Abstract
Oocytes are usually the largest cells in the body and as such offer unique opportunities for single-cell analysis. Unfortunately, these cells are also some of the rarest in the mammalian female, usually necessitating single-cell analysis. In cases of infertility in humans, determining the quality of the oocyte is often restricted to a morphological analysis or to the study of cellular behaviors in the developing embryo. Minimally invasive approaches could greatly assist the clinician to prioritize oocytes for fertilization or following fertilization, which embryo to transfer back into the woman. Transcriptomics of human and mouse oocytes may have great utility, and recently it was learned that the polar body faithfully reflects the transcript prevalence in the oocyte. The polar body may thus serve as a minimally invasive proxy for an oocyte in the clinic. In the mouse, the transcriptomes of oocytes from mice of the same strain are markedly similar; no significant differences are apparent in transcript prevalence or identity. In human oocytes however, the transcript pool is highly variable. This is likely the result of different histories of each oocyte, in the age of the donor woman, the different hormonal exposures and the prolonged time from specification of the primary oocyte to the fully grown and ovulated egg. This variability in human oocytes also emphasizes the need for cell-by-cell analysis of the oocytes in vitro; which oocytes have a better potential for fertilization and development? To this end, new imaging capabilities are being employed. For example, a single-cell analytical device for oocytes (the simple perfusion apparatus, or SPA) enables investigators to load multiple oocytes in individual wells, to visualize them on the microscope and to use controlled temperature and media flow by perfusion for optimal clinical applications. Recently, developed Raman microspectroscopy approaches suggest that this imaging modality may enable more in-depth analysis of the molecular characteristics of an oocyte that, in combination with the SPA and transcriptomic approaches, might assist the clinician to prioritize more effectively human oocytes and embryos for transfer into women. This review is intended to update the reader on the status of the examination of single oocytes from a variety of approaches and to emphasize areas that may be primed for advancement in the near future.
Keywords: oocyte, transcriptome, simple perfusion apparatus, single-cell analysis, oocyte gene expression, Raman microspectroscopy
Introduction
Nearly, 400 years ago William Harvey wrote De Generatione Animalium, which revolutionized the world of reproduction with his declaration of ex ovo Omnia (from the egg, everything!). His seminal text started the ovist movement that was in direct conflict with spermists who defended the idea of the homunculus, that sperm had a preformed miniature human and needed only the nutritive environment of the egg to materialize. Harvey's work thus stimulated thinking in reproduction for years to come. The modern era of reproduction has been shaped by several key discoveries; e.g. the likes of Jacques Loeb who performed the initial studies that showed a calcium rise in an egg initiated development (Loeb, 1915), Frank Lillie who posed the fertilizin hypothesis that was the initial model for a molecular mechanism of sperm binding to an egg (Lillie, 1919), M.C. Chang who improved culture conditions to enable in vitro fertilization in mammals, and who later worked on the oral contraceptive with Gregory Pincus (Florman et al., in press), Miriam Menkin and John Rock who performed the first documented IVF in humans (Marsh and Ronner, 2008), Ryuzo Yanagimachi, who contributed sentinel work for sperm capacitation, fertilization, embryo cloning and the original ICSI experiments (Yanagimachi, 2009, 2012), Patrick Steptoe, a British gynecologist who developed a laparoscopic approach to oocyte retrieval and Robert Edwards, a physiologist who developed methods to fertilize human eggs in the lab followed by transfer back into the woman (Steptoe and Edwards, 1978). All of these scientists contributed directly to human IVF that made the birth of Louise Brown in 1978 a reality. The broader ramifications of this progress on society has been enormous; in 2012 over 150 000 ART procedures in the USA resulted in more than 65 000 live births (1.5% of total live births in the USA) (Sunderam et al., 2015). However, the current technology that is utilized in assisted reproduction, has not evolved significantly from these original investigations and still largely employs the same techniques that have been used for decades. The field of assisted reproduction remains relatively stagnant despite all of our advances, perhaps because the human oocyte is such a rare cell in the body, and its study challenges many ethical considerations.
Investigations of mammalian oocytes are generally hindered by the very low yield of oocytes obtained from each laboratory animal (Fig. 1). Ovulation induction in mice may yield on average 10–50 oocytes depending on the strain (Luo et al., 2011; Pfeiffer et al., 2015), whereas in rhesus monkeys the yield is highly variable—from a few to over 100 (Nusser et al., 2001), bovine abattoir animals have ∼12 oocytes per ovary (Hamano and Kuwayama, 1993) and humans only one every 28–30 days normally, or 8–15 oocytes on average upon stimulation. In the case of human eggs, most research is performed on discarded oocytes, which may be of poor quality or immature, and therefore findings are highly variable, and may have limited applicability to fertilizable, mature oocytes. While some states in the USA permit payment of egg donors for the purpose of research, this practice is not widespread and may present bioethical issues. In as much as mammalian oocytes are rare and technically challenging to obtain, they are also ethereal with a very limited window of time for study. These challenges make single-cell analysis of the oocyte essential for studying these precious cells.
Figure 1.

Egg yields in some of the many animals investigators use for studying oocyte biology and developmental potential. Listed are sample species and yield, with or without hormonal stimulation to maximize oocyte yield.
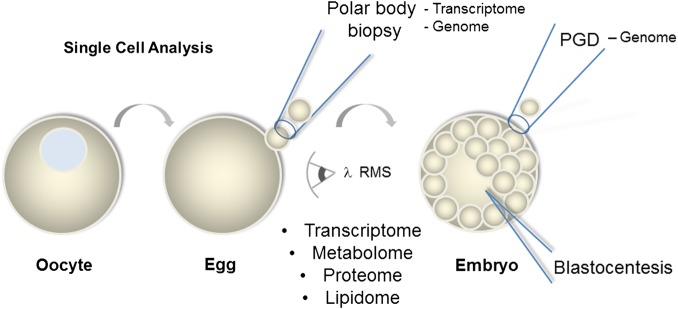
While the oocyte is among the rarest cells in a human, it is also amongst the largest cells of the human body. As such, single-cell analysis in oocytes may actually yield more information than otherwise possible in somatic cells. Here we highlight the approaches that are currently being used to study human oocytes, which include: single oocyte gene expression, lipid profiling, structural analysis, gene methylation, microfluidic devices and microspectroscopy approaches (Fig. 2).
Figure 2.
Oocyte (immature egg), eggs (fertilizable) and early embryos (blastocyst stage of mammals) can each be examined by a variety of single-cell approaches. RMS, Raman microspectroscopy; PGD, preimplantation genetic diagnosis.
Visualizing Single Oocytes and Test Platforms for Experimentation
By virtue of their large size, the oocyte is usually easy to detect, but its size is also a burden. These large, round, and rare cells are sometimes difficult to keep in place, and to visualize in 3D. Currently, in the clinic, individual oocytes are retained in single droplets of media with a mineral oil covering to prevent evaporation. While useful for the clinic, this arrangement is cumbersome, especially for the experimentalist. Ideally, one would like to be able to visualize, manipulate, have each oocyte in an individual compartment for experimentation, and then recover the oocytes following the experimental protocol for additional analysis.
Microfluidic Devices for Oocyte Experimentation
The oocyte simple perfusion apparatus (SPA) is a new instrument to study individual mammalian oocytes. Angione et al. (2015) described a hydrodynamic trap within a perfusion platform to easily perform individual oocyte and embryo experiments, while still having the ability for high-resolution imaging (Fig. 3). The authors describe the apparatus as a multipurpose device that can be used for immunohistochemistry, viability assays, fertilization and even embryo culture. The major advantage of this microfluidic device is that there is no risk of losing the oocyte during manipulation, experimentation and imaging. Furthermore, the trapping mechanism is based on fluid flow, not on pressure constraints that might otherwise harm the cell or change its physiology. The device is fabricated using non-toxic material that can be incubated at a range of temperatures providing for prolonged longitudinal studies without needing to transfer these precious cells. The oocytes and embryos can be observed one-by-one as separate entities, allowing for cell-by-cell analysis, for testing variables under exacting conditions without requiring a high number of oocytes available for each experiment. This device has the potential to be used clinically with human in vitro fertilization; the authors successfully obtained fertilization of sea star eggs by flowing a sperm suspension across the oocytes, then washing the excess sperm out of the well. They also observed prolonged development of these same embryos, all without removing them from the chamber and all while on a temperature controlled microscope stage. This device may also be re-purposed to permit the streamlining of oocyte cryopreservation en masse through microfluidics that could gradually and slowly change media conditions required for vitrification. This device could help further the cause of low cost IVF options.
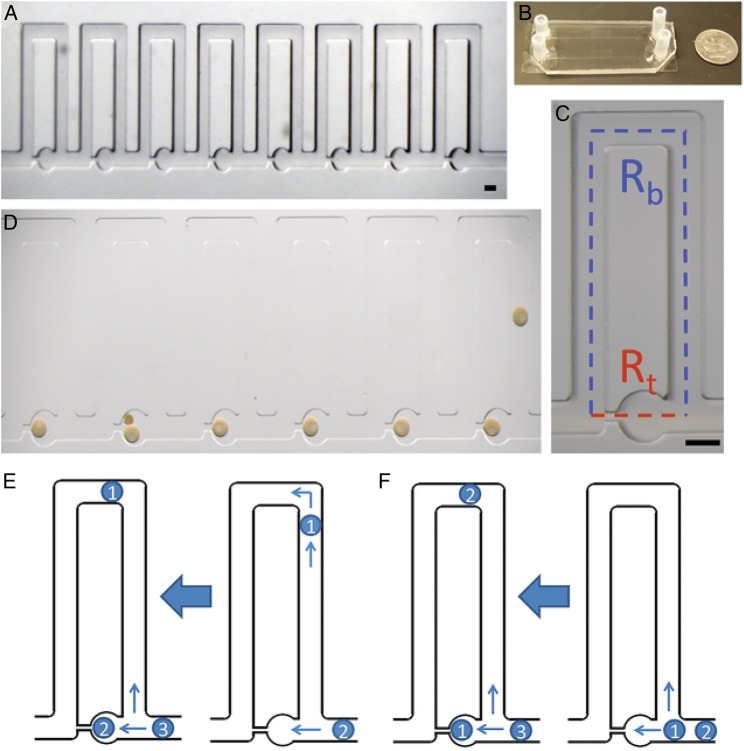
Figure 3.
The simple perfusion apparatus (SPA) using a hydrodynamic trap array: (A) images of the 8-trap hydrodynamic array. Oocytes flow in and load into the wells from right to left, which is the direction of fluid flow. (B) Macroscale view of hydrodynamic trap arrays. Image displays two parallel arrays bonded to a standard size microscope slide coverslip. (C) An individual trap. This consists of a trapping channel (red) and a bypass channel (blue). The resistances of each segment dictate the trapping behavior of the device. The trapping region consists of a circular trap (320 μm in diameter) and a high resistance channel (40 μm width), which connects to the ends of the bypass segment. (D) Immature sea star oocytes within the chip. Oocytes load into the chip from right to left for the six traps displayed. The nucleus and nucleolus are visible in each oocyte which is ∼200 μm. Double trapping can occur with smaller oocytes, as seen in trap five. Scale bars = 200 μm. (E) Indirect trapping. Occurs when the trap resistance is greater than the bypass channel resistance (Rt:Rb = 1.56) causing the first oocyte to be directed into the bypass channel. (F) Direct trapping. Occurs when the trap resistance is lower than the bypass resistance (Rt:Rb = 0.8), directing the first oocyte into the trap. Subsequent oocytes can be directed into the trap causing double trapping, or directed into the bypass channel depending on the change in resistance due to the trapped oocytes for either method (from Angione et al., 2015).
A significant advantage of this type of device in experimental approaches over current oil-drop cultures is that the oocyte can be challenged with a variety of reagents—fluorescent indicators, small molecule inhibitors or activators, different media conditions or even rapid temperature changes—all the while imaging on a confocal microscope, an epifluorescence stage, or perhaps by Raman microspectroscopy and without fear of losing the sample by transfers. The perfusion capabilities of the SPA enable precise control of condition changes without disturbance of the position of the cell.
Despite all of the useful features of this device its limitation is that the oocyte cannot be sampled or analyzed molecularly without compromising the cell. Seemingly normal appearing oocytes and embryos can still harbor significant molecular abnormalities that cannot be discerned just by improved visualization and tracking, and which may not be manifest until during development. Therefore, it would be advantageous to be able to conduct single-cell analysis of the molecular constituents of the cell with non-invasive approaches.
Recent advances in Raman microspectroscopy may permit single oocyte molecular analysis for improved clinical prioritizations for egg fertilizations, and embryo transfers (Mallidis et al., 2014). Raman spectroscopy uses non-invasive, and non-phototoxic light waves to measure inelastic scattering of pre-determined test wavelengths that reveal specific molecular fingerprints within a sample (Matthaus et al., 2008). When coupled with an appropriate microscope, Raman microspectroscopy has been used to identify characteristics in the frog (Xenopus) oocyte (Rusciano et al., 2010), and sheep oocyte (Bogliolo et al., 2012). Raman microspectroscopy has also been used to non-invasively interrogate the zona pellucida (Bogliolo et al., 2012) and cortical actin of sheep oocytes following vitrification (Bogliolo et al., 2012, 2015). Since many of the molecular signatures of a cell are distinct (e.g. lipids, carbohydrates, nucleic acids), one may be able to profile, non-invasively, molecular characteristics of a human oocyte. Coupled with the SPA for oocyte handling, and with subsequent genetic testing, the interrogation of human oocytes may yield far better insight into the developmental potential of the cell.
Genomics of the Oocyte
Single oocyte genome analysis is essential for future experimental and clinical analysis. Were this approach feasible, then clinicians could more carefully prioritize oocytes for fertilization, and make embryos only in cases where the oocytes have a high chance of successful embryonic and fetal development. Currently, the technologies are limited for this approach since most protocols require lysis of the cell for genome analysis. Oocytes do, however, extrude polar bodies during meiotic resumption, and these meiotic products have been successfully analyzed as a metric for the oocyte (Reich et al., 2011, 2012; Montag et al., 2013). The polar bodies have even been used for mitochondrial genetic analysis, which may become more important in the near future with the advent of mitochondrial replacement therapy (Wolf et al., 2015). The polar body can be procured, even prior to fertilization, and may enable earlier detection of genetic errors, but since the polar body contains the DNA alleles that the oocyte now does not have, it is also difficult to definitely identify the oocyte genetic makeup, even with two polar body analyses containing the three meiotic product alleles. The alternative to polar body analysis is to fertilize the egg and then use blastocyst biopsies for detection of inherited diseases, maternal age-related aneuploidy and chromosomal aberrations. Traditionally, blastocyst biopsies require embryo freezing until the cytogenetic analyses have been completed on a single cell of an embryo. However, this approach increases the risk of complications to an embryo by the freezing protocol, increases the time of culture necessary for the embryo, and is not necessarily definitive in the genetic analysis; many cells of the blastocyst embryo have genetic aberrations that may give false-positive, or false-negative results for the embryo.
In the past, whole-genome analyses of single cells has been fraught with poor sequence uniformity. However, Hou et al. (2013) have successfully performed single human oocyte genome analysis using a new approach—that of multiple annealing and a looping-based amplification cycle (MALBAC) sequencing technology. Specifically, this method uses a DNA polymerase derived from Bacillus stearothermophilus (Bst polymerase) and specialized primers to form circular DNA fragments/amplicons which then prevents them from being further amplified in subsequent cycles of MALBAC (Zong et al., 2012). This technique yields both copy number information and single nucleotide variants, but generates extremely high false-positive error rates, making it more suitable for copy number profiling such as with aneuploidy in an oocyte or a fetal cell from amniocentesis. Hou et al. (2013) sequenced single polar bodies (first and second) and female pronuclei from oocytes retrieved from healthy, parous volunteers who had already conceived naturally. The researchers found that the polar bodies accurately deduced the ploidy and allele makeup of the female pronuclei.
These exciting developments in single-cell genomics may soon make clinical diagnostics in assisted reproduction and obstetrics a much more efficient process. However, the genomic information does not identify the numerous epigenetic modifications that give rise to unique transcription pathways and that include acetylation and phosphorylation of histones, as well as methylation and hydroxymethylation of the DNA.
Epigenetics of the Oocyte
The DNA of eggs and sperm are the workshop of evolution, and changes in this information may occur at a single nucleotide to wide-scale chromosomal recombination events. Any non-lethal changes in this blueprint are then subject to test driving in the wild for its impact in fitness. Recently, it has become obvious that heritable changes may also arise not in changes in the sequence of the DNA, but how the DNA is treated. Such heritable, epigenetic changes are an important consideration especially in oocytes of humans since these cells may last for several decades in the human female, all while in a non-dividing state. How, or even if, the environment of the female alters the epigenome of the oocytes over decades is of great importance for reproductive health.
Does the oocyte epigenome even change over the timeframe of reproduction? Work in mouse and in non-mammals documents that multiple types of epigenetic changes occur in the lifetime of an oocyte. These changes include methylation of the DNA and post-translational modifications to the histones attached to the DNA. Differential DNA methylation occurs throughout the oocyte genome, and changes between early and late oocytes. These modifications result in differential promoter usage within the oocyte between young and mature oocytes, and may result in imprinted marks on genes whose function is only apparent in the embryo or fetus. Remarkably, the order of imprinting genes in the genome during oocyte growth appears to be conserved and are significantly different than the marks found in developing sperm. Thus, stage-dependent and gamete-dependent DNA methylation appears dynamic during gamete development and changes in these patterns have dramatic consequences on the fetus (Clarke and Vieux, 2015; Ge et al., 2015). Mutations in genes responsible for these DNA modifications have dramatic results on the developmental success of the embryo and fetus. For example, deletion of one of the DNA methyl-transferases in the mouse (DNMT3A or DNMT3L) results in severe methylation deficiencies and death at mid-gestation (Bourc'his et al., 2001).
Changes in modifications to histones in chromatin are even more dramatic during oocyte development. Using antibodies to a broad series of histone modifications to label oocytes in situ, documents, that in mice the histone complement of epigenetic modifications is under constant flux. These include both transcriptionally repressive marks and activating marks during oocyte growth, maturation and meiosis. Many of the marks detected reach a peak in the germinal vesicle of the fully grown GV stage, and then decrease quickly at germinal vesicle breakdown (reviewed in (Clarke and Vieux, 2015)). It would be great were investigators able to test this marker activity by a different approach, e.g. immunoblots, to control for simple epitope exposure differences in this in situ approach during development, but such experiments require large numbers of synchronized oocytes for detection. Importantly though, testing of some of the dynamics of histone modifications demonstrate that these changes are critical for oocyte function. For example, deletion selectively in oocytes of the histone deacetylase genes, Hdac1 and Hdac2, results in decreased transcriptional activity and arrest of oocyte growth (Ma et al., 2012).
Since both chromatin activation and repressive marks appear to be dynamic in the growing oocyte, it is important to identify what regions of the chromatin are regulated by what histone modifications. Normally, an immunoprecipitation of the histone modification linked to the DNA is performed and sequenced, but the problem is that it takes thousands of cells, oocytes in this case, to obtain sufficient material for these analyses. Even in mice, these approaches are extremely consuming and may await more sensitive approaches, or new technologies to identify these genomic loci.
It is currently difficult to see how one might visualize such detailed biochemical modifications while retaining oocyte viability although Lefevre and Blachere (2015) have developed a protocol optimized for DNA methylation studies on individual oocytes and cleavage-stage embryos. The protocol features bisulfite treatment to detect methylation sites followed by amplification steps and DNA sequencing. For research purposes, this single-cell methylation approach appears to be the most sensitive protocol currently, although it still requires lysis of the cell. Alternatively, a transcriptomic profile of an oocyte's sibling polar body may enable the best, genetic-based test of reproductive potential that reflects an accumulation of all the transcriptional regulation experienced by the oocyte (see below). Although minimally invasive, it too has many limitations, including that the oocyte has completed first meiosis.
Single Oocyte Transcriptomics
Genetic analysis is essential in many cases to test for candidate gene mutations and eventually for more global screening. With a single cell and current technologies though, it means destroying the cell first, diminishing the protocol's utility in the clinic. For study though, often the overall readout of the genome is even more useful—that is—the consequence of all gene activities, epigenetic modifications, miRNA and piRNA functions and mRNA stability.
The transcriptome can reveal the overall, steady-state result of gene activities in the cell, and perhaps even with abundance measurements, be capable of determining which genes are critical to the oocyte and how those genes might differ between fertile and subfertile phenotypes. Reich et al. (2011) described the variation between individual oocytes of the same wild-type genotype and those from a premature ovarian insufficiency (POI) mouse model, the Taf4b hypomorphs. Taf4b is a core transcriptional element enriched in the follicle and is essential for fertility in the mouse (Grive et al., 2014). Taf4b-deficient mice are known to make oocytes that appear morphologically normal but are of poor quality with regards to successful embryogenesis. Taf4b-null animals are viable as adults, but the oocytes they make are depleted prematurely at birth leading to a POI phenotype (Grive et al., 2014), and any oocytes that do mature and are fertilized do not develop past the two- to four-cell stage (Falender et al., 2005; Lovasco et al., 2010). Their hypothesis was that the transcriptome of the Taf4b-deficient oocyte differs significantly from that of the wild-type oocyte. Reich et al. tested the variation in steady-state levels of mRNAs (oocytes are transcriptionally inactive during late germinal vesicle stage until post-MII) in mouse oocytes to establish what they refer to as a baseline of ‘normal’ variation. They then compared those results to the mRNA levels of individual oocytes of poor quality (Taf4b-hypomorphs), all by high throughput DNA sequencing following transcriptome amplification. These samples were then compared within and between genotypes to determine the variance (Reich et al., 2011). The authors learned that the transcriptomes of good/bad oocytes under these conditions were readily discernable, and that the biological variability of transcriptomes can be quantified between single cells within a genotype. The developmental potentials of the oocytes were therefore prioritized accurately based on the cognate transcriptomes.
In examining mRNA between oocytes of the same strain of mice, Reich et al. (2011) found striking similarities between oocytes both in terms of transcript identity and in ranked transcript abundance. A similar study in human oocytes, however, revealed enormous variations between oocytes (Reich et al., 2011). This may not be surprising though when considering that the human oocytes were from different individuals (as were the mouse oocytes) and from different ages (not as in the mouse example). However, the extreme variation differences between mouse and human may reflect the combination of challenges confronting the clinician in examining human oocytes—they are all different! Age, environment, culture conditions, pathologies, nutrition and different protocols for ovarian stimulation—all likely play a role in the huge diversity of mRNA profiles in a human oocyte. These undoubtedly also contribute to the oocyte quality, and subsequent embryo developmental capability, whereas the highly controlled conditions of the mouse females likely contributes to the uniformity of those oocytes. It might be worth testing the similarity of oocytes of wild-type mice collected from the field, and differences in multiple mouse strains, with variable nutrition, age, stress and genetic background. This type of testing paradigm may more accurately reflect the human condition, and serve as a better testing ground for the clinic. Now with transcript analysis protocols from single oocytes becoming more routine (Xue et al., 2013), this analysis may provide a rich intersection of information for basic and clinical applications.
The transcriptome may also reveal oocyte gene expression after epigenetic modifications from differing environmental conditions. Many studies have revealed that lifestyle and environmental factors influence the expression of genes in the oocyte. Chaffin et al. (2014) demonstrated that oocytes from rhesus macaques exposed to low-dose sucrose intake over a 6-month period significantly affected resumption of oocyte meiosis with subsequent effects on the early embryo. However, to date there are no published studies that examine transcript differences that could occur as a result of the supportive or destructive milieu provided to the oocyte. Perhaps the nurture (diet, lifestyle, smoking and obesity) versus the nature (genome) of the oocyte could lead to metabolic perturbations that would distress the routine function of the oocyte. Single-cell analysis of the metabolic processes, perhaps detectable by Raman microspectroscopy, could be a potential avenue to separate those oocytes that have superior functionality versus those who might lead to embryonic arrest or poor implantation.
Oocyte Metabolomics
The cumulus oocyte complex is an intricate interdependent metabolic unit where each component—the specialized granulosa cells and the oocyte—has specific and vital responsibilities to make a mature ovulatory oocyte. However, the oocyte alone in this mix is responsible for the functional metabolic processes of the future embryo. Therefore, it seems imperative to select metabolically fit oocytes for fertilization and implantation. Koek et al. (2010) reported on the diverse metabolic profile of a single Xenopus laevis oocyte which contained organic acids, fatty acids, alcohols and sugars utilizing gas chromatography coupled to mass spectrometry. More recently, Sutton-McDowall and colleagues followed metabolic changes as a result of hormone exposure in cumulus-oocyte complexes. For example, supplementation of media with BMP15 modulated reductive metabolism within the oocyte whereas FSH-stimulation decreased the oocytes' ability to regulate cellular stress. Further, the metabolome has been seen to change with age and stress factors in several consistent and predictable ways that may explain the decreased embryo viability seen from aged eggs (Dumesic et al., 2015). Unfortunately, single oocyte metabolomics is a largely untapped research field that could have direct clinical correlations needed to select the ‘best’ oocytes obtained from an in vitro cycle.
Proteomics of the Oocyte
The protein footprint of an oocyte may present another option for the evaluation of the best oocyte for in vitro fertilization. Although preliminary success has been seen in the very large Xenopus oocytes by detection of almost 4000 proteins (Sun et al., 2014), the protein profile of an individual mammalian oocyte has yet to be described, and the technology may not yet be available to identify the majority of oocyte proteins. A significant representation of proteins from as few as five oocytes has been seen using radiolabeling approaches (Xu et al., 1997), although this does not definitively identify the labeled peptides. The most abundant proteins in the mammalian oocyte are the zona pellucida proteins, and these have been identified from individual oocytes. Researchers were able to detect an electrophoretic mobility shift in ZP2 correlated with fertilization and their sensitivity following biotinylation and avidin blotting of the ZP proteins was the equivalent of ¼ of a zona pellucida (Gross et al., 1996). Recently, literature has documented just over 2010 groups of proteins from pooled mouse oocytes (Pfeiffer et al., 2011, 2015; Schwarzer et al., 2014). These investigators discovered that a subset of regulatory and catalytic proteins of the epigenome varied by donor strain and this variation may correlate with quality of embryo development (Pfeiffer et al., 2015). More detailed analysis of the protein profiles would aid investigators, especially with proteins secreted from the oocyte, as possible candidates for analysis with minimal or non-invasive approaches.
Lipid Profiling of Single Cells
Different from the protein content within an oocyte in which the ∼16 k expressed genes (Reich et al., 2011) are making perhaps 50–100 000 different variations in the protein products, any one form in low abundance, lipids within the cell are likely less diverse. In this case, the more abundant and invariant lipids may be more observable from single cells. For example, triglycerides are the most common lipid in the oocyte and provide an energy source (Homa et al., 1986; Ferguson and Leese, 1999; Kim et al., 2001; Lefevre and Blachere, 2015). These molecules may be candidates for single-cell analysis although lipid profiles are dynamic and likely change to accommodate the needs of the maturing egg. Pirro et al. (2014) documented lipid profiling of single oocytes in pigs. They compared immature oocytes to 24-h and 44-h in vitro matured oocytes. Their data were acquired by using desorption electrospray ionization-mass spectrometry and utilized low- and mid-level data fusion strategies with the aim of describing the vast amount of chemical information contained in the two mass spectrometric lipid profiles. They reported free fatty acids, phospholipids, cholesterol-related molecules, di- and triacylglycerols among immature and in vitro matured porcine oocytes. The authors reported that the most significant changes in this time-frame were seen in triacylglycerol composition, as well as increases in fatty acid metabolism, although changes in overall membrane complexity were evident throughout maturation of the oocyte. This is another area where Raman microspectroscopy may be employed for specific, non-invasive, analysis (Mallidis et al., 2014). Analysis of longitudinal dynamics on a single-cell level could then provide insight into routine, metabolic and membrane processes of the cell and its ability to fertilize and produce a viable embryo. What may be important now is to identify specific lipid compositions, differences and their dynamics in an oocyte, so that the lipid profile becomes a metric for a healthy oocyte.
Clinical Application of Single-Cell Analysis and the Emergence of the Egg Bank
Infertility care for many is moving toward third party assisted reproduction by using donor egg banks instead of directed individual donors. These egg banks have extensive medical, psychological and genetic screening of the egg donors and the donor's past successes with fertility and development. However, individual oocytes from a cohort of retrieved oocytes are not routinely analyzed for reproductive potential. The oocytes are simply deemed immature or mature and subsequently cryopreserved. Historical performance of a donor's oocytes can indicate good quality, yet it is widely accepted that considerable variability exists in oocytes from one stimulated cycle to the next, and within oocytes of each cycle (see, e.g. single-cell transcriptome analysis above (Reich et al., 2012). Additionally, the donor herself may have environmental exposures that may compromise her oocyte quality, but currently we have no reported clinically diagnostic on single-cell analysis even though this is a very important metric for the improved efficiency of eggs both in general, and for eggs stored in banks. It is clear and apparent that single-cell analysis is increasing in importance in both the clinical and laboratory setting.
Conclusions
Although clinical technology in the field of IVF has not evolved significantly over the past few decades, the many advances in research technology and in applications are primed to impact current clinical care. A driving force in the field is in singleton pregnancies since serious complications and obstetric and neonatal risks increase significantly from higher order births. Therefore, the clinician needs to be able to effectively evaluate oocytes before fertilization and to determine which one(s) might be best suited to transition into an embryo, and then to transfer the best embryo into the recipient. Currently, the evaluative process is a strictly morphological metric—the uniformity of cells, the adherence of cells to each other and the size and shape of the blastocoele. This metric, known as the Gardner scale (Gardner, 1999) is limiting, not standard, and very subjective, especially in a climate of technological change in molecular sensitivity never before seen. The single-cell detection approaches listed above, in combination, may offer the best solution to oocyte and embryo grading.
What makes oocytes special? Clearly, the major answer is their ability to support the development of an embryo and all the potential of a new organism. These are also one of only two cells in a human body that undergo meiosis, and they have a variety of organelles unique to this cell, e.g. cortical granules. A close second though is the size, rarity and challenge facing those dedicated to understanding the functionality of these special cells.
Authors' roles
L.M.B. topic research and analysis, preparation and writing. G.W. preparation, writing, editing and topic selection.
Funding
We gratefully acknowledge support from the National institutes of Health (US) 2R01HD028152 to G.M.W. and the RSDP program to L.M.B.
Conflict of interest
None declared.
Acknowledgements
We are grateful for the illustrations crafted by Jessica Poon. We are also grateful to Reproductive Scientist Development Program (L.M.B.) and NIH (G.M.W.).
References
- Angione SL, Oulhen N, Brayboy LM, Tripathi A, Wessel GM. Simple perfusion apparatus for manipulation, tracking, and study of oocytes and embryos. Fertil Steril 2015;103:281–290 e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogliolo L, Ledda S, Innocenzi P, Ariu F, Bebbere D, Rosati I, Leoni GG, Piccinini M. Raman microspectroscopy as a non-invasive tool to assess the vitrification-induced changes of ovine oocyte zona pellucida. Cryobiology 2012;64:267–272. [DOI] [PubMed] [Google Scholar]
- Bogliolo L, Murrone O, Piccinini M, Ariu F, Ledda S, Tilocca S, Albertini DF. Evaluation of the impact of vitrification on the actin cytoskeleton of in vitro matured ovine oocytes by means of Raman microspectroscopy. J Assist Reprod Genet 2015;32:185–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science 2001;294:2536–2539. [DOI] [PubMed] [Google Scholar]
- Chaffin CL, Latham KE, Mtango NR, Midic U, VandeVoort CA. Dietary sugar in healthy female primates perturbs oocyte maturation and in vitro preimplantation embryo development. Endocrinology 2014;155:2688–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke HJ, Vieux KF. Epigenetic inheritance through the female germ-line: the known, the unknown, and the possible. Semin Cell Dev Biol 2015;43:106–116. [DOI] [PubMed] [Google Scholar]
- Dumesic DA, Meldrum DR, Katz-Jaffe MG, Krisher RL, Schoolcraft WB. Oocyte environment: follicular fluid and cumulus cells are critical for oocyte health. Fertil Steril 2015;103:303–316. [DOI] [PubMed] [Google Scholar]
- Falender AE, Shimada M, Lo YK, Richards JS. TAF4b, a TBP associated factor, is required for oocyte development and function. Dev Biol 2005;288:405–419. [DOI] [PubMed] [Google Scholar]
- Ferguson EM, Leese HJ. Triglyceride content of bovine oocytes and early embryos. J Reprod Fertil 1999;116:373–378. [DOI] [PubMed] [Google Scholar]
- Florman H. MC Chang special issue. Mol Reprod Dev 2016. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Wells D. Aneuploidy in the human blastocyst. Cytogenet Genome Res 2011;133:149–159. [DOI] [PubMed] [Google Scholar]
- Gardner D. Towards Reproductive Certainty: Fertility and Genetics Beyond. New York: Parthenon Publish, 1999. [Google Scholar]
- Ge ZJ, Schatten H, Zhang CL, Sun QY. Oocyte ageing and epigenetics. Reproduction 2015;149:R103–R114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianaroli L, Magli MC, Pomante A, Crivello AM, Cafueri G, Valerio M, Ferraretti AP. Blastocentesis: a source of DNA for preimplantation genetic testing. Results from a pilot study. Fertil Steril 2014;102:1692–1699 e1696. [DOI] [PubMed] [Google Scholar]
- Grive KJ, Seymour KA, Mehta R, Freiman RN. TAF4b promotes mouse primordial follicle assembly and oocyte survival. Dev Biol 2014;392:42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross V, Dubey A, Penzias AS, Layman L, Reindollar R, Ducibella T. Biochemical study of individual zonae from human oocytes that failed to undergo fertilization in intracytoplasmic sperm injection. Mol Hum Reprod 1996;2:959–965. [DOI] [PubMed] [Google Scholar]
- Hamano S, Kuwayama M. In vitro fertilization and development of bovine oocytes recovered from the ovaries of individual donors: a comparison between the cutting and aspiration method. Theriogenology 1993;39:703–712. [DOI] [PubMed] [Google Scholar]
- Homa ST, Racowsky C, McGaughey RW. Lipid analysis of immature pig oocytes. J Reprod Fertil 1986;77:425–434. [DOI] [PubMed] [Google Scholar]
- Hou Y, Fan W, Yan L, Li R, Lian Y, Huang J, Li J, Xu L, Tang F, Xie XS et al. Genome analyses of single human oocytes. Cell 2013;155:1492–1506. [DOI] [PubMed] [Google Scholar]
- Kim JY, Kinoshita M, Ohnishi M, Fukui Y. Lipid and fatty acid analysis of fresh and frozen-thawed immature and in vitro matured bovine oocytes. Reproduction 2001;122:131–138. [PubMed] [Google Scholar]
- Koek MM, Bakels F, Engel W, van den Maagdenberg A, Ferrari MD, Coulier L, Hankemeier T. Metabolic profiling of ultrasmall sample volumes with GC/MS: from microliter to nanoliter samples. Anal Chem 2010;82:156–162. [DOI] [PubMed] [Google Scholar]
- Lefevre A, Blachere T. Methylation of specific regions: bisulfite-sequencing at the single oocyte or 2-cell embryo level. Methods Mol Biol 2015;1222:209–226. [DOI] [PubMed] [Google Scholar]
- Lillie FR. Problems of Fertilization. Chicago, IL: The University of Chicago press, 1919. [Google Scholar]
- Loeb J. Calcium in permeability and irritability. J Biol Chem 1915;23:423–430. [Google Scholar]
- Lovasco LA, Seymour KA, Zafra K, O'Brien CW, Schorl C, Freiman RN. Accelerated ovarian aging in the absence of the transcription regulator TAF4B in mice. Biol Reprod 2010;82:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Zuniga J, Edison E, Palla S, Dong W, Parker-Thornburg J. Superovulation strategies for 6 commonly used mouse strains. J Am Assoc Lab Anim Sci 2011;50:471–478. [PMC free article] [PubMed] [Google Scholar]
- Ma P, Pan H, Montgomery RL, Olson EN, Schultz RM. Compensatory functions of histone deacetylase 1 (HDAC1) and HDAC2 regulate transcription and apoptosis during mouse oocyte development. Proc Natl Acad Sci USA 2012;109:E481–E489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magli MC, Jones GM, Gras L, Gianaroli L, Korman I, Trounson AO. Chromosome mosaicism in day 3 aneuploid embryos that develop to morphologically normal blastocysts in vitro. Hum Reprod 2000;15:1781–1786. [DOI] [PubMed] [Google Scholar]
- Mallidis C, Sanchez V, Wistuba J, Wuebbeling F, Burger M, Fallnich C, Schlatt S. Raman microspectroscopy: shining a new light on reproductive medicine. Hum Reprod Update 2014;20:403–414. [DOI] [PubMed] [Google Scholar]
- Marsh M, Ronner W. The Fertility Doctor. Baltimore, MD, US: Johns Hopkins University Press, 2008. [Google Scholar]
- Matthaus C, Bird B, Miljkovic M, Chernenko T, Romeo M, Diem M. Chapter 10: infrared and Raman microscopy in cell biology. Methods Cell Biol 2008;89:275–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montag M, Köster M, Strowitzki T, Toth B. Polar body biopsy. Fertil Steril 2013;100:603–607. [DOI] [PubMed] [Google Scholar]
- Nusser KD, Mitalipov S, Widmann A, Gerami-Naini B, Yeoman RR, Wolf DP. Developmental competence of oocytes after ICSI in the rhesus monkey. Hum Reprod 2001;16:130–137. [DOI] [PubMed] [Google Scholar]
- Pfeiffer MJ, Siatkowski M, Paudel Y, Balbach ST, Baeumer N, Crosetto N, Drexler HC, Fuellen G, Boiani M. Proteomic analysis of mouse oocytes reveals 28 candidate factors of the ‘reprogrammome’. J Proteome Res 2011;10:2140–2153. [DOI] [PubMed] [Google Scholar]
- Pfeiffer MJ, Taher L, Drexler H, Suzuki Y, Makalowski W, Schwarzer C, Wang B, Fuellen G, Boiani M. Differences in embryo quality are associated with differences in oocyte composition: a proteomic study in inbred mice. Proteomics 2015;15:675–687. [DOI] [PubMed] [Google Scholar]
- Pirro V, Oliveri P, Ferreira CR, Gonzalez-Serrano AF, Machaty Z, Cooks RG. Lipid characterization of individual porcine oocytes by dual mode DESI-MS and data fusion. Anal Chim Acta 2014;848:51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich A, Klatsky P, Carson S, Wessel G. The transcriptome of a human polar body accurately reflects its sibling oocyte. J Biol Chem 2011;286:40743–40749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich A, Neretti N, Freiman RN, Wessel GM. Transcriptome variance in single oocytes within, and between, genotypes. Mol Reprod Dev 2012;79:502–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusciano G, Pesce G, Salemme M, Selvaggi L, Vaccaro C, Sasso A, Carotenuto R. Raman spectroscopy of Xenopus laevis oocytes. Methods 2010;51:27–36. [DOI] [PubMed] [Google Scholar]
- Schoolcraft WB, Katz-Jaffe MG. Comprehensive chromosome screening of trophectoderm with vitrification facilitates elective single-embryo transfer for infertile women with advanced maternal age. Fertil Steril 2013;100:615–619. [DOI] [PubMed] [Google Scholar]
- Schwarzer C, Siatkowski M, Pfeiffer MJ, Baeumer N, Drexler HC, Wang B, Fuellen G, Boiani M. Maternal age effect on mouse oocytes: new biological insight from proteomic analysis. Reproduction 2014;148:55–72. [DOI] [PubMed] [Google Scholar]
- Scott RT Jr, Upham KM, Forman EJ, Zhao T, Treff NR. Cleavage-stage biopsy significantly impairs human embryonic implantation potential while blastocyst biopsy does not: a randomized and paired clinical trial. Fertil Steril 2013;100:624–630. [DOI] [PubMed] [Google Scholar]
- Steptoe PC, Edwards RG. Birth after the reimplantation of a human embryo. Lancet 1978;2:366. [DOI] [PubMed] [Google Scholar]
- Sun L, Bertke MM, Champion MM, Zhu G, Huber PW, Dovichi NJ. Quantitative proteomics of Xenopus laevis embryos: expression kinetics of nearly 4000 proteins during early development. Sci Rep 2014;4:4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunderam S, Kissin DM, Crawford SB, Folger SG, Jamieson DJ, Warner L, Barfield WD. Assisted reproductive technology surveillance—United States, 2012. Morbidity and mortality weekly report. Surveillance Summaries (Washington, D.C.: 2002) 2015;64(Suppl 6):1–29. [PubMed]
- Wolf DP, Mitalipov N, Mitalipov S. Mitochondrial replacement therapy in reproductive medicine. Trends Mol Med 2015;21:68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Abbott A, Kopf GS, Schultz RM, Ducibella T. Spontaneous activation of ovulated mouse eggs: time-dependent effects on M-phase exit, cortical granule exocytosis, maternal messenger ribonucleic acid recruitment, and inositol 1,4,5-trisphosphate sensitivity. Biol Reprod 1997;57:743–750. [DOI] [PubMed] [Google Scholar]
- Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 2013;500:593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagimachi R. Germ cell research: a personal perspective. Biol Reprod 2009;80:204–218. [DOI] [PubMed] [Google Scholar]
- Yanagimachi R. Fertilization studies and assisted fertilization in mammals: their development and future. J Reprod Dev 2012;58:25–32. [DOI] [PubMed] [Google Scholar]
- Yang Z, Liu J, Collins GS, Salem SA, Liu X, Lyle SS, Peck AC, Sills ES, Salem RD. Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: results from a randomized pilot study. Mol Cytogenet 2012;5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science 2012;338:1622–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]