follows.
Results
Determining a bioinformatic profile of CYP27A1 variants reported to cause CTX
We conducted a survey of all variants reported pathogenic for CTX that included searches of public databases and medical literature. Table 1 shows the mutations identified and included in this study as pathogenic CTX-causing mutations. A total of 72 variants were identified, 57 single nucleotide variants (SNVs) and 16 frameshift variants. Ten single nucleotide and three frameshift variants were identified in the literature that were not present in ClinVar and these have subsequently been deposited into ClinVar. Of the 57 SNVs, 13 were nonsense, 25 missense, and 19 suspected or shown to affect splicing.
Table 1.
Allele frequency of CYP27A1 variants that are predicted to have functional impact.
| SOURCE | Mutation Type | cDNA | Protein | SIFT | PolyPhen2 | CADD Phred | GERP | PhyloP | EUR | FIN | AFR | AMR | EAS | SAS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Literature | Missense | T2C | M1T | 1 | 0.81 | 13 | 3.90 | 1.61 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00132 |
| ExAC | Nonsense | C211T | Q71X | 0.6 | - | 36 | 4.39 | 2.27 | 0.00000 | 0.00016 | 0.00000 | 0.00000 | 0.00000 | 0.00000 |
| ExAC | Missense | G277A | G93S | 1 | 1 | 29 | 5.67 | 2.68 | 0.00000 | 0.00000 | 0.00010 | 0.00000 | 0.00000 | 0.00000 |
| ExAC | Missense | G356T | R119L | 1 | 1 | 36 | 5.67 | 2.68 | 0.00000 | 0.00015 | 0.00000 | 0.00000 | 0.00000 | 0.00000 |
| ExAC | Missense | C374G | P125R | 1 | 1 | 32 | 5.67 | 2.68 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00006 |
| ClinVar | Missense | C379T | R127W | 1 | 1 | 32 | 4.75 | 1.30 | 0.00000 | 0.00000 | 0.00019 | 0.00000 | 0.00012 | 0.00000 |
| ClinVar;ExAC | Missense | G380A | R127Q | 1 | 1 | 33 | 5.67 | 2.68 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 |
| Literature | Nonsense | G399A | W133X | 0 | - | 39 | 5.63 | 2.67 | - | - | - | - | - | - |
| ClinVar;ExAC | Missense | C409T | R137W | 1 | 1 | 35 | 5.63 | 2.67 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 0.00012 | 0.00000 |
| ClinVar;ExAC | Missense | G410A | R137Q | 0.99 | 1 | 35 | 5.63 | 2.67 | 0.00000 | 0.00000 | 0.00010 | 0.00009 | 0.00023 | 0.00000 |
| Literature | Missense | T425C | L142P | 0.82 | 1 | 23 | 5.63 | 2.16 | 0.00017 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 |
| ClinVar | Missense | G433A | G145R | 1 | 1 | 29 | 5.63 | 2.67 | - | - | - | - | - | - |
| ClinVar | Missense | G434A | G145E | 1 | 1 | 27 | 5.63 | 2.67 | - | - | - | - | - | - |
| ClinVar | Splice | G435T | G145G | - | - | 3 | −0.62 | −0.44 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00069 | 0.00000 |
| ClinVar | Splice | 446+1G>A | - | - | - | 32 | 5.63 | 2.67 | - | - | - | - | - | - |
| ExAC | Missense | C472T | R158C | 1 | 1 | 34 | 5.93 | 2.83 | 0.00003 | 0.00000 | 0.00010 | 0.00000 | 0.00000 | 0.00006 |
| ExAC | Missense | G473A | R158H | 1 | 1 | 28 | 5.93 | 2.83 | 0.00002 | 0.00000 | 0.00000 | 0.00009 | 0.00000 | 0.00000 |
| ClinVar;ExAC | Nonsense | C475T | Q159X | 0.45 | - | 38 | 5.93 | 2.83 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 |
| ExAC | Nonsense | C562T | R188X | 0 | - | 15 | 4.11 | 0.82 | 0.00000 | 0.00000 | 0.00010 | 0.00078 | 0.00000 | 0.00000 |
| ClinVar | Nonsense | G583T | E195X | 0.42 | - | 34 | 2.69 | 0.83 | - | - | - | - | - | - |
| ExAC | Nonsense | C601T | Q201X | 0.71 | - | 13 | 3.02 | 0.35 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 |
| ExAC | Nonsense | G643T | E215X | 1 | - | 36 | 5.91 | 2.82 | 0.00000 | 0.00000 | 0.00000 | 0.00009 | 0.00000 | 0.00000 |
| ClinVar;ExAC | Splice | G646C | A216P | 0.97 | 1 | 32 | 5.91 | 2.82 | 0.00004 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 |
| Literature | Splice | 647-1G>A | - | - | - | 32 | 5.91 | 2.82 | - | - | - | - | - | - |
| ClinVar | Splice | 647-1G>T | - | - | - | 30 | 5.91 | 2.82 | - | - | - | - | - | - |
| ExAC | Missense | G674A | R225H | 1 | 1 | 29 | 5.91 | 2.82 | 0.00000 | 0.00000 | 0.00000 | 0.00017 | 0.00000 | 0.00000 |
| ExAC | Missense | G683A | C228Y | 1 | 1 | 21 | 5.91 | 2.82 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 |
| Literature | Nonsense | C688T | Q230X | 0.71 | - | 34 | −0.46 | −0.01 | - | - | - | - | - | - |
| ClinVar | Nonsense | C691T | R231X | 0.62 | - | 35 | 1.95 | 0.36 | - | - | - | - | - | - |
| ClinVar;ExAC | Nonsense | C745T | Q249X | 0 | - | 36 | 0.58 | 0.14 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00012 |
| ClinVar | Nonsense | C752A | S251X | 0 | - | 38 | 5.28 | 1.61 | - | - | - | - | - | - |
| ClinVar | Missense | A776G | K259R | 0.42 | 0.01 | 17 | 2.22 | 0.52 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 |
| ClinVar | Nonsense | G779A | W260X | 0 | - | 39 | 6.15 | 2.93 | - | - | - | - | - | - |
| ExAC | Missense | T802C | W268R | 1 | 1 | 20 | 6.15 | 2.36 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00042 |
| Literature | Missense | G804T | W268C | 1 | 1 | 30 | 6.15 | 2.93 | - | - | - | - | - | - |
| ClinVar;ExAC | Nonsense | C808T | R270X | 0 | - | 35 | 2.35 | 0.16 | 0.00001 | 0.00000 | 0.00010 | 0.00000 | 0.00000 | 0.00006 |
| ClinVar | Splice | 844+1G>A | - | - | - | 32 | 6.15 | 2.93 | - | - | - | - | - | - |
| ExAC | Splice | 844+1G>C | - | - | - | 27 | 6.15 | 2.93 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00006 |
| Literature | Splice | 844+1G>T | - | - | - | 31 | 6.15 | 2.93 | - | - | - | - | - | - |
| ClinVar | Splice | 845-1G>A | - | - | - | 29 | 5.44 | 2.83 | 0.00010 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 |
| ClinVar;ExAC | Nonsense | A850T | K284X | 0 | - | 44 | 5.44 | 2.29 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 |
| ExAC | Nonsense | C886T | Q296X | 0.38 | - | 10 | 2.30 | 0.73 | 0.00000 | 0.00000 | 0.00010 | 0.00000 | 0.00000 | 0.00000 |
| Literature | Missense | C1004T | A335V | 0.99 | 1 | 33 | 5.44 | 2.83 | - | - | - | - | - | - |
| ExAC | Missense | T1010C | V337A | 1 | 1 | 25 | 5.44 | 2.29 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 |
| ClinVar;ExAC | Splice | C1016T | T339M | 1 | 1 | 33 | 5.44 | 2.83 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 0.00012 | 0.00007 |
| ClinVar | Splice | G1017C | T339T | - | - | 10 | 4.56 | 1.54 | - | - | - | - | - | - |
| Literature | Missense | C1028G | T343R | 1 | 1 | 23 | 5.55 | 2.60 | - | - | - | - | - | - |
| ExAC | Missense | C1028T | T343M | 1 | 1 | 29 | 5.55 | 2.60 | 0.00018 | 0.00000 | 0.00010 | 0.00000 | 0.00023 | 0.00012 |
| ClinVar | Missense | A1061G | D354G | 0.96 | 0.15 | 21 | 0.49 | −0.14 | - | - | - | - | - | - |
| ExAC | Nonsense | C1072T | Q358X | 1 | - | 39 | 5.76 | 2.71 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00046 | 0.00000 |
| ExAC | Missense | T1148A | M383K | 1 | 1 | 23 | 5.76 | 2.19 | 0.00000 | 0.00000 | 0.00000 | 0.00078 | 0.00000 | 0.00000 |
| ClinVar;ExAC | Missense | C1151T | P384L | 1 | 1 | 21 | 5.76 | 2.71 | 0.02373 | 0.00333 | 0.00433 | 0.00846 | 0.00023 | 0.03046 |
| ClinVar;ExAC | Splice | C1183A | R395S | 1 | 1 | 22 | 5.76 | 2.71 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 |
| ClinVar;ExAC | Splice | C1183T | R395C | 1 | 1 | 22 | 5.76 | 2.71 | 0.00021 | 0.00030 | 0.00010 | 0.00017 | 0.00000 | 0.00006 |
| ClinVar;ExAC | Splice | G1184A | R395H | 1 | 1 | 23 | 5.49 | 2.56 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 |
| Literature | Splice | G1184T | R395L | 1 | 1 | 22 | 5.49 | 2.56 | - | - | - | - | - | - |
| ClinVar | Splice | 1184+1G>A | - | - | - | 23 | 5.49 | 2.56 | 0.00009 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00067 |
| ClinVar | Splice | 1185-1G>T | - | - | - | 22 | 5.75 | 2.69 | - | - | - | - | - | - |
| ClinVar | Missense | C1202G | P401R | 0.99 | 1 | 22 | 4.86 | 1.37 | - | - | - | - | - | - |
| ClinVar | Missense | C1209G | N403K | 0.91 | 1 | 18 | 0.96 | 0.09 | - | - | - | - | - | - |
| ClinVar | Missense | C1213T | R405W | 1 | 1 | 21 | 3.72 | 1.37 | 0.00001 | 0.00000 | 0.00000 | 0.00009 | 0.00000 | 0.00006 |
| ClinVar;ExAC | Missense | G1214A | R405Q | 1 | 1 | 22 | 5.75 | 2.69 | 0.00003 | 0.00000 | 0.00010 | 0.00009 | 0.00046 | 0.00000 |
| ClinVar | Nonsense | G1222T | E408X | 0.82 | - | 17 | 5.75 | 2.69 | - | - | - | - | - | - |
| ClinVar | Missense | T1238A | V413D | 1 | 1 | 18 | 5.75 | 2.18 | - | - | - | - | - | - |
| ClinVar | Splice | 1263+1G>A | - | - | - | 21 | 5.75 | 2.69 | 0.00006 | 0.00000 | 0.00000 | 0.00009 | 0.00012 | 0.00000 |
| ClinVar | Splice | 1263+5G>T | - | - | - | 17 | 5.75 | 2.69 | 0.00001 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 |
| ClinVar | Splice | 1264-1G>A | - | - | - | 22 | 5.23 | 2.41 | - | - | - | - | - | - |
| ExAC | Missense | C1336T | P446S | 1 | 1 | 29 | 5.23 | 2.41 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00012 |
| ExAC | Missense | C1342A | R448S | 1 | 1 | 25 | 5.23 | 2.41 | 0.00000 | 0.00000 | 0.00000 | 0.00017 | 0.00000 | 0.00000 |
| ExAC | Missense | C1342T | R448C | 1 | 1 | 24 | 5.23 | 2.41 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00006 |
| ExAC | Missense | G1343A | R448H | 1 | 1 | 33 | 5.23 | 2.41 | 0.00000 | 0.00000 | 0.00031 | 0.00000 | 0.00000 | 0.00006 |
| Literature;ExAC | Nonsense | C1381T | Q461X | 0.68 | - | 9 | 4.26 | 2.41 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 |
| ClinVar | Missense | C1402T | P468S | 0.98 | 1 | 23 | 5.08 | 2.33 | - | - | - | - | - | - |
| ClinVar;ExAC | Missense | G1415C | G472A | 1 | 1 | 22 | 5.25 | 2.44 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00104 | 0.00000 |
| ClinVar;ExAC | Missense | C1420T | R474W | 1 | 1 | 22 | 5.25 | 2.44 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 0.00012 | 0.00000 |
| ClinVar;ExAC | Missense | G1421A | R474Q | 1 | 1 | 23 | 5.25 | 2.44 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 0.00012 | 0.00000 |
| ClinVar | Missense | C1435T | R479C | 1 | 1 | 22 | 4.36 | 1.16 | 0.00002 | 0.00000 | 0.00000 | 0.00000 | 0.00012 | 0.00000 |
| ClinVar | Missense | C1435G | R479G | 1 | 0.96 | 22 | 4.36 | 1.16 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 |
| Literature | Nonsense | C1573T | Q525X | 0.2 | - | 18 | 2.40 | 1.15 | - | - | - | - | - | - |
| ExAC | Nonsense | C1582T | Q528X | 0.74 | - | 38 | 1.39 | 0.14 | 0.00003 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 |
| ExAC | Nonsense | C1588T | Q530X | 0.75 | - | 38 | 2.40 | 1.15 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00006 |
Literature and database searches were conducted to identify variants in CYP27A1 that may be pathogenic. Bioinformatic assessment using SIFT, PolyPhen2, CADD, GERP++, and PhyloP was utilized to predict pathogenicity and those variants meeting criteria for being pathogenic are listed here. These variants are shown along with the results of the bioinformatic algorithms and their allele frequencies in five global populations according to the ExAC database. EUR = 33,370 Non-Finnish Europeans, FIN = 3,307 Finnish Europeans, AFR = 5,203 Africans, AMR = 5,789 Latino, EAS = 4,327 East Asians, and SAS = 8,256 South Asians. Source refers to where a variant was identified wether it be from the literature, ClinVar, and/or ExAC.
All CTX-causing missense variants were assessed for potential to perturb protein function using CADD, Sift, PolyPhen, GERP++, and PhyloP. CADD was conceived to combine information from multiple algorithms and as such represents a composite of 63 points of predictive data (Kircher et al., 2014). Rather than offering a categorical determination of pathogenic or not, CADD gives a Phred-scaled score. The average CADD score for variants demonstrated to be pathogenic appears to vary depending on the particular disease (Kircher et al., 2014). The average CADD score for CTX missense variants was 23 (Figure 1 and Table 1). SIFT and PolyPhen are widely utilized algorithms for predicting pathogenic effects of variants. We plotted the SIFT and PolyPhen scores, normalized from 0 to 1 with 1 being maximally damaging, for all missense CTX pathogenic variant. The average scores were 0.99 and 0.98 respectively (Figure 1 and Table 1).
Figure 1. Bioinformatic analyses of variants reported pathogenic in CTX patients.
Missense variants reported pathogenic in CTX patients were assessed bioinformatically and the resulting scores were plotted as violin plots with histograms superimposed. Outliers are plotted as red dots. The metric studied are (A) CADD Phred (B) SIFT (C) PolyPhen2 (D) PhyloP (E) GERP++. A similar assessment was conducted for variants within plus or minus two base pairs from an exon-intron boundary and any variant reported to affect splicing. Results of analysis of these splice variants are shown in (E) CADD Phred (F) PhyloP (G) GERP++.
Evolutionary conservation was measured using PhyloP and GERP++. PhyloP predicts departures from neutral evolution with a range of scores from min = −13.9 to max = 2.9 and genome-wide mean = 0.03 (Siepel, 2006). The mean PhyloP score for CTX-causing missense variants was 1.98, a clear elevation from the genome-wide average. GERP++ approximates evolutionary constraint at a locus by maximum likelihood estimation (Cooper et al., 2005b; Davydov et al., 2010b). The genome-wide average GERP++ score is −0.125 with a maximum GERP++ score of 6.18 (Davydov et al., 2010a). Likewise, the GERP++ score for CTX-causing missense variants showed an elevated mean of 5.22 (Figure 1 and Table 1).
Two variants had scores that were outliers for three of the bioinformatic metrics: SIFT, PolyPhen2, GERP++. These variants, g.2:219677404, c.776A>G, p.K259R and g.2:219678787, c.1061A>G, p.D354G, were both originally reported in the same study (Verrips et al., 2000). This study was conducted prior to the advent of these bioinformatics predictors and prior to the existence of large-scale public databases of genetic variation and thus could not receive the same vetting as current studies.
In a separate analysis, variants that have been reported to affect splicing and those within 2 bases of an exon-intron boundary and were considered for the potential to affect splicing. These variants were evaluated for conservation and constraint as high conservation or constraint may indicate a functional role such as contribution to splicing. Figure 2 shows the distribution of CADD, PhyloP and GERP++ scores for these splicing variants. After removal of outliers, bioinformatic predictors showed higher means for the group of splice variants than for the group of missense variants (Table 2).
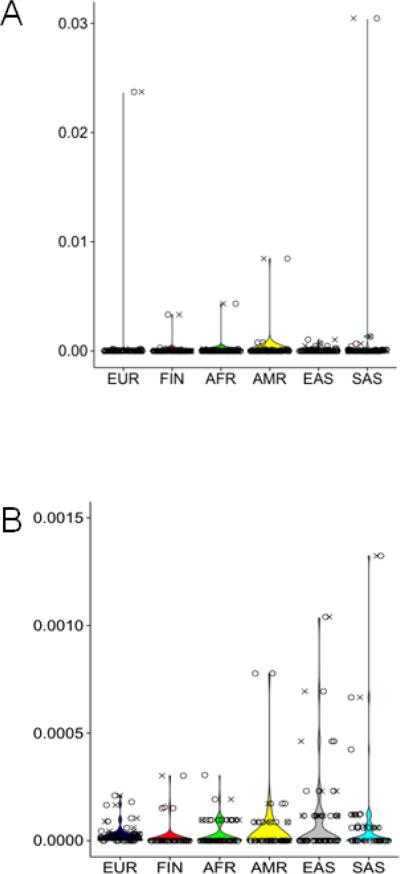
Figure 2. Population allele frequency distributions of variants reported pathogenic in CTX patients and those predicted pathogenic.
The allele frequency of every variant found through literature or database searches is shown in X and the allele frequency for every variant identified in ExAC passing bioinformatics criteria is shown in O. Each population is represented individually: EUR is Non-Finnish European, FIN is Finnish, AFR is African, AMR is Latino, EAS is East Asian, and SAS is Southeast Asian. Plot A shows all data and B shows the same exact dataset, but B uses a Y scale maximum value of 0.0015 in order to allow display of lower frequency variants.
Table 2.
Bioinformatic predictors of pathogenicity for variants reported to cause CTX
| MISSENSE | SPLICE | |||
|---|---|---|---|---|
| Mean | Std.Dev | Mean | Std.Dev | |
| CADD | 23 | 7.4 | 26 | 5.2 |
| SIFT | 0.99 | 0.04 | 1 | 0.01 |
| PolyPhen2 | 0.98 | 0.04 | 1 | 0 |
| PhyloP | 1.98 | 0.89 | 2.72 | 0.14 |
| GERP++ | 5.22 | 0.65 | 5.64 | 0.37 |
Within the splice site group, CADD, GERP++ and PhyloP identified two variants with outlier scores by all three metrics. One of these, g.2:219674479G>T, c.435G>T, p.G145G, is 12 base pairs upstream from the exon 2 3’ exon-intron boundary. The original report of this variant included sequencing of mRNA from a patient homozygous for this variant and this analysis showed ~90% of mRNA sub-cloned from patient fibroblasts showed skipping of exon 2 or partial deletion of exon 2 (Chen et al., 1998). The other outlier variant lies in the last base of exon 5, g.2:219679439C>T, c.1435C>T, p. R479G and was reported as compound heterozygous with a missense variant and no exploration of the variant's affect on splicing was conducted (Wallon et al., 2010).
Bioinformatically predicting pathogenic variants in healthy carriers
Operating under the hypothesis that not all CTX-causing variants have been reported, we interrogated CYP27A1 sequences in the ExAC cohort of approximately 60,000 unrelated adults none of whom are reported to carry a diagnosis of CTX. These individuals represent global populations and include 33,370 Non-Finnish Europeans, 3,307 Finnish Europeans, 5,203 Africans, 5,789 Latinos, 4,327 East Asians, and 8,256 South Asians. All variants present in the CYP27A1 sequence data from this cohort were ascertained and analyzed for CADD, Sift, PolyPhen, GERP++, and PhyloP. Using the bioinformatics profile generated for CTX-causing variants as a benchmark, we determined thresholds for classifying variants not previously reported as pathogenic as ‘potentially pathogenic’. Missense variants were considered potentially pathogenic if they have a value for CADD, Sift, PolyPhen, GERP++, and PhyloP that is equal to or higher than the mean value for four out of five of these metrics with the all scores being higher than the 1st quartile in the CTX causing missense variant group (Table 2). Likewise, variants within 2 bases plus or minus of an exon-intron boundary were considered potentially pathogenic if they had a value for CADD, GERP++, and PhyloP that was equal to or higher than the mean value for two out of three of those metrics in the CTX causing splice variant group (Table 2). All nonsense and frameshift variants were considered ‘potentially pathogenic’.
A total of 51 variants in the ExAC cohort meet the bioinformatics profile of CTX-causing variants (23 missense, 6 splice, 13 nonsense, and 9 frameshift) (Table 1). Of these 51 variants present in ExAC, 29 had not already been reported as causing CTX. Conversely, of the 72 total variants reported as causing CTX, 32 were observed in this cohort of healthy adults. All 13 nonsense variants found in ExAC were considered ‘potentially pathogenic’, and 5 of these had been observed in CTX patients. There were 9 frameshift variants found in the ExAC cohort all of which we considered ‘potentially pathogenic’. Two of these frameshift variants have been reported in CTX patients.
Allele frequencies of CYP27A1 disease-causing variants show significant global variation
We determined the frequency of pathogenic and ‘potentially pathogenic’ CYP27A1 variants in the ExAC cohort of approximately 60,000 unrelated adults. Allele frequency distribution of both groups of variants is highly similar with most variants observed in < 1% in all populations (Figure 2). Notably, there is one variant reported as pathogenic in a CTX patient that is higher frequency. This variant, c.C1151T p.P384L, has been reported as pathogenic in the literature and in ClinVar and occurs in 0.02 in Europeans, 0.004 in Africans, 0.009 in American, 0.0002 East Asians, and 0.03 South Asians. The high frequency of this variant triggered closer inspection of the bioinformatic support that it may be pathogenic and in fact it scored very high in all metrics utilized: SIFT = 1, PolyPhen2 = 1, CADD = 21.3, GERP = 5.76, and PhyloP = 2.71 (Table 1).
Outside of p.P384L the most common pathogenic variants were observed only in the Asian population (Figure 2B). The top four most frequent East Asian variants range in frequency from 0.0005 to 0.001 and only one of these has been observed in other populations. The top three most frequent have been reported in patients and the fourth is a nonsense variant observed in the ExAC cohort. South Asia's most common variant, aside from p.P384L, had allele frequency 0.0013 and was reported in one patient. The variant appears in the first codon and had very low coverage in the ExAC database for all populations; it is not uncommon for first exons to have lower coverage in exome capture experiments. South Asia has one more variant with higher allele frequency 0.0007 and does not appear in other populations. Most common American variant, aside from P384L, was a nonsense variant with allele frequency 0.0008. European population did not have any variant with allele frequency higher than 0.0002.
Estimation of CTX disease incidence reveals potential under-diagnosis
We sought to generate an accurate estimate of CTX disease incidence based on carrier frequencies in adult populations who do not carry a diagnosis of CTX. Studying carrier frequencies, using both the pathogenic and ‘potentially pathogenic’ variants in a large cohort, provides a disease estimate that should not be significantly influenced by ascertainment bias. Disease incidence was estimated based on the combined group of pathogenic and potentially pathogenic variants. These results show an exceedingly high incidence for CTX with an incidence of 1:1,623 in Europeans, 1:28,845 in Africans, 1:8,311 in Americans, 1:49,188 in East Asians and 1:882 in South Asians. The primary driver of these incidence estimates is the one high allele frequency variant, p.P384L. To generate a more conservative estimate of disease incidence we calculated incidence using all variants except p.P384L and found incidences of 1:461,358 in Europeans, 1:468,624 in Africans, 1:148,914 in Americans, 1:64,712 in East Asians and 1:75,601 in South Asians (Table 3).
Table 3.
CTX disease incidence based on known and predicted pathogenic variants.
| MODEL | EUR | FIN | AFR | AMR | EAS | SAS |
|---|---|---|---|---|---|---|
| LIT-ExAC | 461,358 | 2,734,062 | 468,624 | 148,914 | 64,712 | 75,601 |
| p.P384L-NULL + LIT-ExAC | 134,970 | 729,083 | 263,222 | 71,677 | 64,268 | 36,072 |
Considering the strong bioinformatics support for pathogenicity of high frequency variant p.P384L, we wanted to consider a genetic model that includes the possibility that this allele may contribute to disease burden in a non-classical Mendelian inheritance. There are many examples of exceptions to classic Mendelian inheritance within disorders typically thought of as single gene Mendelian diseases. In particular, there is a precedent for higher frequency variants causing recessively inherited inborn errors of metabolism only when present in combination with certain other alleles (van Maldegem et al., 2010). We hypothesized that p.P384L could cause a classic CTX clinical presentation only when present in trans with a null allele. Incidence was calculated that included all pathogenic and potentially pathogenic variants with p.P384L only in combination with nonsense and frameshift alleles. This calculation showed an incidence of 1:134,970 in Europeans, 1:263,222 in Africans, 1:71,677 in Americans, 1:64,267 East Asians, and 1:36,072 South Asians.
Discussion
We created a bioinformatic framework to assess the potential pathogenicity of genetic alleles segregating in a large cohort of unrelated adults and subsequently generate an unbiased estimate of disease incidence for CTX. In addition to the previously reported 57 CTX-causing alleles we identified 29 genetic variants with strong bioinformatics support of pathogenic potential. Disease incidence calculations based on these variants show CTX is under-diagnosed incidence ranging from 1:134,970 – 461,358 in Europeans, 1:263,222 – 468,624 in Africans, 1:71,677 – 148,914 in Americans, 1:64,267 – 64,712 in East Asians and 1:36,072 – 75,601 in South Asians. Evaluation of these alleles identified one variant with frequency ≥ 1% that motivated further exploration of possible alternate genetic paradigms for CTX as observed in other IEMs such as biotinidase deficiency that exhibit higher frequency alleles that reduce enzymatic activity and lead to disease only when in combination with other, specific alleles.
Typically, bioinformatics studies focused on identifying disease causing variants take into account some metrics that strive to determine if a variant is ‘damaging’ or not, independent of what is known about the bioinformatics profile of variants previously demonstrated to cause the specific disease. More recently the CADD score was developed to provide a quantitative measure and does not specifically attempt to ascribe pathogenicity, but the higher a score the more likely it is to be damaging. In the initial study describing CADD, the authors noted that the average CADD score varied depending on disease (Kircher et al., 2014). We elected to construct a bioinformatics framework specifically around the known CTX causing variants in order to tailor our bioinformatics analyses to this disease. We determined the bioinformatics profile of CTX variants for commonly used metrics SIFT, PolyPhen2, PhyloP, and GERP++ plus CADD and then determined what threshold to use to select potentially pathogenic variants. We elected to take a conservative approach in selection criteria in order to obtain variants with the likeliest possiblilty of being pathogenic. In addition, when selecting criteria, we took into consideration the possibility that historically reported variants that preceed contemporary technology and databases may not necessarily be truly pathogenic. The thresholds utilized were calibrated to obtain 50-75% of the previously reported pathogenic CTX variants. A resulting 29 variants from ExAC that were either missense or splice passed these criteria, 15 of these were already noted in patients.
Another notable aspect of this study approach is that this framework was applied without regard to variant allele frequency. This was done with the knowledge that higher frequency alleles can contribute to disease burden particularly in IEMs. The list of variants noted to cause CTX includes a higher frequency variant, p.P384L, which has frequency ranging from 1 – 3 % in global populations. We identified three additional variants with frequency ≥ 0.1% in the Asian population. All other variants were observed in less than 0.1% in any population. The Asian populations have higher frequency variants that are not present in other populations, consequently, this population also has the highest incidence of CTX. This could be due to better ascertainment given an early awareness of CTX in the Japanese population or some population genetics factors such as balancing selection that are currently undescribed.
Incidence estimates show CTX is more frequent than previously appreciated. No previous population based studies of the incidence of CTX have been reported to our knowledge. Instead, incidence has been presumed rare since few patients have been described. One study asserted the incidence of CTX may be more common, but this study was based on only 115 healthy control individuals(Lorincz et al., 2005). Utilizing the ExAC cohort of ~60,000 individuals empowers this study to yield improved disease estimates. Even when applying a conservative bioinformatics approach and totally excluding variant p.P384L from calculations, we arrived at incidence rates higher than currently appreciated: 1:461,358 in Europeans, 1:468,624 in Africans, 1:148,914 in Americans, 1:64,712 in East Asians and 1:75,601 in South Asians. When considering the possibility that p.P384L may cause disease when combined with a null mutation (nonsense or frameshift) incidence is significantly lower: 1:134,970 in Europeans, 1:263,222 in Africans, 1:71,677 in Americans, 1:64,267 East Asians, and 1:36,072 in South Asians. While this work may be the largest population based study of incidence of CTX reported, given that the predicted incidence of CTX ranges from 1:263,222 to 1:36,072 the resolution of this study is limited by the number of individuals in this study (~60,000).
Biotinidase deficiency is a well-characterized IEM that harbors a common variant, BTD p.Asp444His, for which genotype-phenotype studies and extensive biochemical profiling has been conducted. This variant is present in the ExAC Non-Finnish Europeans at allele frequency 4%. It is documented that individuals who are homozygous for this variant have reduced biotinidase activity to 50% of normal and do not manifest clinical symptoms of biotinidase deficiency(Wolf, 1993). However, this allele causes partial biotinidase deficiency when in trans with a variant that causes severe reduction in biotinidase deficiency(Wolf, 2015). Further, when p.Asp444His is in cis with a second BTD variant, p.Ala171Thr, it behaves as a severe deficiency allele and causes profound biotinidase deficiency when this compound allele (p.Asp444His; p.Ala171Thr) is in trans with another severe allele(Wolf, 1993). This sets a precedent that must be considered for other alleles and is especially relevant to other IEMs like CTX. This, plus strong bioinformatics support of pathogenicity, provides the basis for inclusion of the high frequency variant, CYP27A1 p.P384L, in our disease estimates as a pathogenic variant only in combination with null alleles. Functional studies characterizing enzymatic activity of this allele remain to be conducted. A thorough functional assessment of this allele and others identified bioinformatically as potentially disease causing is required before definitive conclusions can made for diagnostic and other purposes.
Creating greater awareness of CTX and better screening for diagnosis is needed given the findings of this study; this is especially relevant in a disease like CTX where significant diversity in clinical presentation is observed. There are several factors contributing to the under-diagnosis of CTX. Significant variability in clinical presentation presents a challenge to diagnosis. Not only do clinical features vary among patients but the age of onset of the individual components of CTX varies as well(Mignarri et al., 2014). The most common clinical feature is bilateral cataracts, but not all cases of CTX have cataracts, and the age of onset of cataracts varies from the first decade of life to appearance in the 60s. Additionally, the fact that the various manifestations of CTX can appear decades apart may obfuscate that there is a common underlying cause to the observed constellation of features, and the diagnosis of CTX may never be made in some cases. Since acquired bilateral cataracts are the most common feature of CTX and because juvenile onset cataracts are often caused by genetic disorders, these cataract cases should receive genetic screening for all genes that cause cataracts including CYP27A1. Additional pediatric signs of CTX may include unexplained chronic diarrhea, neonatal cholestatic jaundice and intellectual impairment. These symptoms are less common than cataracts, but since treatment is available and is more effective when started before the age of 25(Yahalom et al., 2013), early diagnostic testing for CTX either through genetic or biochemical screening is worthwhile as it directly impacts patient treatment and disease course. Any clinical consequence due to impairment of this enzyme is worth detecting since treatment is available.
Highlights.
Cerebrotendinous Xanthomatosis (CTX) is under-diagnosed
Data extraction and modeling estimates incidence based on ~60,000 subjects
CTX incidence is 1:134,970 – 1:461,358 in Europeans
Genetic screening in juvenile cataract patients may increase diagnosis of CTX
Acknowledgements
Research reported in this publication was supported by National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award number R01NS083726 to PEB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berginer VM, Salen G, Patel SB. Cerebrotendinous Xanthomatosis. In Rosenberg's Molecular and Genetic Basis of Neurological and Psychiatric Disease. In: Rosenberg RN, Pascual JM, editors. Academic Press; Elsevier: 2015. pp. 589–598. [Google Scholar]
- Bonnen PE, Yarham JW, Besse A, Wu P, Faqeih EA, Al-Asmari AM, Saleh MA, Eyaid W, Hadeel A, He L, Smith F, Yau S, Simcox EM, Miwa S, Donti T, Abu-Amero KK, Wong LJ, Craigen WJ, Graham BH, Scott KL, McFarland R, Taylor RW. Mutations in FBXL4 cause mitochondrial encephalopathy and a disorder of mitochondrial DNA maintenance. Am J Hum Genet. 2013;93:471–481. doi: 10.1016/j.ajhg.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Kubota S, Teramoto T, Nishimura Y, Yonemoto K, Seyama Y. Silent nucleotide substitution in the sterol 27-hydroxylase gene (CYP 27) leads to alternative pre-mRNA splicing by activating a cryptic 5′ splice site at the mutant codon in cerebrotendinous xanthomatosis patients. Biochemistry. 1998;37:4420–4428. doi: 10.1021/bi972940a. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Stone EA, Asimenos G, Program NCS, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005a;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Stone EA, Asimenos G, Program NCS, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005b;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol. 2010a;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol. 2010b;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz MT, Rainier S, Thomas D, Fink JK. Cerebrotendinous xanthomatosis: possible higher prevalence than previously recognized. Arch Neurol. 2005;62:1459–1463. doi: 10.1001/archneur.62.9.1459. [DOI] [PubMed] [Google Scholar]
- Mignarri A, Gallus GN, Dotti MT, Federico A. A suspicion index for early diagnosis and treatment of cerebrotendinous xanthomatosis. J Inherit Metab Dis. 2014;37:421–429. doi: 10.1007/s10545-013-9674-3. [DOI] [PubMed] [Google Scholar]
- Mitchell JJ. Phenylalanine Hydroxylase Deficiency. 1993 doi: 10.1097/GIM.0b013e3182141b48. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Pollard KS, Haussler D. New methods for detecting lineage-specific selection. Proceedings of the 10th International Conference on Research in Computational Molecular Biology. (RECOMB) 2006:190–205. [Google Scholar]
- van Maldegem BT, Wanders RJ, Wijburg FA. Clinical aspects of short-chain acyl-CoA dehydrogenase deficiency. J Inherit Metab Dis. 2010;33:507–511. doi: 10.1007/s10545-010-9080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrips A, Hoefsloot LH, Steenbergen GC, Theelen JP, Wevers RA, Gabreels FJ, van Engelen BG, van den Heuvel LP. Clinical and molecular genetic characteristics of patients with cerebrotendinous xanthomatosis. Brain. 2000;123(Pt 5):908–919. doi: 10.1093/brain/123.5.908. [DOI] [PubMed] [Google Scholar]
- Wallon D, Guyant-Marechal L, Laquerriere A, Wevers RA, Martinaud O, Kluijtmans LA, Yntema HG, Saugier-Veber P, Hannequin D. Clinical imaging and neuropathological correlations in an unusual case of cerebrotendinous xanthomatosis. Clin Neuropathol. 2010;29:361–364. doi: 10.5414/npp29361. [DOI] [PubMed] [Google Scholar]
- Wolf B. Biotinidase Deficiency. 1993 [Google Scholar]
- Wolf B. Why screen newborns for profound and partial biotinidase deficiency? Mol Genet Metab. 2015 doi: 10.1016/j.ymgme.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Yahalom G, Tsabari R, Molshatzki N, Ephraty L, Cohen H, Hassin-Baer S. Neurological outcome in cerebrotendinous xanthomatosis treated with chenodeoxycholic acid: early versus late diagnosis. Clin Neuropharmacol. 2013;36:78–83. doi: 10.1097/WNF.0b013e318288076a. [DOI] [PubMed] [Google Scholar]